Abstract
An investigation of the influence of five DNA polymerase-buffer systems on real-time PCR showed that the choice of both DNA polymerase and the buffer system affected the amplification efficiency as well as the detection window. The analytical repeatability of the data for different systems changed clearly, leading us to conclude that basing quantitative measurements on single-data-set standard curves can lead to significant errors.
Sequence-specific nucleic acid quantification in areas such as diagnostic PCR and molecular biology has been greatly improved by the introduction of real-time PCR technology (9). While this technology has tremendous potential for accurate and sensitive quantification, further studies addressing the quantification aspect of this technology are required before it can be widely implemented. Previous results from this laboratory (5), in which the range of detection of real-time PCR was modeled in a pure system using two different DNA polymerases, gave an indication that DNA polymerases and their buffer systems influence the performance of PCR by affecting the detection window and linear range of amplification. The aim of this work was to systematically study the effect of five DNA polymerase-buffer systems on absolute quantification using the LightCycler instrument (Roche Diagnostics, Mannheim, Germany).
A primer set, Y1 and Y2, for Yersinia enterocolitica was used (6). To a commercial LightCycler kit (LCTaq) (Roche Diagnostics), a 0.4 mM concentration of each primer was added together with 4 mM MgCl2. Sterile Millipore water was added to a volume of 16 μl and complemented with 4 μl of Y. enterocolitica DNA. The concentration of DNA was fluorimetrically determined using a TD-700 fluorimeter (Turner Designs, Sunnyvale, Calif.), and the DNA was diluted to appropriate concentrations in sterile Millipore water. The four other master mixtures, contained 2.5 U of DNA polymerase and 1× associated buffer, 4 mM MgCl2, 0.4 ml of each primer, 0.2 mM (each) deoxynucleoside triphosphate, 10,000-fold-diluted SYBR Green I, and 4 μl of Y. enterocolitica DNA in a total volume of 20 μl. The following DNA polymerases were used: DyNazyme II (FINNZYMES OY, Espoo, Finland), rTth (Applied Biosystems, Foster City, Calif.), and Taq (Roche Diagnostics) and Tth (Roche Diagnostics). Each amplification started with a denaturation step of 1 min at 95°C, followed by 40 cycles of 0.1 s of denaturation at 95°C, 5 s of annealing at 60°C, and elongation for 15 s at 72°C, followed by a single fluorescence measurement and finally 25 s of final elongation. Amplification was followed by melting curve analysis between 65 and 95°C and finally cooling for 1 min at 40°C. The quantification data, in terms of the crossing point value (ROM) (which is expressed as the fractional cycle number and is the intersection of the log-linear fluorescence curve with a threshold crossing line), were determined using the second derivative method of the LightCycler Software, version 3 (Roche Diagnostics).
Amplification efficiency and analytical repeatability.
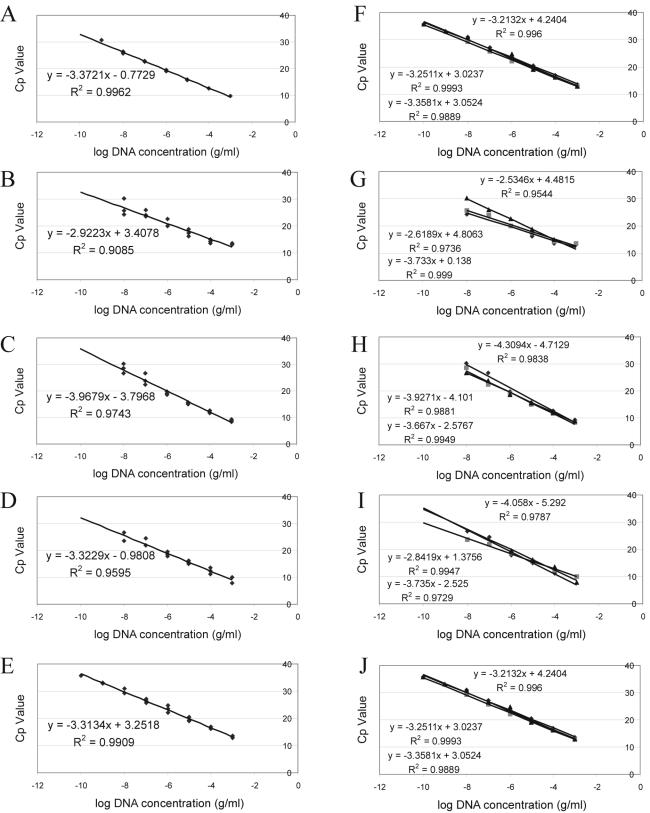
Independent triplicates of 10-fold dilutions of Y. enterocolitica DNA, from 1 mg/ml to 1 fg/ml, were used to obtain standard curves for each polymerase-buffer system (Fig. 1). After amplification, results from the melting curve were analyzed, and the ROM values of all samples that gave a positive specific product peak between 88 and 92°C were plotted against the log of the initial DNA concentration. From this slope the amplification efficiency was calculated using the equation E = (10−1/slope) − 1 (4). Figure 1A to E shows the slope through all generated data, while Fig. 1F to J shows independent analysis of the triplicates for each DNA polymerase. Assuming that all slopes should be −3.32 (which would lead to the optimal amplification efficiency of 1), differences can be seen between different DNA polymerases in Fig. 1A to E (P = 0.053). In particular, Taq, Tth, and DyNazyme II have amplification efficiencies very close to 1. When looking at Fig. 1F to J, it is clear that the different DNA polymerases show differences in repeatability, as this is defined as the intralaboratory variability. In particular, Taq and LCTaq show great variation between different runs.
FIG. 1.
Standard curves generated for five different DNA polymerase-buffer systems after two different types of analysis of the same independent triplicate data. Panels A to E show standard curves and corresponding equations when all three sets of data for each DNA polymerase were used for one analysis of DyNazyme II (A), LCTaq (B), rTth (C), Taq (D), and Tth (E). Panels F to J show standard curves and corresponding equations when the three sets of data were used for independent analysis of DyNazyme II (F), LCTaq (G), rTth (H), Taq (I), and Tth (J).
Detection window and detection probability.
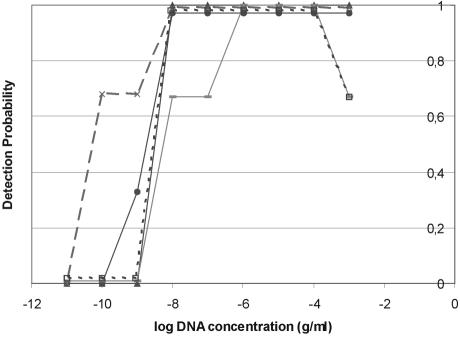
From the triplicate analysis it is possible to create a detection probability graph showing the number of detectable points at each DNA concentration (Fig. 2). A significant difference can be seen between the results from the different DNA polymerase-buffer systems. Thus, the detection window for Tth is the broadest, with at least 67% detection probability over a window of 8 log units, compared to Taq and LCTaq, with a window of 6 log units. This confirms previous indications about a possible difference between the performance of different DNA polymerase-buffer systems in real-time PCR, as proposed by Knutsson et al. (5). The results for both amplification efficiency and detection window were confirmed by repeating this test with another primer pair, coding for a 0.6-kb region of the Yersinia virulence gene yadA (6), which showed the same trends (data not shown). However, other factors, such as the thermal cycler model and the probe system, may affect the PCR per- formance.
FIG. 2.
Detection probability using different DNA polymerase-buffer systems. The detection probability for each DNA concentration was determined by checking the amount of data points/total amount of analysis. Data were determined after independent triplicate experiments. —•—, DyNazyme II; --□--, LCTaq; —▴—, rTth; ——, Taq; and ---×: Tth.
Impact on nucleic acid quantification.
The effect of the DNA polymerase-buffer system on DNA quantification was demonstrated by quantifying four standardized DNA samples (Table 1). In particular, Taq and LCTaq generated less-accurate quantification data. The main reason for this is the narrower detection window and the greater deviations in the standard curve.
TABLE 1.
The effect of the DNA polymerase and the accompanying buffer on quantitative real-time PCR
| DNA polymerase | DNA concn (mg/ml) (± SD)a from samples containing DNA concn (mg/ml)b of:
|
|||
|---|---|---|---|---|
| 0.5 | 0.5 × 10−2 | 0.5 × 10−4 | 0.5 × 10−6 | |
| DyNazyme II | 0.20 ± 0.21 | 0.38 × 10−2 (±0.14 × 10−2) | 0.49 × 10−4 (±0.47 × 10−4) | 0.03 × 10−6 (±0.02 × 10−6) |
| LCTaq | 0.03 ± 0.03 | 0.26 × 10−2 (±0.02 × 10−2) | 0.49 × 10−4 (±0.07 × 10−4) | 0.35 × 10−6 (±0.08 × 10−6) |
| rTth | 0.24 ± 0.29 | 0.14 × 10−2 (±0.11 × 10−2) | 0.34 × 10−4 (±0.16 × 10−4) | 0.07 × 10−6 (±0.02 × 10−6) |
| Taq | 0.07 ± 0.02 | 0.90 × 10−2 (±0.27 × 10−2) | 0.38 × 10−4 (±0.02 × 10−4) | 0.03 × 10−6 (±0.03 × 10−6) |
| Tth | 0.56 ± 0.01 | 0.62 × 10−2 (±0.56 × 10−2) | 0.76 × 10−4 (±0.02 × 10−4) | 0.69 × 10−6 (±0.15 × 10−6) |
DNA concentrations determined by quantitative real-time PCR with different DNA polymerases in three independent measurements. Standard curves from Fig. 1A to E were used for analysis of the unknown data points.
DNA concentration as determined by fluorescence measurements.
The effect of buffer composition.
The influence of buffer components on the amplification efficiency and the detection window was determined for Taq and Tth (Table 2). From the data it is clear that at least for Tth, the buffer composition affects the detection window, since with increasing complexity of the buffer the detection window becomes wider. However, comparing data between Taq and Tth, it can be seen for all buffers that the use of Tth improves the PCR performance. This implies that both buffer composition and DNA polymerase can influence the results.
TABLE 2.
The influence of buffer components on amplification efficiency and detection window using Taq polymerase and Tth DNA polymerase
| Buffer | Amt of buffer componenta
|
Amplification efficiencyc
|
Detection windowd
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Tris-HCl (mM) | KCl (mM) | MgCl2 (mM) | Tween 20 (vol [%]) | BSA (μg/ml) | Taq | Tth | Taq | Tth | |
| 1 | 100 | 500 | 15 | 1.01 | 1.00 | 5 | 5 | ||
| 2 | 100 | 500 | 15 | 500 | 0.98 | 0.99 | 5 | 6 | |
| 3 | 100 | 500 | 15 | 0.5 | 0.61 | 0.94 | 4 | 5 | |
| 4 | 100 | 500 | 15 | 0.5 | 500 | 1.04 | 1.01 | 5 | 7 |
| 5 | 100 | 1,000 | 15 | 0.5 | 500 | 0.96 | 0.99 | 5 | 8 |
| 6b | 100 | 1,000 | 15 | 0.5 | 500 | 1.01 | 1.00 | 5 | 8 |
Components in the 10-fold-concentrated buffers.
Buffer 6 is the commercially available Tth buffer.
The amplification efficiency is calculated from triplicate data using all data points (similarly to Fig 1A to E).
The detection window is defined as the number of log units in initial DNA concentration that gives a detection probability of 67% and above.
Correct data analysis.
There seems to be no consensus on the correct way to analyze quantitative data and to create standard curves for absolute quantification with real-time PCR. Most published data show standard curves constructed from one data set (2, 7, 8), while others analyze and use multiple data sets to calculate the amplification efficiency (1, 3). The data shown in Fig. 1 indicate that especially when using certain DNA polymerases, such as Taq polymerase or rTth, the intralaboratory variation can differ between data sets and that it is of great importance to perform multiple analyses. Furthermore, the linear range of amplification, the area of the detection window in which a linear relationship is obtained between the log DNA concentration and the Cp value, does not always seem to match the detection window. For example, the lowest DNA concentration for LCTaq seems to have such a great variation between the points that it may be questioned whether these data points should be used to calculate the amplification efficiency. In conclusion, this study has shown that the DNA polymerase-buffer system used for quantitative analysis can impact the performance of the system, and when used to quantify unknown samples it affects the accuracy of the data. Furthermore, it has indicated a need for consensus on the correct way to analyze quantitative PCR data in order to be able to compare the performance of different assays.
Acknowledgments
This work was financially supported by the Commission of the European Communities within the program “FOOD-PCR,” QLK1-1999-00226, and the Swedish Research Council for the Environment, Agricultural Sciences and Spatial Planning, 2001-4068.
REFERENCES
- 1.Brinkman, N. E., R. A. Haugland, L. J. Wymer, M. Byappanahalli, R. L. Whitman, and S. J. Vesper. 2003. Evaluation of a rapid, quantitative real-time PCR method for enumeration of pathogenic Candida cells in water. Appl. Environ. Microbiol. 69:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hein, J., U. Schellenberg, G. Bein, and H. Hackstein. 2001. Quantification of murine IFN-gamma mRNA and protein expression: impact of real-time kinetic RT-PCR using SYBR green I dye. Scand. J. Immunol. 54:285-291. [DOI] [PubMed] [Google Scholar]
- 3.Ibekwe, A. M., and C. M. Grieve. 2003. Detection and quantification of Escherichia coli O157:H7 in environmental samples by real-time PCR. J. Appl. Microbiol. 94:421-431. [DOI] [PubMed] [Google Scholar]
- 4.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 5.Knutsson, R., C. Löfström, H. Grage, J. Hoorfar, and P. Rådström. 2002. Modeling of 5′ nuclease real-time responses for optimization of a high-throughput enrichment PCR procedure for Salmonella enterica. J. Clin. Microbiol. 40:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lantz, P. G., R. Knutsson, Y. Blixt, W. A. Al Soud, E. Borch, and P. Rådström. 1998. Detection of pathogenic Yersinia enterocolitica in enrichment media and pork by a multiplex PCR: a study of sample preparation and PCR-inhibitory components. Int. J. Food Microbiol. 45:93-105. [DOI] [PubMed] [Google Scholar]
- 7.Malinen, E., A. Kassinen, T. Rinttila, and A. Palva. 2003. Comparison of real-time PCR with SYBR Green I or 5′-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology 149:269-277. [DOI] [PubMed] [Google Scholar]
- 8.Nogva, H. K., A. Bergh, A. Holck, and K. Rudi. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmittgen, T. D. 2001. Real-time quantitative PCR. Methods 25:383-385. [DOI] [PubMed] [Google Scholar]