SUMMARY
High energy production in mitochondria is essential for maintaining cardiac contraction in the heart. Genes regulating mitochondrial function are commonly downregulated during heart failure. Here we show that both PPARα and Sirt1 are upregulated by pressure overload in the heart. Haploinsufficiency of either PPARα or Sirt1 attenuated pressure overload-induced cardiac hypertrophy and failure, whereas simultaneous upregulation of PPARα and Sirt1 exacerbated the cardiac dysfunction. PPARα and Sirt1 coordinately suppressed genes involved in mitochondrial function that are regulated by estrogen related receptors (ERRs). PPARα bound and recruited Sirt1 to the ERR response element (ERRE), thereby suppressing ERR target genes in an RXR-independent manner. Downregulation of ERR target genes was also observed during fasting, and this appeared to be an adaptive response of the heart. These results suggest that suppression of the ERR transcriptional pathway by PPARα/Sirt1, a physiological fasting response, is involved in the progression of heart failure by promoting mitochondrial dysfunction.
INTRODUCTION
Heart failure is a leading cause of death worldwide. The heart is an organ with high energy production and consumption because it must constantly pump blood against blood pressure. Cardiac myocytes harbor a high volume of mitochondria, which play a major role in mediating energy production in these cells. Cardiac hypertrophy is induced by increased mechanical load such as high blood pressure. Hypertrophic growth is initially a compensatory response that helps maintain cardiac function by reducing wall stress, but it eventually leads to the development of heart failure. This progression is well demonstrated by an experimental model in which high blood pressure is induced in the heart, termed transverse aortic constriction (TAC). The left ventricle (LV) is subjected to high blood pressure by constriction of the aorta, which provokes cardiac hypertrophy and then heart failure. Heart failure represents a complex phenotype that includes reduced myocardial contractility, increased myocyte cell death and myocardial fibrosis. During heart failure, mitochondria display ultrastructural and bioenergetic defects, which lead to insufficient energy production, increased oxidative stress and cell death(Russell et al., 2005). However, the mechanism mediating these pathological changes in mitochondria during the development of cardiac disease is not fully understood.
Nuclear receptors form a superfamily of transcription factors that orchestrate a wide range of biological functions, including development, immune responses, cell growth and metabolism(Glass, 2006). DNA binding sites of nuclear receptors, known as hormone response elements (HREs), consist of a common hexad sequence, such as AGGTCA or AGAACA, which can be configured into direct repeats, inverted repeats, and everted repeats (Mangelsdorf et al., 1995). In addition, the single hexad sequence is recognized by several nuclear receptors, including estrogen related receptors (ERRs)(Sladek et al., 1997).
ERRs consist of 3 different isoforms ERR-α, -β, and -γ. ERRs are activated by transcription coactivators such as PGC-1α and PGC-1β, crucial regulators of mitochondrial function(Huss et al., 2002). The ERR response elements (ERREs), whose consensus sequence is TNAAGGTCA (where N is any nucleotide), are frequently found in the promoter regions of genes regulating mitochondrial function, such as those encoding components of the respiratory chain (Giguere, 2008). In addition, ERRs directly regulate transcription of modulators of cardiac contraction, namely Atp2a2 and Hrc, and important transcription factors, including ERRα itself, PGC-1α, Jarid2 and Irx4, and, therefore, ERRs are recognized as pleiotropic regulators of heart function (Dufour et al., 2007; Wang et al., 2005). Gene disruption of mouse ERRα causes heart failure with impaired mitochondrial ATP production under pressure overload conditions (Huss et al., 2007). Since downregulation of the PGC1α-ERRα axis and consequent development of both mitochondrial and contractile dysfunction are frequently observed in heart failure patients (Karamanlidis et al., 2010; Sihag et al., 2009), impaired ERR target gene expression is considered to be a hallmark of failing hearts.
Peroxisome proliferator-activated receptors (PPARs), comprising PPARα, -β/δ and -γ, belong to the nuclear receptor superfamily. PPARs form heterodimers with Retinoid X receptors (RXRs) and bind to a response element called direct repeat 1 (DR1) or PPAR response element (PPRE), whose consensus sequence is AGGTCANAGGTCA (Schoonjans et al., 1996), with PPAR and RXR occupying the 5' and 3' AGGTCA sequences, respectively. PPARα is activated by fatty acid ligands to induce expression of genes promoting fatty acid uptake and oxidation, whereas PPARγ is an important regulator of adipogenesis and fat storage (Spiegelman, 1998). In the heart, artificial ligands for PPARα, such as fenofibrate, protect against endothelin-induced cardiac hypertrophy and failure (Irukayama-Tomobe et al., 2004), and cardiac function is severely damaged in PPARα null mice during pressure overload (Smeets et al., 2008). Cardiac-specific overexpression of PPARα also results in hypertrophy and failure in association with intracellular accumulation of neutral lipids, termed lipotoxicity (Finck et al., 2002). Thus, PPARα both positively and negatively regulates cardiac function. The molecular mechanism through which PPARα exerts dichotomic effects on the heart remains to be elucidated.
Silent information regulator 1 (Sirt1) plays an important role in gene silencing and life span extension under starvation conditions (Bordone and Guarente, 2005). Cellular responses to nutrient starvation, including metabolic adaptation to fasting, are mediated by Sirt1 through its protein-protein interaction with and post-translational modification of PGC-1α and PPAR (Rodgers et al., 2008). However, Sirt1-mediated regulation of PPAR is complex. For example, fatty acid mobilization from adipose tissues is enhanced by Sirt1 through inhibition of PPARγ (Picard et al., 2004), whereas Sirt1 activates PPARα to promote fatty acid oxidation in the liver (Purushotham et al., 2009).
We here demonstrate that PPARα negatively regulates expression of genes involved in mitochondrial respiration and cardiac contraction through direct interaction with a single HRE hexad DNA motif, such as ERRE, in the heart under pressure overload. The negative regulation of a single hexad motif of HREs by PPARα takes place independently of RXR but in a Sirt1-dependent manner in the heart in response to pressure overload, as well as under physiological fasting conditions. These results suggest that binding of PPARα/Sirt1 to the single hexad motif of HREs plays an important role in mediating downregulation of the ERR target genes, which in turn promotes heart failure. Although this mechanism could mediate physiological adaptation of the heart in response to starvation, inappropriate activation of the same mechanism could lead to impaired energy production and contractility, thereby facilitating progression of heart failure during pressure overload, when the heart requires high energy to maintain contraction against an elevated afterload.
RESULTS
PPARα and Sirt1 mediate pressure overload-induced cardiac hypertrophy
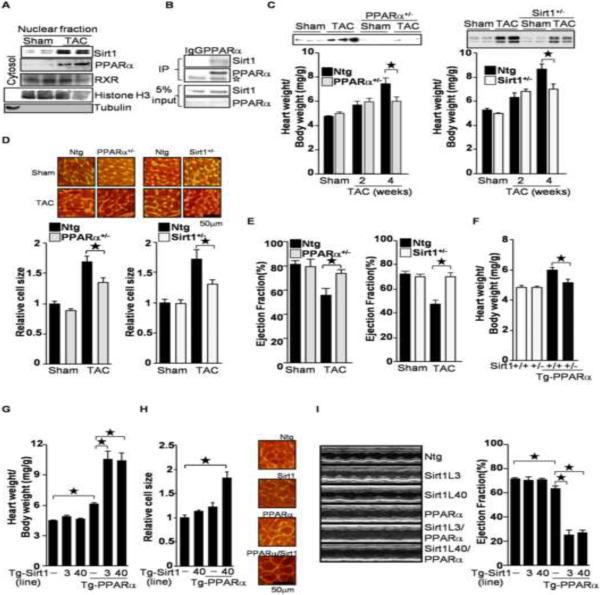
The level of Sirt1 in the nuclear fraction of the heart was upregulated in response to pressure overload imposed by transverse aortic constriction (TAC), consistent with our previous report(Alcendor et al., 2007). TAC also upregulated PPARα (Figure 1A) with activation of luciferase reporter gene driven by 3 repeats of the PPAR response element in vivo (Tg-PPRE-luc) (Figure S1A). PPARα interacts with Sirt1 in cardiac myocytes (Figure 1B). Haploinsufficiency of either PPARα (PPARα+/−) or Sirt1 (Sirt1+/−) attenuated cardiac hypertrophy after 4 weeks, but not 2 weeks, of TAC, as characterized by heart weight/body weight (Figure 1C), heart weight/tibia length (Figure S1B and S1C), and myocyte size (Figure 1D). Echocardiographic analyses showed that TAC-induced systolic dysfunction, characterized by a decreased ejection fraction, was prevented in PPARα+/− and Sirt1+/− mice (Figure 1E). Thus, PPARα and Sirt1 play an important role in mediating TAC-induced cardiac hypertrophy and failure. Transgenic mice with cardiac-specific PPARα overexpression (Tg-PPARα) show moderate cardiomyopathy characterized by hypertrophy and systolic dysfunction. However, the PPARα-induced cardiac hypertrophy was ameliorated by downregulation of Sirt1 (Tg-PPARα/Sirt1+/−) (Figure 1F and S1D), suggesting that endogenous Sirt1 is required for the cardiac hypertrophy observed in the Tg-PPARα. Taken together, these results indicate that PPARα and Sirt1, in concert, facilitate pressure overload-induced cardiac dysfunction and hypertrophy. Since both PPARα and Sirt1 were upregulated in the heart under pressure overload conditions, in order to test whether simultaneous upregulation of PPARα and Sirt1 is sufficient for mediating cardiac hypertrophy, we crossed 2 lines each of Tg-Sirt1 and Tg-PPARα to generate bigenic mice overexpressing both PPARα and Sirt1 (Figure S1E). The PPARα transgene was Flag-tagged (Finck et al., 2002), and bound to both endogenous and exogenous Sirt1 in the heart (Figure S1F). We did not detect any acetylation of PPARα even in Tg-PPARα with a Sirt1−/− background (Figure S1G). As shown previously, mild cardiac hypertrophy was observed in Tg-PPARα. However, cardiac hypertrophy was more severe in the Tg-PPARα/Tg-Sirt1 bigenic mice (Figure 1G, 1H and S1H). Lung congestion was more prominent in the Tg-PPARα/Tg-Sirt1 bigenic mice (Figure S1I). Systolic function was impaired in the Tg-PPARα/Tg-Sirt1 bigenic mice, as evidenced by a decreased ejection fraction and fractional shortening (Figure 1I and Table S1). These results suggest that coupregulation of Sirt1 and PPARα is sufficient to induce cardiac hypertrophy and failure, even without pressure overload.
Figure 1.
PPARα and Sirt1 are involved in pressure overload(PO)-induced cardiac hypertrophy. (A) PO induces co-upregulation of PPARα and Sirt1. Nuclear fractions were prepared from wild type mice subjected to transverse aortic constriction (TAC) for 4 weeks. Anti-histone H3 and tubulin antibodies were used to show nuclear and cytosolic fractions, respectively. (B) Sirt1 was co-immunoprecipitated with PPARα from primary cultured cardiac myocytes. Open asterisk represents IgG. (C–E) Haploinsufficiency of PPARα and Sirt1 attenuates PO-induced cardiac hypertrophy (C–D) and systolic dysfunction (E). PPARα+/− and Sirt1+/− mice were subjected to 4 weeks of TAC. (C) The heart weight/body weight ratio was measured. Heart lysates were subjected to Western blot analyses with anti-PPARα and anti-Sirt1 antibodies. (D) After 4 weeks of TAC, cell size was measured using WGA staining. Representative images are shown. (E) After 4 weeks of TAC, ejection fraction was measured by echocardiography. (F) Haploinsufficiency of Sirt1 normalizes PPARα-induced cardiac hypertrophy. (G–H) Co-overexpression of PPARα and Sirt1 induces cardiac hypertrophy. Heart weight/body weight ratio and myocytes cell size were measured. (I) Ventricular dysfunction in Tg-PPARα/Tg-Sirt1 bigenic mice was evaluated by echocardiographic measurements. Representative M-mode echocardiographic images of the left ventricle and ejection fraction are shown. The numbers of mice examined in each experimental group were: 4–12 (C), 3–8(D), 6–14 (E), 3–6 (F), 9–21 (G), 3 (H), and 7–19 (I). Error bars represent S. E. M.
Sirt1 attenuates PPARα-induced lipotoxicity
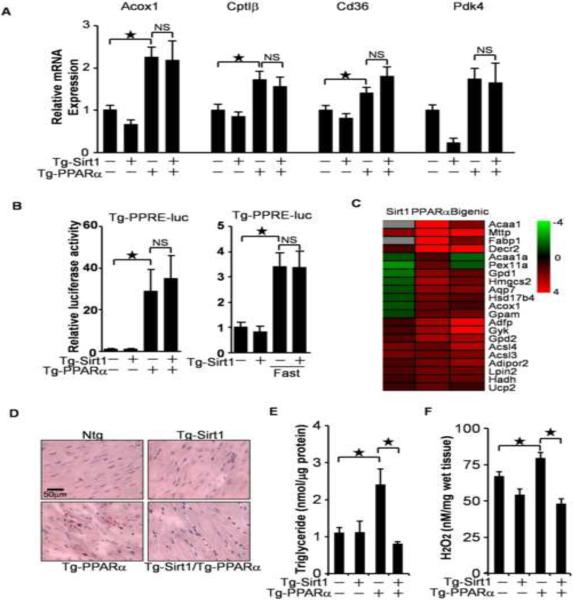
Sirt1 has been reported to activate PPARα (Purushotham et al., 2009). The cardiomyopathic phenotype in the Tg-PPARα/Tg-Sirt1 bigenic mice could be due to enhancement of PPARα-mediated transcriptional activation by Sirt1. To test this hypothesis, expression levels of PPARα target genes were evaluated. As expected, known target genes of PPARα, such as Acox1, CptIβ, Cd36 and Pdk4, were upregulated in Tg-PPARα. However, expression of those genes was not significantly changed in the Tg-PPARα/Tg-Sirt1 bigenic mice compared to in Tg-PPARα (Figure 2A). To elucidate the effect of Sirt1 on PPARα-induced PPRE activation in vivo, the Tg-PPRE-luc mice were crossed with Tg-PPARα and Tg-Sirt1. As shown in Figure 2B, PPARα-induced activation of the PPRE-luc activity was not significantly altered in the Tg-PPARα/Tg-Sirt1 bigenic mice. Furthermore, Sirt1 overexpression did not affect PPRE-luc activity induced by fasting, a physiological stimulus of PPARα. Thus, overexpression of Sirt1 does not affect PPARα-induced PPRE activation in the heart. Microarray analyses demonstrated that a subset of the PPAR target genes was upregulated, while the rest of the target genes were either downregulated or unaffected in the bigenic mice (Figure 2C). Thus, the effect of Sirt1 on PPARα-induced target gene expression was non-uniform in terms of directionality, rather than exhibiting simple enhancement, a result which does not support the involvement of PPRE-regulated genes in the cardiomyopathic phenotype observed in the Tg-PPARα/Tg-Sirt1 bigenic mice.
Figure 2.
The effect of Sirt1 on PPARα activity. (A) Relative mRNA expression of PPARα target genes in transgenic mice. (B) The effect of Sirt1 and PPARα on PPRE-luciferase reporter activity in vivo. Tg-PPRE-luc mice were crossed with Tg-PPARα, Tg-Sirt1, Tg-PPARα/Tg-Sirt1 or control mice (Left). Mice were subjected to 24 hours of fasting (Right). Luciferase assays were performed using heart lysates prepared from these mice. (C) A heat map of known PPARα target genes that were upregulated in Tg-PPARα in microarray analyses. Gray indicates undetectable expression. (D–E) PPARα-induced neutral lipid accumulation was attenuated by Sirt1. Oil red O staining (D) and triglyceride quantification (E) were conducted in each mouse genotype. (F) PPARα-induced increases in H2O2 were attenuated by Sirt1. The numbers of mice examined in each experimental group were: 4–8 (A), 4–7 (B), 4–5 (E) and 6–9 (F). Error bars represent S. E. M.
Although accumulation of neutral lipids, termed lipotoxicity, is thought to be a major cause of PPARα-induced cardiomyopathy (Finck et al., 2002), PPARα-induced triglyceride accumulation was actually suppressed by Sirt1 (Figure 2D and 2E). Lipotoxicity is frequently accompanied by increased oxidative stress. PPARα-induced increases in H2O2 were significantly attenuated in the presence of Sirt1 (Figure 2F). Thus, Sirt1-induced exacerbation of PPARα-induced cardiomyopathy in the Tg-PPARα/Tg-Sirt1 bigenic mice is not due to the enhancement of lipotoxicity.
The target genes of the estrogen related receptor (ERR) are downregulated in Tg-Sirt1/Tg-PPARα bigenic mice
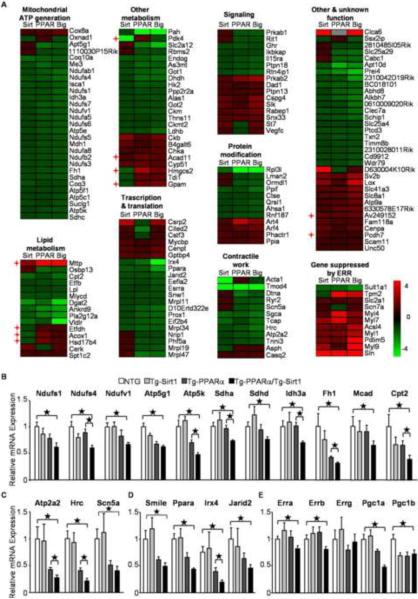
Despite the varied effects of Sirt1 on expression of PPARα target genes, surprisingly, microarray analyses revealed that PPARα and Sirt1 cooperatively downregulated expression of many, if not all, known and predicted target genes of ERRs (Figure 3A). Although a subset of the known ERR target genes, such as Mttp and Acox1, was not suppressed but rather upregulated, those genes are also known to be direct targets of PPARα. Furthermore, genes reported to be upregulated in ERRα knockout mice, such as myosin light chains (Dufour et al., 2007), were upregulated in Tg-PPARα/Tg-Sirt1 bigenic mice, indicating that the bigenic mice have impaired ERR function. The expression level of representative ERR target genes regulating mitochondrial energy metabolism (Figure 3B), cardiac contraction (Figure 3C) and transcription (Figure 3D) was further assessed by quantitative PCR. Overall, these ERR target genes were downregulated in Tg-PPARα and in Tg-Sirt1, and they were further downregulated in the Tg-PPARα/Tg-Sirt1 bigenic mice. These results suggest that expression of ERR target genes was coordinately downregulated by PPARα and Sirt1 in the heart. We also examined the expression levels of ERRs and PGC-1s, since ERRα and PGC-1α are reported to be ERR target genes (Dufour et al., 2007; Wang et al., 2005). These genes were downregulated in Tg-PPARα/Tg-Sirt1 bigenic mice, except for ERRγ (Figure 3E).
Figure 3.
Gene expression profiles of ERR target genes in transgenic mice. (A) A heat map representing the expression profile of ERR target genes in transgenic mice. Known PPAR target genes are indicated by cross mark. Relative gene expression levels of ERR target genes regulating (B) mitochondrial metabolism, (C) contractile work, (D) transcription factors and (E) PGC-1 and ERR isoforms in the indicated transgenic animals are shown (N=8–16). Error bars represent S. E. M.
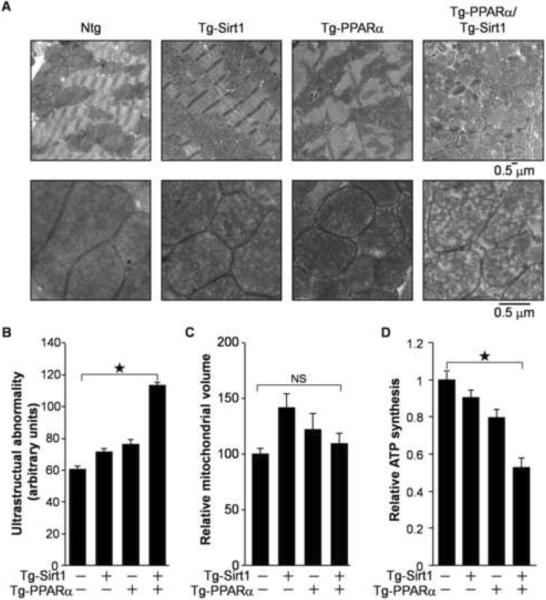
Mitochondria in the hearts of the Tg-PPARα/Tg-Sirt1 bigenic mice exhibited structural abnormalities, such as reduced cristae density (Figure 4A and 4B), but normal mitochondrial volume (Figure 4C). In addition, ATP production was impaired in mitochondria isolated from the Tg-PPARα/Tg-Sirt1 bigenic mice (Figure 4D). Notably, downregulation of ERR target genes regulating myocardial contraction coincided with systolic dysfunction in Tg-PPARα/Tg-Sirt1 bigenic mice (Figure 1I). Taken together, these results suggest that downregulation of the ERR target genes is well correlated with the functional defects in both mitochondria and contractility in the Tg-PPARα/Tg-Sirt1 bigenic mice.
Figure 4.
Mitochondrial function was impaired in Tg-PPARα/Tg-Sirt1 bigenic mice. (A) Electron microscopic analysis of the heart in transgenic mice. (B) Quantitative morphometric analyses of cardiac mitochondrial ultrastructural abnormalities. (C) Mitochondrial volume of the transgenic mice. (D) ATP synthesis in cardiac mitochondria isolated from the transgenic mice (N=7–10). Error bars represent S. E. M.
PPARα and Sirt1 suppress ERRE-mediated gene expression
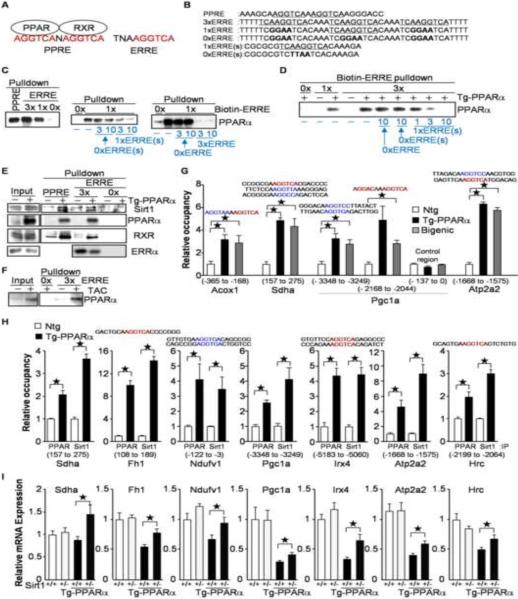
Since the ERRE contains a whole binding sequence of PPARα as a monomeric unit (AGGTCA) (Figure 5A), we hypothesized that PPARα suppresses gene expression through direct interaction with the ERRE. To investigate whether PPARα binds to the ERRE, double-stranded DNA pull-down assays were performed with biotin-labeled double-stranded DNA (Figure 5B) and bacterially expressed recombinant PPARα. As shown in Figure 5C and Figure S2A, PPARα was pulled down with biotin-labeled double-stranded DNA comprising a PPRE, three repeats of ERRE with a 4 bp spacer (3×ERRE) or a single ERRE (1×ERRE), but not with that comprising a 3×ERRE mutant (0×ERRE). The interaction between PPARα and biotin-labeled 1× and 3×ERRE was competitively inhibited by biotin-free double-stranded DNA containing ERRE(s), but not by that with the ERRE mutant(s), suggesting that PPARα directly binds to the ERRE in a sequence-specific manner. In heart lysates prepared from Tg-PPARα, PPARα was pulled down with biotin-labeled 1× and 3×ERRE. The interaction between PPARα and 3×ERRE was competitively inhibited by biotin-free 1×ERRE (Figure 5D and Figure S2B). Thus, PPARα is able to bind to a single hexad motif within the ERRE. As shown in Figure 5E and Figure S2C, Sirt1 was pulled down with an oligonucleotide containing a PPRE, as well as with one containing ERREs, more effectively from Tg-PPARα heart lysates than from non-transgenic heart lysates, suggesting that PPARα recruits Sirt1 to both the PPRE and the ERRE. The amount of RXR pulled down with the ERRE was less than that with the PPRE, suggesting that PPARα and RXR form a dimer more effectively on the PPRE than on the ERRE. ERRα was pulled down with a biotin-labeled ERRE, but the interaction was competitively inhibited in the presence of PPARα, indicating that PPARα and ERRα compete for the common DNA binding site (Figure 5E). Endogenous PPARα was recovered with biotin-labeled 3×ERRE more effectively from the heart lysate prepared after 4 weeks of TAC than from the control heart lysate (Figure 5F and Figure S2D). Recombinant PPARα bound to ERREs even in the presence of 20- and 60-fold excesses of non-biotin labeled polydeoxy (Inosinate-Cytidylate) acid as a non-specific competitor (Figure S2E). To examine whether PPARα binds to the AGGTCA motif in vivo, chromatin immunoprecipitation (ChIP) assays were performed. As expected, genomic flanking regions of the PPRE, such as those in the promoter regions of Acox1 and PGC-1α (−2168 to −2044), were significantly precipitated with anti-Flag antibody when Flag-tagged PPARα was expressed in the hearts of Tg-PPARα and Tg-PPARα/Tg-Sirt1 bigenic mice (Figure 5G). Flag-tagged PPARα also bound to flanking regions of single AGGTCA motifs, which are not configured as a typical PPRE but are found either within an ERRE or as an isolated AGGTCA motif, such as those in the promoter regions of Sdha, PGC-1α (−3348 to −3249), and Atp2a2 (Figure 5G). Increased expression of PPARα resulted in increased DNA binding of both PPARα and Sirt1 on flanking regions of the single hexad motif in the promoter regions of PPARα/Sirt1 suppressed genes (Figure 5H). There was no typical PPRE located within the 1kb flanking region of all promoters we tested by ChIP assays, except in those of Acox1 and PGC1α (Figure 5G), suggesting that PPARα recruits Sirt1 to the single hexad motif in the heart. The downregulation of the ERR target genes observed in Tg-PPARα was partially normalized by haploinsufficiency of Sirt1 (Figure 5I). Thus, Sirt1 recruited by PPARα negatively regulates gene expression directed by the single hexad motif.
Figure 5.
PPARα binds to the single hexad motif within the ERRE. (A) Schematic representation of topology of PPAR and RXR on the PPRE, and comparison of the PPRE and the ERRE. (B–E) Recombinant PPARα (C) and PPARα in Tg-PPARα heart lysate (D–E) binds to both the PPRE and the ERRE. PPARα was pulled down using biotin-labeled double-stranded DNA with the indicated sequences (B). Non-biotin labeled competitors (1 to 10-fold of biotin-labeled DNA) are shown in blue (C–D). (F) Endogenous PPARα was pulled down with 3×ERRE after 4 weeks of TAC. (G) PPARα binds to the single hexad motif in the heart. ChIP assays were performed with homogenates from Tg-PPARα and Tg-PPARα/Tg-Sirt1 hearts and anti-Flag antibody. Target single hexad motifs are shown in red (AGGTCA and AGGACA) and blue (one nucleotide redundancy in the last 3 letters from AGGTCA or AGGACA) with proximal 7bp sequences. Genomic regions amplified by quantitative PCR are indicated by nucleotide number from first codon under target genes (N=6–12). (H) PPARα recruits Sirt1 to the flanking region of the single hexad motif. ChIP assays were performed in Tg-PPARα hearts using anti-PPARα and anti-Sirt1 antibodies (N=8). (I) Sirt1 recruited by PPARα suppresses gene expression. Quantitative PCR was performed using cDNA prepared from the indicated transgenic mice (N=5−20). Error bars represent S. E. M.
PPARα suppresses the ERRE in a Sirt1-dependent and RXR-independent manner
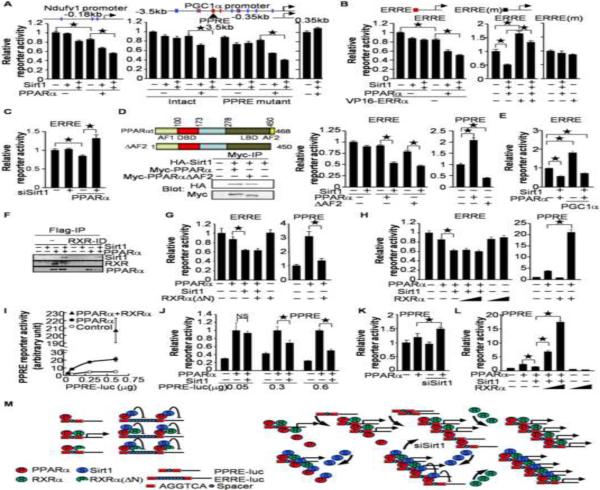
To investigate whether PPARα and Sirt1 suppress promoter activity through interaction with the ERRE, reporter gene assays were performed in cultured myocytes. As shown in Figure 6A, PPARα and Sirt1 cooperatively suppressed reporter activity driven by the promoter region of Ndufv1 containing 3 hexad motifs. The −3.5kb to −0.35kb promoter region of PGC1α also contains multiple common hexad motifs. PPARα and Sirt1 cooperatively suppressed reporter activity driven by a 3.5kb, but not by a 0.35kb, promoter region. The PGC-1α promoter contains a typical PPRE. PPARα may displace the other PPARs, preventing them from binding to the PPRE and thereby suppressing the promoter activity. However, PPARα and Sirt1 cooperatively suppressed reporter gene activity driven by the PGC-1α promoter with a PPRE mutation. Thus, the PPRE is not required for PPARα/Sirt1-mediated suppression of PGC-1α promoter activity. PPARα and Sirt1 also suppressed reporter activity driven by 3 repeats of the ERRE connected to the thymidine kinase promoter, whereas VP16-ERRα, a transcriptional activation domain fused to ERRα, normalized both the reporter gene activity and ERR target gene expression, which were suppressed by PPARα/Sirt1 in cultured myocytes (Figures 6B and S3A). However, co-expression of PPARα and Sirt1 did not effectively suppress, and VP16-ERRα failed to activate, the reporter activity when the ERRE sequences were mutated (Figure 6B). Thus, the single hexad motif of HREs, including that in the ERRE, plays an essential role in mediating transcriptional suppression by PPARα and Sirt1. PPARα-mediated suppression of ERRE-reporter activity was attenuated by knockdown of Sirt1 (Figure 6C), suggesting that endogenous Sirt1 is a crucial partner for the PPARα-mediated suppression of the ERRE.
Figure 6.
The role of RXR in PPARα/Sirt1-mediated ERRE suppression. (A) PPARα and Sirt1 cooperatively suppress ERR target gene promoters. (B) The ERRE is an important interface for PPARα/Sirt1-mediated gene suppression. (A–B) Schematic representations of each promoter are shown in these panels. The common hexad motifs of HREs are indicated by a red line (AGGTCA and AGGACA), blue line (one nucleotide redundancy in the last 3 letters from AGGTCA or AGGACA) or black line (ERRE mutant: TCGGAATCA). (C) Sirt1 is a crucial partner for PPARα-induced ERRE suppression. (D) Transcriptional activation of PPARα is not essential for ERRE suppression. Schematic representation of PPARα and PPARαΔAF2 are shown. AF1: Activation function 1, DBD: DNA binding domain, LBD: ligand binding domain, AF2: activation function 2. HA-Sirt1 was co-immunoprecipitated with both Myc-PPARα and Myc-PPARαΔAF2. (E) PGC-1α fails to normalize PPARα/Sirt1-mediated ERRE suppression. (F) RXR is not necessary for interaction between PPARα and Sirt1. RXR was removed from heart lysates by immunodeprivation. PPARα was then immunoprecipitated with anti-Flag-antibody. (G) The effect of an RXRα mutant lacking the DNA binding domain (RXRα(ΔN)) on PPARα-induced PPRE activation and ERRE suppression. (H) The effect of full-length RXRα on PPARα-induced PPRE activation and ERRE suppression. (I) The dosage effect of PPRE-reporter on PPARα-induced reporter activity. The indicated amounts of pPPRE-luc reporter plasmids were transfected together with PPARα and RXRα expression vectors (0.3 μg). (J) The dosage effect of PPRE-reporter on PPARα/Sirt1-mediated transcriptional regulation. The mean value for activity with PPARα expression alone was expressed as 1. (K) Knockdown of Sirt1 enhances PPARα-induced PPRE activation. (L) RXRα counteracts PPARα/Sirt1-mediated suppression of PPRE reporter activity. (A–F and G–L) Luciferase assays were performed after 1 (A–B, D–E, and G–L) and 3 days (C and K) of transduction and transfection with the indicated constructs (N=6–12). Error bars represent S. E. M. (M) Schematic representation of the data shown in this figure. PPARα-induced PPRE activation is enhanced by RXRα but prevented by RXRα(ΔN). In contrast, PPARα/Sirt1-mediated ERRE suppression is not affected by RXRα or RXRα(ΔN) (Left). When RXR does not cover the half core site of the PPRE, the PPARα/Sirt1 complex suppresses transcription through the single hexad unit within the PPRE (Right).
PPARα-induced PPRE activation may contribute to the suppression of the ERRE through recruitment of transcription activation machinery from the ERRE to the PPRE, termed squelching. To test this possibility, we examined the effect of a PPARα mutant (PPARαΔAF2), which cannot bind to the coactivators, on ERRE-reporter gene activity. The PPARα mutant retained the ability to bind to Sirt1 and suppressed ERRE reporter gene activity in the presence of Sirt1 (Figure 6D). Thus, PPARα-mediated squelching is not required for the ERRE suppression.
Since PGC-1α and PGC-1β activate ERRs, downregulation of PGC-1s (Figure. 3E) may be a major mechanism for PPARα/Sirt1-induced downregulation of ERR target genes. However, suppression of ERRE, Naufv1 and PGC-1α reporter genes by PPARα/Sirt1 was observed even in the presence of PGC-1α overexpression (Figure 6E and S3B). Furthermore, overexpression of PGC-1α failed to normalize ERR target gene suppression by PPARα/Sirt1 (Figure S3C). Thus, downregulation of PGC1α is not essential for PPARα/Sirt1-mediated ERRE suppression.
RXRs play an important role in mediating DNA binding and transcriptional activation of PPARα through the PPRE. However, an isolated single hexad motif, such as that in the ERRE, would not provide a binding site for RXR. Therefore, we investigated the role of RXR in mediating the function of PPARα and Sirt1 at the ERRE. To examine the role of RXR in the interaction between PPARα and Sirt1, immunodeprivation of RXRs was performed with an anti-RXR antibody recognizing all RXR isoforms. Sirt1 was co-precipitated with Flag-PPARα after immunodeprivation of RXR, suggesting that RXR is not required for interaction between PPARα and Sirt1 in the heart (Figure 6F). To assess the role of RXR in PPARα/Sirt1-mediated ERRE suppression, we examined the effects of RXRα and an RXRα mutant lacking the N-terminal DNA binding domain (RXRα (ΔN)) on ERRE reporter activity. Although PPARα-induced PPRE activation was significantly suppressed by RXRα (ΔN), PPARα/Sirt1-mediated ERRE suppression was not affected (Figure 6G). Furthermore, although RXRα significantly enhanced PPRE reporter activity in the presence of PPARα, it did not affect PPARα/Sirt1-mediated ERRE suppression (Figure 6H). These results suggest that RXR is a crucial factor for PPARα-induced PPRE activation, but not for PPARα/Sirt1-mediated ERRE suppression (Figure 6M left).
Since PPRE/DR1 contains the single hexad motif (Figure 5A), the PPARα/Sirt1 complex may suppress transcription under circumstances where the level of RXRs is insufficient on the PPRE/DR1. To test this hypothesis, we introduced different doses of the PPRE reporter genes, together with PPARα and Sirt1, into myocytes. PPARα-induced luminescence increased sharply in response to increased amounts of the PPRE reporter gene in the low dosage range, but became saturated in the high dosage range. The saturated reporter activity was augmented, however, by overexpression of RXRα, suggesting that the amount of endogenous RXRs forming dimers with exogenous PPARα on the PPRE is limited (Figure 6I). PPARα-induced PPRE activation was not significantly changed by overexpression of Sirt1 when a smaller amount of PPRE reporter plasmid was introduced, but Sirt1 suppressed the reporter activity when a higher amount of PPRE reporter was cotransfected into the cells (Figure 6J). The PPARα-induced PPRE reporter activation was enhanced by knocking down Sirt1 (Figure 6K), suggesting that activation of the PPRE reporter gene by PPARα may be determined by the balance between PPARα/RXR-mediated transcriptional activation and PPARα/Sirt1-mediated transcriptional suppression when a high level of PPRE reporter is used. PPARα/Sirt1-mediated PPRE suppression was reversed by overexpression of RXRα (Figure 6L). These results suggest that PPARα/Sirt1 suppresses transcription at the PPRE when RXRs are insufficient to cover the other side of the half core site of PPRE (Figure 6M right). Thus, PPARα facilitates both activation of the PPRE and suppression of the single hexad motif, like in the ERRE, with RXR and Sirt1, respectively.
PPARα and Sirt1 suppress ERR target gene expression during pressure overload in the heart
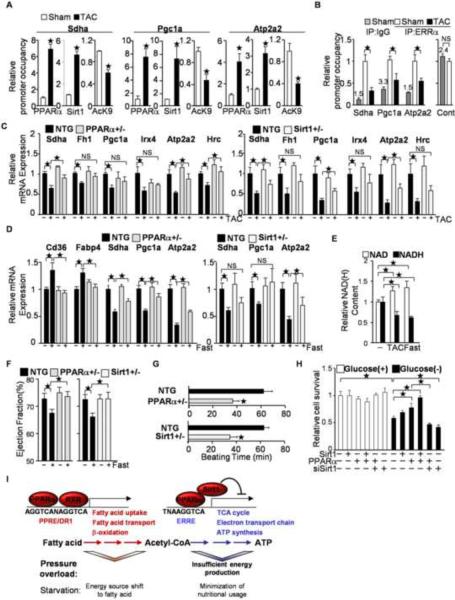
To examine whether PPARα and Sirt1-mediated gene suppression takes place in the heart subjected to pressure overload, ChIP assays were performed. Binding of PPARα and Sirt1 to the flanking region of the single hexad motif was significantly increased after TAC in the promoters of Shda, PGC-1α and Apt2a2 after 2 weeks (Figure S4A) and 4 weeks of TAC (Figure 7A). The level of histone H3 acetylation at lysine 9, which is deacetylated by Sirt1, was decreased in the promoter regions of the ERR target genes after TAC. The occupancy of ERRα on the ERREs was decreased by pressure overload (Figure 7B) and by overexpression of PPARα (Figure S4B). On the other hand, knocking down ERRα did not affect ERRE reporter gene activity or recruitment of PPARα to the ERRE in cultured cardiac myocytes (Figure S4C to S4E), despite the fact that the occupancy of ERRα on the flanking region of ERREs was significantly reduced (Figure S4F) and that expression of several ERR target genes was significantly attenuated by downregulation of ERRα (Figure S4G). Thus, downregulation of ERRα is not sufficient to induce PPARα/Sirt1-mediated ERRE suppression. TAC-induced downregulation of the ERR target genes was attenuated in PPARα+/− and Sirt1+/− mice (Figure 7C and Figure S4H). Thus, endogenous PPARα and Sirt1 are both involved in the suppression of the ERR target genes in the heart during pressure overload.
Figure 7.
Involvement of PPARα/Sirt1-mediated ERRE suppression in pressure overload(PO)-induced cardiac hypertrophy and fasting. (A–B) After 4 weeks of TAC, ChIP assays were performed with anti-Sirt1, anti-PPARα and anti-histone H3 acetyllysine 9 specific antibodies (A) or anti-ERRα antibody (B) (N=4–8). PPARα and Sirt1 bind to the single hexad motifs of HREs in response to PO in the heart (A), whereas the occupancy of ERRα on the ERRE decreases (B). (B) The control region corresponds with Pgc1a (−137 to 0). The number above the control IgG column represents raw data of the copy number (×1000) (C) Haploinsufficiencies of PPARα and Sirt1 attenuate PO-induced downregulation of ERR target gene expression. The expression levels of the indicated genes were examined by quantitative PCR (N=4–12). (D) PPARα/Sirt1-mediated ERRE suppression is a fasting response. PPARα+/− and Sirt1+/− mice were subjected to fasting for 24 hours. The expression levels of the indicated genes were examined (N=8–20). (E) The NAD/NADH ratio is increased by 24 hours of fasting and 4 weeks of TAC (N=4–8). (F) Fasting-induced attenuation of systolic function is mediated by PPARα and Sirt1. Ejection fraction was measured in PPARα+/− and Sirt1+/− mice after 24 hours of fasting. (N=18–20). (G) Hearts were isolated from PPARα+/− and Sirt1+/− mice and subjected to the Langendorff perfused heart experiment without any nutrients in the perfusate. The duration of consecutive beating was evaluated in each heart (N=7–9). (H) PPARα and Sirt1 prolong survival of cultured cardiac myocytes under glucose-free conditions. The indicated expression or knockdown was introduced with adenovirus vectors. Cell viability was evaluated with Trypan blue dye exclusion following incubation with complete or glucose-free medium for 72 hours (N=3). Error bars represent S. E. M. (I) Schematic representations of the regulatory roles of PPARα, Sirt1 and RXR on the PPRE and the ERRE during pressure overload and fasting. PPARα shifts the energy source to fatty acids through PPRE activation, and minimizes nutritional usage through ERRE suppression, with the functional binding partners RXR and Sirt1, respectively. PPARα-mediated ERRE suppression is triggered by pressure overload, which exacerbates cardiac dysfunction due to insufficient energy production.
PPARα/Sirt1-induced suppression of ERR target gene expression is adaptive during fasting
In the last part of the study, a physiological role for the gene suppression through PPARα and Sirt1 interaction was investigated. Since both PPARα and Sirt1 are crucial factors that mediate fasting responses, we hypothesized that PPARα/Sirt1-mediated ERR suppression is a fasting response. To test this hypothesis, PPARα+/− and Sirt1+/− mice were subjected to fasting for 24 hours. As shown in Figure 7D, although typical PPAR target genes, including Cd36 and Fabp4, were upregulated following fasting in wild type mice, their expression was reduced in PPARα+/− mice. ERR target genes, such as Sdha, Atp2a2 and PGC1α were downregulated in wild type mice following fasting, but this downregulation was partly prevented in PPARα+/− mice, suggesting that PPARα mediates both PPRE activation and ERRE suppression during fasting. The fasting-induced downregulation of ERR target genes was partly normalized in Sirt1+/− mice. Thus, Sirt1 is required for suppression of ERR target genes during starvation in the heart. Increased NAD+ and decreased NADH levels were observed in the heart following both fasting and TAC, conditions known to effectively promote Sirt1 activation due to an increased NAD/NADH ratio (Figure 7E).
To investigate the role of PPARα and Sirt1 in the heart under fasting, cardiac function was assessed. Cardiac systolic function, such as ejection fraction, was slightly decreased in wild type mice after fasting, but not in either PPARα+/− or Sirt1+/− mice (Figure 7F). Using an isolated working mouse heart model, under nutrient-free conditions, spontaneous beating terminated earlier in hearts isolated from PPARα+/− and Sirt1+/− mice than in hearts from wild type mice, suggesting that PPARα and Sirt1 play an important role in maintaining beating of the heart under starvation conditions (Figure 7G). After 72 hours of incubation with a glucose-free medium, the survival rate was decreased in control cultured myocytes, but was improved by exogenous PPARα. This protective effect was attenuated by knockdown of Sirt1, suggesting that Sirt1 is required for the PPARα-mediated prolongation of myocyte survival under starvation conditions (Figure 7H). These results suggest that ERRE suppression by the PPARα/Sirt1 is a fasting response to prolong cardiac working time and cell survival under starvation conditions through reduction of mitochondrial and systolic outputs. This is in contrast to the condition of pressure overload, where the heart cannot afford to slow down because of the need to maintain cardiac output against increased afterload. Thus, ERRE suppression by the PPARα/Sirt1 complex is induced by both pressure overload and fasting, but the advantageousness of its function is context-dependent.
DISCUSSION
PPARα binding to the single hexad motif suppresses transcription through the ERRE
We here demonstrate molecular mechanisms leading to heart failure under conditions of mechanical loading which involve suppression of the single hexad DNA binding motif of nuclear receptors, including that in the ERRE, by the PPARα/Sirt1 complex. Although PPARα acts as a transcriptional activator on PPRE, it acts as a transcriptional suppressor on the single hexad motif found in the ERRE. Previously, ChIP-on-chip analyses identified the isolated single hexad motif of HREs as a major binding element of PPARα. Furthermore, DNA binding sequences directly bound by PPARα are found not only in the promoters of 16% of upregulated genes, but also in the promoters of 18% of downregulated genes in the presence of a PPARα ligand (van der Meer et al., 2010). These findings support the possibility that PPARα could negatively regulate transcription through the single hexad motif. Our results not only establish the functional significance of the regulation of the single hexad DNA binding motif by PPARα, but also show that it could contribute to mitochondrial dysfunction and eventual development of heart failure under conditions of mechanical loading in the heart.
Dichotomous functions of PPARα in the heart
The cardiac role of PPARα is controversial because both PPARα knockout and PPARα-overexpression lead to detrimental phenotypes in the mouse heart (Finck et al., 2002; Smeets et al., 2008). The detrimental effect seen in PPARα homozygous knockout mice may result from impaired PPRE-mediated gene expression. Our results suggest that PPARα-mediated ERRE suppression is detrimental to the heart. Since the PPARα/RXR dimer exhibits a higher affinity for the PPRE than for the single hexad motif (data not shown), PPRE activation is probably the primary role of the PPARα/RXR dimer. However, when PPARα (and Sirt1) is upregulated, the PPARα-Sirt1 complex suppresses ERRE independently of RXR. Although PPARα-mediated activation of the PPRE is protective, suppression of ERRE could be detrimental under stress conditions, such as pressure overload. Thus, our results provide a potential explanation for the controversy regarding the cardiac role of PPARα.
Cardiac role of Sirt1
Sirt1 is required for PPARα-induced regulation of target gene expression through the PPRE in the liver (Purushotham et al., 2009), whereas we here show that PPARα and Sirt1 work together, but that they suppress the ERRE in the heart. The direction of the transcriptional activity of the PPARα/Sirt1 complex may depend upon DNA binding sites, requirement of RXR, cell type or possibly other unknown mechanisms.
Sirt1 deacetylates ERRα to enhance its DNA binding in 293 cells (Wilson et al., 2010). Although ERRα was modestly acetylated in the Sirt1−/− heart, ERRα acetylation was below the detection limit in wild type mice both at baseline and after pressure overload (Figure S4I). Even if Sirt1 deacetylates ERRα in the heart under pressure overload, its role in mediating the PPARα/Sirt1-induced suppression of ERRE remains to be elucidated.
Since suppression of the ERR target genes under pressure overload conditions is attenuated in Sirt1+/− mice (Figure 7C), upregulation of endogenous Sirt1 plays an essential role in mediating suppression of the ERRE. The results of the ChIP experiments showed that the acetylation status of histone H3 at lysine 9, a specific deacetylation site of Sirt1, located in flanking regions of the ERRE, was decreased during pressure overload (Figure 7A). Thus, we speculate that upregulation and recruitment of Sirt1 through PPARα may inhibit transcription through the ERRE via histone deacetylation. Alternatively, Sirt1-mediated recruitment of nuclear corepressors (Picard et al., 2004) or histone H1 (Vaquero et al., 2004) may contribute to the suppression of ERRE-mediated transcription. Further investigation is required to elucidate the mechanism by which the PPARα/Sirt1 complex mediates the suppression.
Potential effects of PGC1s, ERRs and the other nuclear receptors on the ERRE in the failing heart
Although PGC-1s are important transcription co-factors for ERRs, downregulation of PGC-1α was not essential for PPARα/Sirt1-mediated ERRE suppression (Figure 6E, S3B and S3C). Knock-down of ERRα does not appear to be sufficient for inducing recruitment of PPARα to ERREs, whereas upregulated PPARα was recruited to the ERRE (Figure S4C). The level of PPARα at baseline may not be sufficient to occupy ERREs in neonatal myocytes. Alternatively, ERRγ or ERRβ may effectively compensate for the downregulation of ERRα, thereby preventing the recruitment of PPARα to ERREs. Interestingly, each monomeric unit of the nuclear receptor commonly recognizes and binds to the single hexad motif, regardless of the overall receptor assembly (McKenna et al., 2009). Therefore, our finding that PPARα can bind to the single hexad motif is potentially applicable to the other nuclear receptors as well. In addition, it is possible that the PPARα/Sirt1complex suppresses targets of other nuclear receptors when their DNA binding sites do not have a typical PPRE, but are more similar to an isolated single hexad-like sequence. More investigations are needed in order to elucidate the role of the PPARα/Sirt1complex in each nuclear receptor element.
Fasting responses involved in the development of cardiomyopathy
Fatty acid catabolism plays a crucial role in maintaining energy homeostasis during fasting, because fat stored under fed conditions becomes a major source of energy during fasting. Plasma fatty acids are transported into mitochondria and then converted to acetyl-CoA through β-oxidation. The acetyl-CoA is consumed by the tricarboxylic acid (TCA) cycle, which couples to ATP synthesis through the electron transport chain. Interestingly, PPARα target genes harboring a PPRE in their promoter regions are mainly found in the upstream metabolic pathway, including those involved in fatty acid transport and β-oxidation (Rakhshandehroo et al., 2007). In contrast, ERR target genes are relatively enriched in the downstream metabolic pathway, including the TCA cycle and the electron transport chain. Here, we showed that fasting-induced ERR target gene suppression is attenuated in PPARα+/− and Sirt1+/− mice (Figure 7D). Thus, although PPARα facilitates fatty acid utilization through PPRE activation in the presence of RXR, together with Sirt1, PPARα can minimize nutritional consumption through ERRE suppression under starvation conditions (Figure 7I). Since the mitochondrion is the major energy-generating organelle coupling with nutritional consumption, and the heart is a major organ consuming energy and nutrition, the PPARα/Sirt1-mediated restriction of the mitochondrial energy metabolism could contribute to survival of cells and organisms by minimizing the consumption of body-stored nutrition during starvation. Reduced metabolic rate is believed to increase survival time in hibernating animals (Staples and Brown, 2008). Starvation leads to decreased cardiac work, such as reduced fractional shortening in animals. We found that the fasting-induced reduction of ejection fraction was abolished in PPARα+/− and Sirt1+/− mice (Figure 7F). Thus, the cardiac fasting response, which slows down cardiac outputs, is mediated by PPARα and Sirt1. Our data suggest that PPARα/Sirt1-mediated ERRE suppression contributes to the delay of cell death as well as termination of cardiac function during starvation conditions. The physiological significance of this mechanism should be further investigated in the heart in vivo.
Our data support the notion that both PPARα and Sirt1 are crucial mediators of the fasting response. Importantly, although such a metabolic adaptation, which minimizes consumption of nutrition, may provide a survival advantage during fasting, it could be inappropriate during pressure overload, when the heart requires energy to maintain contraction against increased mechanical load. It is currently unknown how PPARα and Sirt1 are upregulated in the failing heart. Reduced nutrition and/or ATP resulting from high energy expenditure may be sensed as conditions of reduced nutritional intake in the heart subjected to pressure overload. Indeed, decreased ATP and creatine phosphate have been observed in human heart failure patients (Weiss et al., 2005). In summary, PPARα/Sirt1-mediated downregulation of genes regulated by the single hexad unit, including a subset of ERR target genes, induces mitochondrial dysfunction, which plays an important role in mediating heart failure. We propose that normalizing the level of PPARα and Sirt1 to attenuate ERRE suppression, and to achieve physiological activation of the PPRE by PPARα/RXR, may alleviate the mitochondrial dysfunction commonly observed in heart failure patients with increasing mechanical loading.
EXPERIMENTAL PROCEDURES
Transgenic mice
The following transgenic mice were used for this study: Cardiac-specific overexpression of PPARα (Tg-PPARα) or Sirt1 (Tg-Sirt1), Tg-PPARα/Tg-Sirt1, cardiac-specific Sirt1 heterozygous knockout (Sirt1+/−), Tg-PPARα/Sirt1+/−, PPRE-luciferase transgenic (Tg-PPRE-luc), Tg-PPARα/Tg-PPRE-luc, Tg-Sirt1/Tg-PPRE-luc, Tg-PPARα/Tg-Sirt1/Tg-PPRE-luc, and PPARα heterozygous knockout (PPARα+/−) mice. The generation strategies and genetic backgrounds of these transgenic mice are described in the supplemental information. All procedures involving animals were performed in accordance with protocols approved by the University of Medicine and Dentistry of New Jersey.
Transverse aortic banding
Mice were anesthetized with pentobarbital sodium. The left chest was opened. Aortic constriction was performed by ligation of the transverse artery and left common carotid artery with a 26- or 27-gauge needle.
Isolated working mouse heart model
Mouse hearts were dissected and perfused with a perfusate [120 mM NaCl, 25 mM NaHCO3, 5.9 mM KCl, 1.2 mM MgCl2, 2.5 mM CaCl2, 0.5 mM EDTA]. The termination time of spontaneous beating was defined as the time at which +dP/dt of the isolated heart reached less than 400 mmHg/second, or the beating was paused for more than 0.6 seconds, whichever came first.
Echocardiography
Mice were anesthetized using avertin (Sigma), and echocardiography was performed to take two-dimension guided M-mode of left ventricular (LV) internal diameter. LV ejection fraction was calculated as [(LVEDD)3-(LVESD) 3]/(LVEDD)3 × 100. LVEDD; Left ventricular end-diastolic diameter; LVESD: LV end-systolic diameter.
Microarray analyses
Total RNA from the hearts was extracted using the RNeasy Fibrous Tissue Mini Kit (Qiagen). The RNA was used to produce a biotin-labeled cRNA probe for hybridization to a mouse expression array 430_A2.0 gene chip (Affymetrix, Santa Clara, CA). The expression signal for each probe-set was obtained by the RMA (robust multi-chip analysis) method, and the detection was assessed by the MAS (Microarray Suite) 5.0 method. Expression patterns of regulated genes were graphically represented in a heat map. The clustering and visualization was carried out by the MultiExperiment Viewer (MeV) (http://www.tm4.org/mev.html). PPAR and ERR target genes were selected from published articles. Those target genes overlapping genes whose expression was significantly changed in Tg-PPARα, Tg-Sirt1 and Tg-PPARα/Tg-Sirt1 bigenic mice were extracted to generate heat maps.
ATP synthesis
Mitochondria were freshly isolated from the ventricule using a mitochondria isolation kit (Sigma, MITOISO1). The isolated mitochondria from 50 mg tissue weight was suspended with 50 μl storage buffer [10 mM Hepes pH 7.4, 250 mM Sucrose, 2 mM K2HPO4], resulting in around 100 μl final volume of mitochondrial fraction. ATP production was measured by luminometric assay using an ATP bioluminescent assay kit (Sigma FL-AA). The ATP assay mix was diluted with the dilution buffer (1000 fold dilution). The diluted ATP assay mix (25 μl) was combined with 25 μl substrate buffer [5 mM Hepes pH 7.9, 210 mM Mannitol, 70 mM Sucrose, 10 mM Pyruvate, 10 mM Malate, 4 mM K2HPO4], and then 1 μl mitochondrial solution were added. After 5 minutes incubation at room temperature, the reaction was started by the addition of 1 μl 12.5 mM ADP. The luminescence was measured by luminometer for 30 seconds. The mean value of non-Tg was expressed as 1.
Quantitative RT-PCR
Total RNA was prepared from left ventricles using the RNeasy Fibrous Tissue Mini Kit (Qiagen), and then cDNA was generated using M-MLV Reverse transcriptase (Promega). Real-time RT-PCR was performed using the Maxima SYBR Green qPCR master mix (Fermentas). Ribosomal RNA (15S) was used as an internal control. The oligonucleotide primers used to carry out the PCRs are described in the supplemental information.
Luciferase assay
Luciferase activity was measure with a luciferase assay system (Promega). Dissected ventricles from Tg-PPRE-luc transgenic mice were weighed and lysed with 300 μl Reporter lysis buffer (Promega) per 50 mg tissue weight using a Polytron homogenizer. In cultured mycocytes, reporter and expression plasmids were transfected into neonatal primary myocytes (12 well plate) using LipofectAmine 2000 (Invitrogen). The amounts of expression vectors transfected into cells per well were 0.3 μg PPARα, 0.3 μg Myc-PPARα, 0.3 μg PPARαΔAF2, 0.01 and 0.1 μg Sirt1, 0.1 and 0.3 μg RXRα, 0.6 μg RXRα(ΔN), 0.3 μg Myc-VP16-ERRα, and 0.3 μg luciferase reporter plasmids. The total amount of plasmid vectors was kept constant (1 to 1.3 μg) using the control vector. Alternatively, after overnight culture, adenovirus vectors carrying Sirt1 (0.1 to 3MOI), PPARα (5 MOI), siRNA-Sirt1 (10 MOI), siRNA-ERRα (10 MOI) or siRNA-scramble (10 MOI) were transduced. The total MOI of adenovirus was kept constant using LacZ virus. Cells were lysed with 100 μl Reporter lysis buffer after 1–4 days of transfection or transduction. For both tissue and cultured cell lysates, the luminescence was normalized by protein content.
Immunoblotting
Mouse heart tissue samples (10–30 μg) were analyzed by Western blot using antibodies against PPARα (Cayman), Sirt1 (Millipore), RXR (Santa Cruz), ERRα (Millipore), Histone H3 (Cell Signaling Technology), Acetylated-lysine (Cell Signaling Technology), Tubulin (Sigma) and GAPDH (Sigma).
Double-stranded DNA pull down assay
Mouse heart tissue samples were weighed and lysed with 0.5 ml lysis buffer containing 10 mM Tris pH 8, 3 mM CaCl2, 1% NP-40, 320 mM Sucrose, 0.1 mM EDTA, 1 mM DTT and Protease Inhibitor Cocktail (Sigma) per 50 mg tissue weight. The heart lysate (50 μl) was diluted with an equal volume of 2 × pull down buffer (20 mM Hepes, pH 8, 100 mM KCl, 5 mM MgCl2, 2 mM EDTA, 2 mM DTT and 0.2% NP-40. Biotin-labeled double-stranded DNA (total 15 pmol/100 μl) and 15 μl streptavidin-agarose (Pierce) were added to the lysate and rotated at 4°C for 2 hours. The streptavidin-agarose was washed with 1 ml 1× pull down buffer 3 times. Biotin labeled sense and non-biotin-labeled anti-sense oligonucleotides were purchased from IDT (Integrated DNA Technologies) and annealed using a thermal cycler to generate biotin labeled double-stranded DNA.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assays were performed using the SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology #9002) with several modifications.. Briefly, formaldehydecrosslikned chromatin solution was prepared from ventricles corresponding to 200–1000 mg (from 3–10 mice). The chromatin solution (1 to 5 μg DNA) was used for each immunoprecipitation with 1 μg antibodies and 15 μl protein G-agarose beads. Collected chromatin fragments were measured by quantitative PCR with oligonucleotide primers described in the supplemental information.
Statistical methods and error bars
Statistical comparisons were made using the Student's t test. P<0.05 was defined as statistically significant and indicated by a filled asterisk. P>0.05 was indicated by NS. All error bars represent S. E. M.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Daniela Zablocki for critical reading of the manuscript, Drs. Yasuhiro Maejima and Shouji Matsushima for technical suggestions, and Drs. Daniel P. Kelly and Teresa Leone at Sanford-Burnham Medical Research Institute for providing the Tg-PPARα mice. This work was supported in part by U.S. Public Health Service Grants HL102738, HL67724, HL69020, HL91469, AG23039, and AG27211, and the Foundation Leducq Transatlantic Networks of Excellence.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, Evans RM, Blanchette M, Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and gamma. Cell Metab. 2007;5:345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, Han X, Gross RW, Kozak R, Lopaschuk GD, Kelly DP. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- Glass CK. Going nuclear in metabolic and cardiovascular disease. J Clin Invest. 2006;116:556–560. doi: 10.1172/JCI27913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Imahashi K, Dufour CR, Weinheimer CJ, Courtois M, Kovacs A, Giguere V, Murphy E, Kelly DP. The nuclear receptor ERRalpha is required for the bioenergetic and functional adaptation to cardiac pressure overload. Cell Metab. 2007;6:25–37. doi: 10.1016/j.cmet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- Irukayama-Tomobe Y, Miyauchi T, Sakai S, Kasuya Y, Ogata T, Takanashi M, Iemitsu M, Sudo T, Goto K, Yamaguchi I. Endothelin-1-induced cardiac hypertrophy is inhibited by activation of peroxisome proliferator-activated receptor-alpha partly via blockade of c-Jun NH2-terminal kinase pathway. Circulation. 2004;109:904–910. doi: 10.1161/01.CIR.0000112596.06954.00. [DOI] [PubMed] [Google Scholar]
- Karamanlidis G, Nascimben L, Couper GS, Shekar PS, Del Monte F, Tian R. Defective DNA Replication Impairs Mitochondrial Biogenesis In Human Failing Hearts. Circ Res. 2010 doi: 10.1161/CIRCRESAHA.109.212753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Cooney AJ, DeMayo FJ, Downes M, Glass CK, Lanz RB, Lazar MA, Mangelsdorf DJ, Moore DD, Qin J, et al. Minireview: Evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol Endocrinol. 2009;23:740–746. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhshandehroo M, Sanderson LM, Matilainen M, Stienstra R, Carlberg C, de Groot PJ, Muller M, Kersten S. Comprehensive Analysis of PPARalpha-Dependent Regulation of Hepatic Lipid Metabolism by Expression Profiling. PPAR Res. 2007;2007:26839. doi: 10.1155/2007/26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LK, Finck BN, Kelly DP. Mouse models of mitochondrial dysfunction and heart failure. J Mol Cell Cardiol. 2005;38:81–91. doi: 10.1016/j.yjmcc.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- Sihag S, Cresci S, Li AY, Sucharov CC, Lehman JJ. PGC-1alpha and ERRalpha target gene downregulation is a signature of the failing human heart. J Mol Cell Cardiol. 2009;46:201–212. doi: 10.1016/j.yjmcc.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets PJ, Teunissen BE, Willemsen PH, van Nieuwenhoven FA, Brouns AE, Janssen BJ, Cleutjens JP, Staels B, van der Vusse GJ, van Bilsen M. Cardiac hypertrophy is enhanced in PPAR alpha−/− mice in response to chronic pressure overload. Cardiovasc Res. 2008;78:79–89. doi: 10.1093/cvr/cvn001. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM. PPAR-gamma: adipogenic regulator and thiazolidinedione receptor. Diabetes. 1998;47:507–514. doi: 10.2337/diabetes.47.4.507. [DOI] [PubMed] [Google Scholar]
- Staples JF, Brown JC. Mitochondrial metabolism in hibernation and daily torpor: a review. J Comp Physiol B. 2008;178:811–827. doi: 10.1007/s00360-008-0282-8. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Ueno M, Uno M, Hirose Y, Zenimaru Y, Takahashi S, Osuga JI, Ishibashi S, Takahashi M, Hirose M, et al. Effects of hormone-sensitive lipase-disruption on cardiac energy metabolism in response to fasting and refeeding. Am J Physiol Endocrinol Metab. 2009 doi: 10.1152/ajpendo.91031.2008. [DOI] [PubMed] [Google Scholar]
- van der Meer DL, Degenhardt T, Vaisanen S, de Groot PJ, Heinaniemi M, de Vries SC, Muller M, Carlberg C, Kersten S. Profiling of promoter occupancy by PPAR{alpha} in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Weiss RG, Gerstenblith G, Bottomley PA. ATP flux through creatine kinase in the normal, stressed, and failing human heart. Proc Natl Acad Sci U S A. 2005;102:808–813. doi: 10.1073/pnas.0408962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha. Mol Endocrinol. 2010;24:1349–1358. doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.