Malignant mesothelioma (MM) is an aggressive form of cancer that arises in the lining of the peritoneal, pleural or pericardial cavities. It has a long latency period, and the development of this rare cancer is typically associated with asbestos exposure. Mesothelioma may also be a concern for carbon nanotubes (CNTs), which are under development for biomedical applications[1-3] but have been recently shown to produce asbestos-like pathogenic behavior in mice, suggesting their potential to induce mesothelioma upon chronic exposure.[4-6] The diffuse nature of the tumors makes their surgical removal difficult, and they are resistant to conventional chemotherapy.[7,8] As an alternative strategy, non-conventional approaches involving new chemical agents, and targeting methods minimizing side effects are needed. Recently it has been shown that selenium, in the form of selenite and selenocysteine, can selectively inhibit growth and induce apoptotic cell death in MM cells compared to normal mesothelial cells,[9] i.e. exhibits differential toxicity. Selenium is believed to play a role in cancer prevention,[10,11] and selenium deficiency has been linked with increased cancer incidence.[11-13] The fundamental mechanisms of selenium chemo-prevention are not fully understood. At doses marginally higher than the essential dose (Recommended Daily Allowance = 55 μg/day, No Adverse Effect Level = 800 μg/day), selenium can cause toxicity.[14] The combination of the administered dose and the chemical form co-determine selenium’s activity as toxic or carcinostatic.[10,11] For example, the selenoenzyme glutathione peroxidase can protect cells from oxidative damage[15,16] and the seleno-enzymes, glutathione S-transferases, can repair damaged DNA and prevent mutation.[16] Other forms of selenium, however, can produce reactive oxygen species[10] leading to oxidative stress and cell death. In recent years, elemental nano-selenium (nSe) has gained special attention due to its therapeutic properties.[17,18] As a major advantage over other selenium forms, elemental (zero-valent) nanoparticles are a high-Se-density formulation with the potential for local delivery of high doses into cancer cells. To achieve this in practice requires a nSe formulation that is rapidly internalized by the target cell and is sufficiently stable in the extracellular fluid to reach the target cell intact. Here we synthesize two competing formulations of nano-selenium and evaluate their relative effectiveness for high-dose intracellular delivery. We show that a novel selenium-carbon composite nanostructure (CNPnSe) is highly effective in inducing death in malignant mesothelial cells in vitro and shows promise for the development of targeted therapy for malignant mesothelioma.[12]
The two-component selenium-carbon nanostructures were synthesized by reduction of selenite in the presence of colloidal carbon nanoparticles pre-synthesized from liquid crystalline precursors.[19] These “supramolecular” carbon nanoparticles were previously reported to be biocompatible, hydrophilic carriers for cell delivery, and to have high activity surfaces arising from a high concentration of graphene edge sites unique to the liquid crystal self-assembly process.[19] Here, we compare the cytotoxic effect of this selenium-carbon composite nanoformulation with elemental selenium nanoparticles as reference material, where bovine serum albumin (BSA)[17] is used as a colloidal stabilizing agent (nSeBSA). Elemental selenium was synthesized (Figure 1E) by reducing sodium selenite (Na2SeO3 ) with glutathione (GSH)[20,21] through a selenodiglutathione intermediate (GSSeSG), which releases elemental selenium at alkaline pH (Figure 1E). Similar to the alkaline hydrolysis of disulfide bonds (RSSR) that gives a sulfenic acid (RSOH) and a thiolate (RS−),[22] the hydroxide anion is believed to cleave the selenotrisulphide bond in GSSeSG (see Figure 1E(b)). We hypothesize that the intermediate selenopersulfide anion (GSSe−) adsorbs on nucleation sites on the active carbon surfaces and releases GS− to leave zero-valent Se that grows into bound nanoclusters (Figure 1C, E). This release mechanism is similar to what has been reported for hydrodisulphide anion (RSS−).[23] The resulting thiolate and sulfenic acid can react to form oxidized glutathione (GSSG) at low pH as the equilibrium is favored in this direction.[24] In the case of selenium, there is evidence for the occurrence of selenopersulfide as the initial reaction product.[25] If the growth of nano-selenium is left unchecked, as in absence of any stabilizer, the elemental selenium released in the solution grows over time to form aggregates of black elemental selenium. The speed of aggregation depends on selenium concentration and temperature.
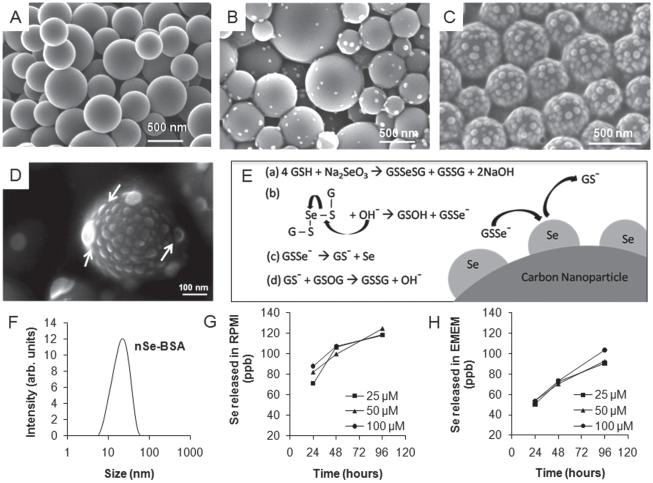
Figure 1.
Synthesis and characterization of selenium/carbon bifunctional nanoparticles. (A) as-produced liquid-crystal-derived carbon nanoparticles synthesized by spray pyrolysis of 0.5-wt% indanthrone disulfonate;[19] (B,C,D) selenium-carbon bifunctional composite nanoparticles with 8.6-wt% Se loading (B) and ~28-wt% Se loading (C); and (D) high-magnifi cation FE-SEM image showing the hemispherical shape of the surface-nucleated nSe clusters. (E) Proposed mechanism: reduction of sodium selenite with glutathione followed by hydroxide ion mediated release of GSSe−1 as a precursor for heterogeneous deposition of self-avoiding elemental selenium nanoclusters; (F) Dynamic light scattering measurement of particle size distribution of nSe stabilized with BSA. (G, H) Release of soluble selenium from nSe over a period of 24, 47 and 96 hours at the doses of 25, 50 and 100 μm in cell culture medium, showing more release in (G) RPMI than (H) EMEM. Note for Se: 1 μM = 78.96 ppb, so only a small fraction of the total Se is mobilized in the extracellular medium after 96 hrs (1–5%).
We perform this reaction in presence of non-agglomerated, near-spherical and sterile supramolecular carbon nanoparticles[19] and are able to nucleate selenium nanoclusters on the highly active surface (Figure 1A–E). The selenium loading density can be readily tuned by varying the ratio of carbon to seleno-reagents (compare Figure 1B, C at 8.6% and 28% selenium loading by mass respectively). The FE-SEM image of hemispherical shape of selenium nanoparticle on the carbon substrate (Figure 1D) strongly suggests a heterogeneous nucleation (on the carbon particle surface) and surface growth model, rather than homogeneous nucleation (in suspension) followed by Se-particle deposition on carbon surfaces. Figure 1C shows that the clusters are distinct and non-overlapping, even under crowded conditions at high Se density. This suggests a growth mechanism that involves self-avoidance (cluster-cluster electrostatic repulsion), consistent with the negative charge on the GSSe− intermediate. Dynamic light scattering measurement (Zetasizer Nano ZS, Malvern Instruments Ltd.) shows a narrow size distribution of selenium nanoparticles synthesized in presence of BSA (Figure 1F). Zeta potential measurements (Zetasizer Nano ZS, Malvern Instruments Ltd.) for CNPnSe (−45.9 ± 2.9 mV) and nSeBSA (−40.3 ± 1.6 mV) indicate acidic surface sites with negative charge on these high-activity surfaces[19] and electrostatic repulsion as the mechanism of colloidal stability. The large negative surface charge on selenium is also consistent with the self-avoiding growth pattern involving the GSSe− intermediate discussed above. XPS (Physical Electronics 5500 Multi-technique Surface Analyzer, URI, USA) analysis shows that the selenium in nano-particulate form exists in its elemental state, giving a binding energy peak at 55.25 eV. XRD (Siemens D5000) analysis shows no signs of crystallinity in the sample confirming the amorphous character of the nano-selenium clusters.
During the Se/C nano-composite synthesis, competition exists between the heterogeneous nucleation of surface nanoclusters and the homogeneous nucleation and growth of free colloidal nSe. We found that lower temperatures favor heterogeneous nucleation leading to surface cluster formation. It was also observed that freshly sterilized carbon nanoparticles were more active for surface Se nanocluster formation than the samples stored for several months. In addition, in a model system, nucleation was observed on as-produced carbon films grown by physical vapor deposition, but not on the annealed surfaces of the same films. Together these observations suggest that carbon active sites provide a reduced activation energy pathway for nucleation of elemental nSe–an observation that can be exploited to optimize synthesis of this therapeutic nanocomposite.
To establish the short-term stability of the selenium clusters, ultra-filtration and ICP were used to quantify soluble selenium released into buffers at lysosomal (pH = 4.5) and physiological (pH = 7.4) pH, and in serum-free cell culture media: EMEM and RPMI (Invitrogen). In the buffered media at both pH values, no measurable release was found from nano-selenium. There was measurable release in cell culture media but only a very small fraction (<2% for 100 μm dose) of the total selenium present (Figure 1G and 1H). Selenium is known to bind to biological thiols (SH)[26] and we note that Se release was higher in RPMI than EMEM, which has a higher content of L-cysteine (324.6 μM vs. 159.7 μM), which is the only thiol-containing amino acid in these two media.
A primary goal of this study was to evaluate the new Se/C bifunctional nanocomposite for chemotherapeutic applications relative to conventional selenium formulations. To do this we exposed MM cells in vitro to sodium selenite, BSA-stabilized nSe (nSeBSA), and the selenium-carbon nanocomposite (CNPnSe), as well as various reactants and by-products from the synthesis as additional controls. All selenium forms induced a cytotoxic effect in a time dependent (data not shown) and dose dependent manner (Figure 2A). The CNPnSe formulation had the highest cytotoxic effect, followed by the conventional form, Na2SeO3 and by simple Se nanoparticles, nSeBSA (Figure 2A). We hypothesized this difference is due to (a) faster or more complete cellular uptake promoted by the delivery vehicle (the CNPs), or (b) faster release of bioavailable, soluble Se, since the CNPnSe surfaces are free of BSA which may passivate surfaces. To assess hypothesis (b), we performed an additional control in which BSA was added to CNPnSe (CNPnSe +BSA) and found no decrease in the CNPnSe potency, indicating that BSA passivation is not the cause of the lower potency of nSe relative to the nano-composite.
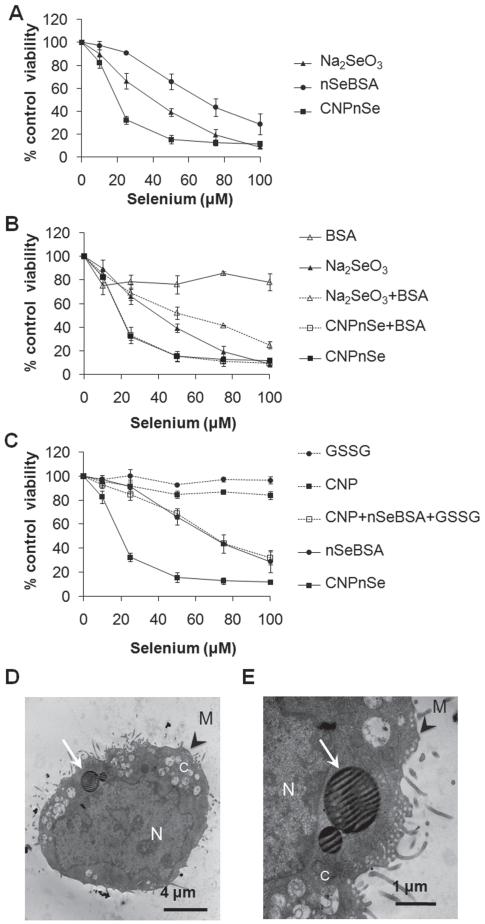
Figure 2.
Relative effectiveness of competing selenium forms for inducing in vitro cytotoxicity in malignant mesothelial cells. (A) CNPnSe induces higher cytotoxicity in MM at lower doses than selenite (Na2SeO3 ) or free selenium nanoparticles (nSeBSA). (B) Control experiments showing only minor effects of the BSA additive, either independently or synergistically, (C) Control experiments to test the independent or synergistic effects of the pure carbon nanoparticles and oxidized glutathione byproduct, GSSG. (D, E) Cellular internalization of CNPnSe. Thin section TEM. N: nucleus. C: cytoplasm. M: cell membrane. White arrows: Selenium-carbon nanocomposite particles within cytoplamic vacuoles. Cells were exposed to increasing μM doses of selenium composites for 72 hrs. Equivalent doses: Se (1 μM) ≡ GSH (4 μM) ≡ GSSG (2 μM) ≡ CNPs (1 μg/ml) ≡ BSA (0.8 μg/ml). Together these results show that the desirable high cytotoxicity of the Se/C composite is a unique bifunctional nanoparticle effect that cannot be explained by the independent contributions of its components.
Figure 2B and C show a large series of control experiments in which the Se/C nanocomposite is tested in comparison to the individual components and by-products (CNP, nSe, BSA, Na2SeO3, GSSG) alone and in combination to understand their contribution to toxicity and their synergistic interactions. The desired high toxicity toward malignant mesothelial cells is seen only for the CNPnSe nanocomposite, which suggests a bifunctional synergy, in which the carbon nanoparticles promote cellular internalization as observed previously,[19] allowing the nSe clusters to deliver biologically active selenium to intracellular targets. Induction of cell death by selenium forms was confirmed by reduction of cellular density, loss of cell symmetry and detachment (Figure 3A–D) as well as typical morphological changes of apoptosis including loss of cellular and nuclear membrane integrity, chromatin condensation and cellular blebs (Figure 3F–H and J–L) compared to untreated cells (Figure 3E and I). These images support the proposed mechanism, which attributes the strong cytotoxic effect of CNPnSe to effective cellular delivery of high-dose selenium.
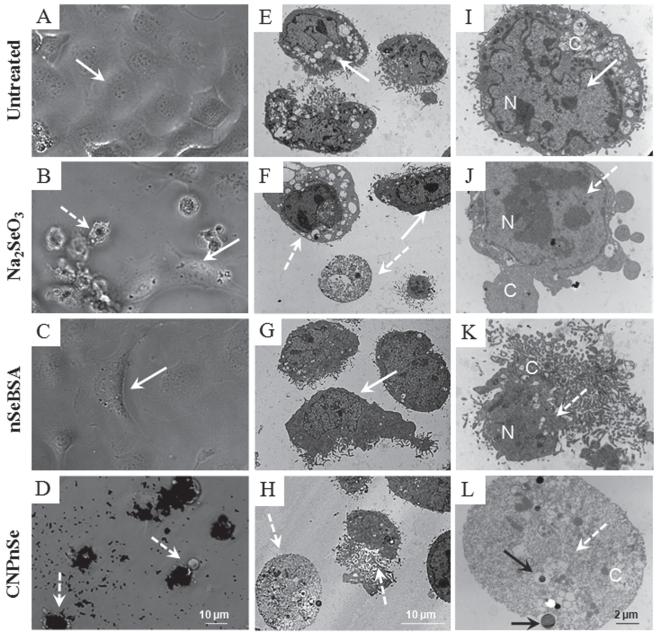
Figure 3.
Morphology of malignant mesothelial cells treated with competing formulations of selenium as a chemotherapeutic agent. Cells were exposed to Na2SeO3, nSeBSA or CNPnSe at the equimolar dose of 25 μM of selenium for 72 hours. (A–D): Phase-contrast optical micro-graphs. In contrast to nSeBSA, where the decrease in cell density may be due to the induction of cell growth arrest together with apoptotic cell death, CNPnSe, at the same dose, decreases cell density by induction of cell death by necrosis, and this effect is stronger than the effect of Na2SeO3 where cell death is induced by both apoptosis and necrosis. (E–L): Thin-section TEM images. Cell death and internalization of CNPnSe. Solid arrows: live cell with intact nucleus (N) and cytoplasm (C). Dashed arrows: dead cell with degraded nucleus and/or cytoplasm. Black arrows: internalized CNPnSe.
At present the toxicity pathways for selenium are only partially understood, but the literature and our present data may provide some insight. It has been proposed that the carcinostatic activity of a selenium form can be attributed to its ability to generate selenide anion (RSe−)[10] which is followed either by oxidation of selenide to elemental selenium within the cells generating byproduct superoxide (O2•−) anion[10,27] or by selenium incorporation in one of the twenty five selenoproteins[15] or in its methylated form methylselenol (CH3SeH).[28] It is likely here that the cytotoxicity of elemental selenium nano-particles is due to carbon-nanoparticle-assisted cellular uptake followed by intracellular release of a biologically active species, such as free selenide or Se bound to proteins or small molecule thiols.
In summary, selenium deserves further consideration for the treatment of malignant mesothelioma, a cancer of the lung and pleural or abdominal linings that is typically associated with asbestos exposure, and may be a concern for carbon nanotube exposure in the future. The present work used in vitro techniques to compare three methods for delivery of selenium to malignant mesothelial cells: conventional selenite salts, colloidal selenium nanoparticles, and a new Se/C composite nanostructure synthesized by heterogeneous nucleation of Se nanoclusters on the high-activity surfaces of liquid-crystal-derived carbon nanoparticles. The nanocomposite was the most effective of the three Se formulations, inducing 70% cell death at 22 μM Se concentration. Separate control experiments show minimal soluble Se release in the extracellular medium, and show only minor effects of the non-Se components (carbon nanoparticles, BSA colloidal stabilizer, and oxidized glutathione) acting either independently or synergistically with the selenium. Together these results show that the desirable high cytotoxicity of the Se/C composite is a unique bifunctional nanoparticle effect. We propose that the high potency of the Se/C bifunctional nanoparticles is due to cellular uptake enhanced by the carbon nanoparticle carrier[19] followed by nSe reduction to active selenide species in intracellular compartments. Nanoparticle-assisted selenium delivery shows promise for further development as a mesothelioma chemotherapeutic agent.
Experimental Section
Materials
Supramolecular CNPs were synthesized as reported previously[19] and sterilized by heating in a tube furnace in nitrogen (400 °C, 30 min). The nanocomposites were fabricated by adding CNPs (1 mg) to 4 ml of 5 mM glutathione (GSH, reduced form, TCI America), and sonicated followed by mixing of 1 ml of 5 mM sodium selenite (Na2SeO3, Alfa Aesar) and then slow addition of 85 μl of 0.2 M sodium hydroxide (NaOH, ACS, FisherChemicals). To prepare BSA stabilized nanoselenium particles, 1 ml of 25 mM sodium selenite solution was mixed with 4 ml of 25 mM glutathione containing 20 mg of BSA[17] followed by an addition of 70 μl of 1 M NaOH thus instantaneously forming red colloidal nano-selenium. The solution was dialyzed for 96 hours using a regenerated cellulose membrane (Spectra Float-A-Lyzer, Spectrum Labs) against a potassium phosphate buffer maintained at physiological pH (7.4).
To test selenium dissolution characteristics, colloidal suspensions of BSA-stabilized nSe were incubated in test media at 37 °C for up to 96 hrs at various doses up to 100 μm. The cultured media was collected and centrifuged (30 min, 4500 rpm) through 5 KD MWCO tube (Amicon Ultra-4, Millipore), which were washed with nanopure water prior to use. The supernatant thus collected was tested for its selenium content using ICP-AES (Jobin-Yvon JY2000 inductively coupled plasma–atomic emission spectrometer). Scanning Electron Microscopy (SEM) was conducted on a LEO 1530 VP using 1–20 kV voltages below 10−5 bar.
Cell viability
For cytotoxicity experiments 40 L murine malignant mesothelial cells[29] were seeded on 96 well plates (Corning, Lowell, MA) at a density of 5000 cells/well in 200 μl RPMI (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), 10 units ml−1 penicillin and 10 μg ml−1 of streptomycin (Invitrogen, Carlsbad, CA) and incubated overnight at 37 °C in humidified 5% CO2. Cells were treated with 10 μM-100 μM equivalent selenium composites as well as precursors and by-products of the synthesis (Figure 2) in RPMI/2% FBS supplemented as above. Cell viability was determined with a CyQuant® Cell Proliferation Assay Kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. CyQuant® dye flourescence is strongly enhanced upon binding to cellular nucleic acids allowing quantification of the total cell number by relative fluorescence units (RFU). Samples were excited at 480 nm and fluorescence emission was measured at 520 nm using a microplate spectrofluorometer (Spectromax 2). Cell viability was expressed as the percentage of RFU from treated cells divided by RFU from untreated cells. Under these conditions the linear detection range of the assay was maintained for all treatments and no interference due to carbon nanomaterials or selenium nanoparticles was observed.
Phase contrast microscopy and transmission electron microscopy (TEM)
Cell uptake and morphology studies were carried out with 40 L cells exposed to 25 μM selenium composites. Phase contrast micrographs of living cells were taken with a Nikon Eclipse E800 microscope. For TEM, cells were fi xed at room temperature with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, rinsed and stored at 4 °C in cacodylate buffer with 8% sucrose for further processing. Samples were processed as previously described.[19] Sections were stained with 1% uranyl acetate in 50% methanol for 10 min and lead citrate for 5 min, placed on copper grids and viewed with a Phillips 410 Transmission Electron Microscope equipped with an Advantage HR CCD camera (Advanced Microscopy Techniques, Danvers, MA). Images were acquired and analyzed with AMT’s imaging software.
Supplementary Material
Acknowledgements
Financial Support was provided by NSF NIRT (R. Hurt) DMI-050661, RO1 ES03721 (A. Kane), and T32 ES07272 (A. Kane). The authors thank Michael Platek at University of Rhode Island and Paula Weston at Brown University for their help with XPS and thin section TEM of cell samples, respectively. The technical contributions of Anthony McCormick and Joseph Orchardo are gratefully acknowledged.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Love Sarin, Division of Engineering, Brown University, Providence, RI 02912 (USA)
Vanesa C. Sanchez, Department of Pathology and Laboratory Medicine, Brown University, Providence, RI 02912 (USA)
Aihui Yan, Department of Chemistry, Brown University, Providence, RI 02912 (USA).
Prof. Agnes B. Kane, Department of Pathology and Laboratory Medicine, Brown University, Providence, RI 02912 (USA)
Prof. Robert H. Hurt, Division of Engineering, Brown University, Providence, RI 02912 (USA).
References
- [1].Yokoyama M. J. Artif. Organs. 2005;8:77. doi: 10.1007/s10047-005-0285-0. [DOI] [PubMed] [Google Scholar]
- [2].McDevitt MR, Chattopadhyay D, Kappel BJ, Jaggi JS, Schiffman SR, Antczak C, Njardarson JT, Brentjens R, Scheinberg DA. J. Nucl. Med. 2007;48:1180. doi: 10.2967/jnumed.106.039131. [DOI] [PubMed] [Google Scholar]
- [3].Liu Z, Davis C, Cai W, He L, Chen X, Dai H. Proc. Nat. Acad. Sci. 2008;105:1410. doi: 10.1073/pnas.0707654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kane AB, Hurt RH. Nat. Nanotechnol. 2008;3:378. doi: 10.1038/nnano.2008.182. [DOI] [PubMed] [Google Scholar]
- [5].Takagi A, Hirose A, Nishimura T, Fukumori N, Ogata A, Ohashi N, Kitajima S, Kanno J. J. Toxicol. Sci. 2008;33:105. doi: 10.2131/jts.33.105. [DOI] [PubMed] [Google Scholar]
- [6].Poland CA, Duffi n R, Kinloch I, Maynard A, Wallace WA, Seaton A, Stone V, Brown S, Macnee W, Donaldson K. Nat. Nanotechnol. 2008;3:423. doi: 10.1038/nnano.2008.111. [DOI] [PubMed] [Google Scholar]
- [7].Nilsonne G, Sun X, Nystrom C, Rundlof AK, Potamitou Fernandes A, Bjornstedt M, Dobra K. Free Radic. Biol. Med. 2006;41:874. doi: 10.1016/j.freeradbiomed.2006.04.031. [DOI] [PubMed] [Google Scholar]
- [8].Holmgren A. Free Radic. Biol. Med. 2006;41:862. doi: 10.1016/j.freeradbiomed.2006.05.024. [DOI] [PubMed] [Google Scholar]
- [9].Apostolou S, Klein JO, Mitsuuchi Y, Shetler JN, Poulikakos PI, Jhanwar SC, Kruger WD, Testa JR. Oncogene. 2004;23:5032. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- [10].Spallholz JE, Palace VP, Reid TW. Biochem. Pharmacol. 2004;67:547. doi: 10.1016/j.bcp.2003.09.004. [DOI] [PubMed] [Google Scholar]
- [11].Letavayova L, Vlckova V, Brozmanova J. Toxicology. 2006;227:1. doi: 10.1016/j.tox.2006.07.017. [DOI] [PubMed] [Google Scholar]
- [12].Levander OA. Annu. Rev. Nutr. 1987;7:227. doi: 10.1146/annurev.nu.07.070187.001303. [DOI] [PubMed] [Google Scholar]
- [13].Jablonska E, Gromadzinska J, Sobala W, Reszka E, Wasowicz W. Eur. J. Nutr. 2008;47:47. doi: 10.1007/s00394-008-0696-9. [DOI] [PubMed] [Google Scholar]
- [14].Yang G, Zhou R. J. Trace Elem. Electrolytes Health Dis. 1994;8:159. [PubMed] [Google Scholar]
- [15].Papp LV, Lu J, Holmgren A, Khanna KK. Antioxidants & Redox Signaling. 2007;9:775. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- [16].Stadtman TC. Ann. Rev. Biochem. 1996;65:83. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- [17].Zhang J-S, Gao X-Y, Zhang L-D, Bao Y-P. Biofactors. 2001;15:27. doi: 10.1002/biof.5520150103. [DOI] [PubMed] [Google Scholar]
- [18].Tran PA, Sarin L, Hurt RH, Webster TJ. J. Biomed. Mater. Res. Part A. 2010;93(4):1417. doi: 10.1002/jbm.a.32631. [DOI] [PubMed] [Google Scholar]
- [19].Yan A, Lau BW, Weissman BS, Külaots I, Yang NYC, Kane AB, Hurt RH. Advanced Materials. 2006;18:2373. [Google Scholar]
- [20].Painter EP. Chem. Rev. 1941;28:179. [Google Scholar]
- [21].Ganther HE. Biochemistry. 1968;7:2898. doi: 10.1021/bi00848a029. [DOI] [PubMed] [Google Scholar]
- [22].Hogg PJ. Trends Biochem Sci. 2003;28:210. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- [23].Koval IV. Russian Chemical Reviews. 1994;63:735. [Google Scholar]
- [24].Andersson LO, Berg G. Biochim Biophys Acta. 1969;192:534. doi: 10.1016/0304-4165(69)90406-1. [DOI] [PubMed] [Google Scholar]
- [25].Ganther HE. Biochemistry. 1971;10:4089. doi: 10.1021/bi00798a013. [DOI] [PubMed] [Google Scholar]
- [26].Haratake M, Fujimoto K, Ono M, Nakayama M. Biochimica et Biophysica Acta (BBA)–General Subjects. 2005;1723:215. doi: 10.1016/j.bbagen.2005.02.002. [DOI] [PubMed] [Google Scholar]
- [27].Spallholz JE. Biomed Environ Sci. 1997;10:260. [PubMed] [Google Scholar]
- [28].Rayman MP. Proc Nutr Soc. 2005;64:527. doi: 10.1079/pns2005467. [DOI] [PubMed] [Google Scholar]
- [29].Miselis NR, Wu ZJ, Van Rooijen N, Kane AB. Molecular Cancer Therapeutics. 2008;7:788. doi: 10.1158/1535-7163.MCT-07-0579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.