Abstract
The application of microarray technology to the study of mammalian organogenesis can provide greater insights into the steps necessary to elicit a functionally competent tissue. To this end, a temporal profile of gene expression was generated with the purpose of identifying changes in gene expression occurring within the developing male and female embryonic gonad. Gonad tissue was collected from mouse embryos at 11.5, 12.5, 14.5, 16.5, and 18.5 days postcoitum (dpc) and relative steady-state levels of mRNA were determined using the Affymetrix MGU74v2 microarray platform. Statistical analysis produced 3693 transcripts exhibiting differential expression during male and/or female gonad development. At 11.5 dpc, the gonad is morphologically indifferent, but at 12.5 dpc, transitions to a male or female phenotype are discernible by the appearance of testicular cords. A number of genes are expressed during this period and many share similar expression profiles in both sexes. As expected, the expression of two well-known sex determination genes, specifically Sry and Sox9, is unique to the testis. Beyond 12.5 dpc, differential gene expression becomes increasingly evident as the male and female tissue morphologically and physiologically diverges. This is evident by two unique waves of transcriptional activity occurring after 14.5 dpc in the male and female. With this study, a large number of transcripts comprising the murine transcriptome can be examined throughout male and female embryonic gonad development and allow for a more complete description of gonad differentiation and development.
Keywords: developmental biology, embryo, gene regulation, ovary, testis
INTRODUCTION
Sex determination, differentiation, and development of the mammalian embryonic gonad have been studied for over half a century. Subsequent to the identification of Sry as the key initiator of sex determination [1, 2], a number of additional genes have been shown to be integral to this complex process. These include Sox8 and Sox9 [1, 3–5], Dmrt1 [6], Dax1 [7], Wt1 [8], and Sf1 [9], among others. The collective actions of the functional units of these genes produce the divergence between the phenotypic male gonad and the default phenotypic female gonad. The function of Sry and its downstream effect, although not completely understood, is fairly well characterized. In contrast, cellular and molecular processes occurring simultaneously or immediately after the establishment of a male or female gonad have not been well characterized. Examining patterns of gene expression at a genomic level during embryonic gonad development is necessary to better understand the processes that result in two distinct reproductive tissues.
The specific expression of Sry in Sertoli cells results in the indifferent gonad progressing toward the male phenotype, specifically the establishment of testicular cords and the regression of the Müllerian ducts induced by anti-Mullerian hormone (AMH), both of which are completed by 13.5 days postcoitum (dpc) [10]. Concurrent with the establishment of testicular cords, cell migration and proliferation occur within the testis and result in substantial growth of the testis [11]. Female gonad development is significantly less dramatic. Clusters of germ cells appear around 13.5 dpc at the basement membrane [10], but a much less dramatic morphological change is evident when compared with the testis at this time period. Germ cells in the embryonic ovary enter meiosis at 13.5 dpc and become arrested in meiosis I around the time of birth at 18.5 dpc [12], unlike male germ cells, which do not enter meiosis until well after birth. Numerous models that describe molecular events and interactions and attempt to order their action in the course of the divergence of the male and female gonad have been proposed [13–15]. With each of these revised and complementary models, new and increasingly more complete models of sex determination have been created.
Much of the work performed to describe molecular events resulting in sex determination has been done using methods such as Northern blots, polymerase chain reaction (PCR), suppression subtractive hybridization, gene knockouts, and hybridization to small, select microarrays. The use of these techniques has allowed for a glimpse of molecular events needed to create differentiated male and female gonads. However, none of these studies have examined sex determination and the subsequent development of the embryonic gonad on a genomic scale. Using microarrays that represent a large percentage of the murine genome, this study was designed to create a broader picture of transcriptional events and components necessary to elicit a separate male and female gonadal phenotype. To this end, the Affymetrix microarray platform was employed to generate a time course of gene expression for the developing gonad of the male and female murine embryo. This report surveys a period of development beginning with an indifferent gonad (11.5 dpc) to just before birth (18.5 dpc) and provides the opportunity for a comprehensive analysis of a large number of transcripts from the murine genome.
In addition to the wide implications in describing sex determination, the period of this study also encompasses two well-documented developmental events, namely initiation of meiosis in the female [16] and gonadotropin-independent steroidogenesis in the male [17], thus providing the opportunity to make comparisons to previously reported expression patterns of specific genes. The independent investigation of these known transcription levels using the current array technology would further support the validity of the array platform and ensure a level of confidence in pursuing the discovery of novel genes. While the quest to determine factors involved in sex determination has been intense, a complete picture of gene expression throughout embryonic gonad development would be a valuable resource. Further comparisons can also be made between the events occurring in the embryonic gonad and during postpartum testicular development. A recent postpartum testicular developmental time course [18] allows for direct comparison of expression levels and specific transcripts involved in the initiation of meiosis between the female (which occurs during the embryonic period of development) and meiosis in the male, gonadotropin-independent versus dependent steroidogenesis, and unique genes and patterns present during these two distinct periods of testis development and maturation.
Creation of male and female gonad development gene expression time courses allows for relatively easy and genome-wide exploration and identification of genes involved in numerous facets of embryonic development, including, but not limited to, sex determination, cellular differentiation, meiosis, and steroid production, and additionally allows for direct comparisons between these activities in the two sexes. Thorough analysis of these time courses will invariably lead to a greater understanding of how changes in the murine transcriptome relate to functional and structural changes in the embryonic gonad.
MATERIALS AND METHODS
Tissue Collection and RNA Preparation
Protocols for the use of animals in this experiment were approved by the Washington State University Animal Care and Use Committee and were in accordance with the National Institute of Health’s standards established by the Guidelines for the Care and Use of Experimental Animals. BL/6–129 mice were maintained and mated in a temperature- and humidity-controlled room with food and water ad libitum. On select days, one male was paired with two females in the afternoon and then removed the following morning. Females with vaginal plugs (0.5 dpc) were placed in separate cages until embryonic gonad tissue was collected at 11.5, 12.5, 14.5, 16.5, and 18.5 dpc. Tail somites were counted to assure embryos were at the proper stage. Because it is impossible to visually identify the sex of 11.5-dpc embryos, undifferentiated gonad tissue from each individual was placed in a separate tube containing 25 μl TRIzol reagent (Invitrogen, Carlsbad, CA) and stored at −75°C until the corresponding backbone and tail tissue could be sexed by Sry genotyping using PCR. Embryonic tissue from 12.5-dpc animals was processed in a similar fashion, as it was not always possible to positively determine the sex of the embryo by visual inspection. Sry genotyping was performed in a standard PCR using the following primers: forward, 5′-CGGGATCCATGTCAAGCGC CCCATGAATGCATTTATG-3′; and reverse, 5′-GCGGAATTCACTTT AGCCCTCCGATGAGGCTGATAT-3′. Upon determination of the sex of each of the collected tissue samples, like tissue (testis or ovary) stored in TRIzol was pooled and total RNA was prepared as per the manufacturer’s instructions. Approximately 70 embryos were processed for a single 11.5-dpc or 12.5-dpc time point, whereas approximately 35 embryos were required for each of the remaining time points. Male and female tissue samples were obtained in duplicate in a similar fashion. Once purified, RNA quality was determined by electrophoretic methods using a denaturing agarose gel or analysis using an Agilent Bioanalyzer 2100 (Palo Alto, CA) and by spectroscopy at 260 and 280 nm. If excessive degradation or protein contamination of the RNA was evident, the sample was not used for microarray hybridization. This study, unlike most others working with exceedingly small tissues, did not use supplemental amplification of the RNA in any fashion.
Microarray Processing
Two to four micrograms of total RNA from each pooled embryonic gonad sample was used to create the cRNA target for the microarray. The target was produced using a reverse transcription reaction to produce cDNA, which was subsequently subjected to in vitro transcription with biotinylated cytidine-5′-triphosphate and uridine-5′-triphosphate using the MEGAScript kit (Ambion, Austin, TX) to produce biotinylated cRNA. The target was then fragmented and hybridized to the MGU74Av2, Bv2, and Cv2 arrays (Affymetrix, Santa Clara, CA) in duplicate using an Affymetrix GeneChip Fluidics Station 400 according to the manufacturer’s standard protocols. The arrays were stained with phycoerythrin-coupled avidin and scanned using an Agilent GeneArray Scanner 2500A. The resultant output was analyzed using Microarray Suite 5.0 (Affymetrix) and examined for excessive background or evidence of RNA degradation. All microarray processing was performed in the Laboratory for Biotechnology and Bioanalysis I at Washington State University, Pullman, WA.
After scanning, all probe sets were scaled to a signal intensity of 125 and relative levels of expression of each transcript (signal) were determined using Microarray Suite 5.0. Microarrays were also examined for physical anomalies and for the presence of excessive background hybridization. Once satisfied with the initial analysis, data were exported and loaded into GeneSpring 6.1 (Silicon Genetics, Redwood City, CA), where statistical and comparative analyses were performed to both verify the data and to isolate specific and novel transcriptional characteristics occurring during embryonic gonad development.
Microarray Analyses
Analyses of the microarray data were primarily performed using GeneSpring. The data comprising the male and female embryonic gonad time courses were normalized in GeneSpring using the default/recommended normalization methods. These included setting of signal values below 0.01 to 0.01, total chip normalization to the 50th percentile, and normalization of each gene to the median based on its measured levels of expression. Duplicate samples were used in every time point in the two time courses and the mean values of the two replicates were used in subsequent analyses. Normalization in this fashion allowed for the analysis and visualization of data based on the relative abundance and resultant expression profile of each transcript independent of a separate control sample. Samples were further delineated between the two sexes to separate the expression profiles occurring within the testis and ovary independent of each other.
Two approaches to the analysis were employed. The first simply identified those transcripts having a raw signal of at least 50 in one tissue (testis or ovary) and present at a level at least twofold greater than the other tissue. This analysis was done for each tissue at each time point and used simple filtering tools within the GeneSpring software. The second approach identified statistically significant, differentially expressed transcripts within each respective time course. To identify these transcripts, one-way ANOVA parametric tests were performed on both the male and the female time courses using a P-value cutoff of 0.05 and using all available error estimates, including statistical calculations made using the Cross-Gene Error Model provided by GeneSpring. Additionally, each transcript expressed in a significant fashion was also required to have a minimum signal of 50 in at least one of the five time points in the respective time course (male or female) and have at least a twofold change occurring within said time course. The signal cutoff was based on the expression level of Sry, which achieved maximum expression at 11.5 dpc in the testis, with a signal of approximately 75 in spite of its integral and necessary role in the sex-determination process. These analyses produced two distinct lists representing transcripts expressed significantly during embryonic testicular and embryonic ovarian development. These lists were sorted within Microsoft Excel (Redmond, WA) based on Unigene number and redundant entries were removed producing a list of nonredundant statistically significant transcripts. Moreover, by comparing these two lists, transcripts expressed significantly and uniquely in both the testis and ovary were identified.
A major goal of this project was to determine the number and identity of transcripts expressed uniquely in male or female gonads at the specific points during embryonic gonad development. Statistical analyses and filtering were used to isolate transcripts that exhibited these characteristics. To accomplish this, the aforementioned lists of significant transcripts in the male and female time courses were subjected to additional one-way ANOVA analyses designed to compare significant differences in expression between the testis and ovary at specific time points in embryonic development (i.e., testis 11.5 dpc was compared statistically with ovary 11.5 dpc). These analyses again employed a P-value cutoff of 0.05 and used all available error estimates derived within GeneSpring. This was performed on each of the five different time points in the time courses and resulted in lists of transcripts that were significantly different between testis and ovary at a given time point. To further dissect the significant transcripts uncovered using the time point comparisons, these lists were subsequently compared with the original lists of significant transcripts in the testis and ovary time courses, resulting in groups of transcripts that were expressed significantly in either the testis or the ovary and at a specific time point. To finalize these lists, comparisons were made within the five lists derived from the ovary time course and the five lists from the testis time courses and transcripts found to be regulated at more than one time point in the testis or ovary time course (i.e., a transcript was found to be regulated at 11.5 and 12.5 dpc in the testis time course) were eliminated. Although this undoubtedly removed some possibly important transcripts, it did allow for a more stringent analysis that focused on precise changes occurring at a specific time point rather than gradual or ambiguous changes in expression over two or more time points.
Cluster Analysis
Cluster analysis was employed to further demarcate the expressional themes and patterns occurring during the development of the embryonic gonad. Hierarchical clustering was used to both cluster transcripts based on similarity of expression between each gene but also to group experimental samples together based on similarity of the overall experimental expression profile. Hierarchical clustering was performed using a smooth correlation coefficient with default separation ratio and distances. Additionally, K-means clustering was used with a smooth correlation to help in the identification of the major patterns of expression occurring within the time courses. Both analyses using clustering were performed using the nonredundant statistically significant transcripts described previously and were performed using GeneSpring.
Functional Characterization
Functional characterization of significantly expressed transcripts was performed using EASE (Expression Analysis Systematic Explorer) from National Institute for Allergy and Infectious Diseases (NIAID) and can be found at http://apps1.niaid.nih.gov/DAVID/[19]. EASE examines the overrepresentation of functional groups within a set of transcripts and provides statistical evaluation of the presence of these transcripts. Statistical significance was based on an EASE score, similar to a P-value. In the analysis, an EASE score of 0.1 or less was considered significant within a 10% margin of error. Analyses were made using the nonredundant transcripts identified previously as expressed significantly and uniquely at a particular time point in either male or female gonad development. The list of all nonredundant statistically significant transcripts was used as the total population for statistical calculations within EASE.
RESULTS
Two distinct time courses were established in this study to examine the transcriptional changes that occur during embryonic testis and ovary development. This study used the Affymetrix MGU74v2 chipset representing nearly 37 000 transcripts, which allowed expression levels for a large percentage of the murine transcriptome to be established. Each time course was comprised of five distinct time points in duplicate, which were chosen to provide a continuous view of gonad development from the indifferent gonad stage (11.5 dpc) to just before birth (18.5 dpc). A total of 60 separate arrays were used and the datasets in their entirety are available through NCBI via the Gene Expression Omnibus (GEO) data repository (http://www.ncbi.nih.gov/geo/), GEO accession numbers GSE1358 and GSE1359. Supplemental data are also available through the Griswold Lab website at http://www.wsu.edu/~griswold/microarray and include data from the statistical and clustering analyses.
A two-pronged approach was taken to analyze the dataset. The first approach identified those transcripts having a raw signal of at least 50 in one tissue (testis or ovary) and present at a level at least twofold greater than the other tissue. This analysis was applied to each tissue at each time point. The number of genes that met these criteria is reported in Table 1. The number of genes increases at each time point and, overall, increases 5.3-fold from 11.5 dpc (491) to 18.5 dpc (2626) in the testis. The number of genes in the ovary increases at each time point, except at 14.5 dpc, where a slight decrease (~22%) is observed. In all time points except 14.5 dpc, the ovary contains more genes present at a level twofold higher than the testis. The greatest differences (ovary versus testis) are observed at 11.5 dpc (703 versus 491) and 12.5 dcp (1312 versus 866). The average raw signal of the 10 genes exhibiting the greatest fold change between tissues at each time point is also listed in Table 1 and a descriptive list of these genes and the fold change observed is reported in Table 2. These genes may or may not exhibit differential expression within the testis or ovary over the time course.
TABLE 1.
Genes expressed at least two-fold or greater than raw signal of 50 in the testis or ovary during development of the embryonic gonad.
| Days postcoitum | Sex | No. total genes | Average raw signal, top 10 | No. differentially expressed genes |
|---|---|---|---|---|
| 11.5 | Testis | 491 | 173 | 47 |
| Ovary | 703 | 86 | 92 | |
| 12.5 | Testis | 866 | 312 | 164 |
| Ovary | 1312 | 171 | 248 | |
| 14.5 | Testis | 1046 | 784 | 256 |
| Ovary | 1021 | 749 | 273 | |
| 16.5 | Testis | 2167 | 1422 | 654 |
| Ovary | 2169 | 797 | 644 | |
| 18.5 | Testis | 2626 | 1931 | 647 |
| Ovary | 2642 | 1485 | 948 |
TABLE 2.
Genes exhibiting the greatest differences in expression between testis and ovary at each time point (all genes must have a raw signal greater than 50).
| Description | Common | Fold change | Genbank no. |
|---|---|---|---|
| 11.5-dpc testis* | |||
| Selected mouse cDNA on the Y | Smcy | 31.5 | AI550379 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | Ddx3y | 26.7 | AJ007376 |
| UI-M-BH0-ajs-d-08-0-UI.s1 NIHpBMAPpMpS1 | 24.3 | A1854280 | |
| Eukaryotic translation initiation factor 2; Y-linked | Eif2s3y | 12.0 | AJ006584 |
| Ribosomal protein L29 | Rp129 | 9.5 | LO8651 |
| EST | 7.7 | AW123383 | |
| UI-M-AO0-ach-c-08-0-UI.s1 NIHpBMAPpMPG | 6.5 | AI839131 | |
| EST | 6.3 | AA821721 | |
| EST Stratagene mouse testis (#937308) | 6.2 | AI429274 | |
| EST Mus musculus C57BL/6J ES cell | Hspb1 | 5.9 | AV101961 |
| 11.5-dpc ovary | |||
| EST Mus musculus C57BL/6J 10–11-day embryo | 35.0 | AV147230 | |
| EST | 18.0 | AV349132 | |
| EST | 17.6 | AV237402 | |
| EST RIKEN cDNA 2810037C03 gene | 12.7 | AV323801 | |
| EST | 11.6 | AV045457 | |
| EST RIKEN full-length enriched, 8-day embryo | Pold1 | 8.5 | AV300905 |
| EST | 8.4 | AI449086 | |
| EST | 8.4 | AV213536 | |
| EST | Stard10 | 8.2 | AV278409 |
| 12.5-dpc testis | |||
| EST | 27.1 | AI854280 | |
| Cystatin 9 | Cst9 | 19.9 | Y18243 |
| Endometrial bleeding-associated factor | Ebaf | 18.8 | AV215669 |
| Glutathione S-transferase, mu 6 | Gstm6 | 17.1 | AI326397 |
| EST | 15.8 | AI853427 | |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | Ddx3y | 14.7 | AJ007376 |
| Vanin 1 | Vnn1 | 13.1 | AV377573 |
| Endometrial bleeding-associated factor | Ebaf | 12.6 | AI643778 |
| RIKEN full-length enriched, 8-day embryo | 11.4 | AV307368 | |
| Leukocyte cell-derived chemotaxin 1 | Lect1 | 10.8 | AV140849 |
| 12.5-dpc ovary | |||
| EST Mus musculus inactive × specific transcripts | Xist | 30.5 | L04961 |
| A disintegrin and metalloprotease domain 8 | Adam8 | 22.3 | X13335 |
| Follistatin | Fst | 18.4 | Z29532 |
| EST | 14.0 | AW260027 | |
| O-linked N-acetylglucosamine (GlcNAc) transferase | Ogt | 12.4 | AW045187 |
| EST | 12.1 | AV146216 | |
| EST | 11.7 | AW048759 | |
| Achaetescute complex homolog-like 1 (Drosophila) | Asc11 | 11.5 | AA672916 |
| EST | 11.3 | AV014646 | |
| Hypothetical protein 4732472I07 | 10.7 | AI877199 | |
| 14.5-dpc testis | |||
| Renin (Ren-1-d); mouse renin (Ren-1-d) gene | Ren1 | 186.8 | M32352 |
| Cytochrome P450, family 17, subfamily a | Cyp17a1 | 167.4 | M64863 |
| Hydroxyacid oxidase (glycolate oxidase) 3 | Hao3 | 103.9 | AI648067 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | Ddx3y | 79.8 | AJ007376 |
| Endometrial bleeding-associated factor | Ebaf | 74.8 | AV215669 |
| Cystatin 9 | Cst9 | 61.9 | Y18243 |
| EST | 61.8 | AI854280 | |
| UI-M-BH2.2-aoq-f-11-0-UI.s1 NIHpBMAPpMpS3.2 | Cyp11a | 48.7 | AW121619 |
| Sclerostin domain containing 1 | Sostdc1 | 37.5 | AI842529 |
| Alcohol dehydrogenase | Adh1 | 24.4 | M22679 |
| 14.5-dpc ovary | |||
| Stimulated by retinoic acid gene 8 | Stra8 | 254.4 | Z75287 |
| A disintegrin and metalloprotease domain 11 | Adam11 | 93.1 | AV256444 |
| RIKEN cDNA 9430532D21 gene | 57.1 | AV260742 | |
| RIKEN cDNA 1700026D08 gene | 43.7 | AW047638 | |
| UI-M-AN1-afb-f-10-0-UI.s1 NIHpBMAPpMBGpN | 41.9 | AI842886 | |
| Placenta-specific homeobox 1 | Psx1 | 40.6 | AF017453 |
| Mus musculus transcribed sequences | Ugt8 | 34.7 | AI850488 |
| Placenta-specific homeobox 2 | Psx2 | 27.3 | AV224043 |
| Homeobox, msh-like 2 | Msx2 | 23.4 | X59252 |
| RIKEN cDNA C030048H21 gene | 23.0 | AU017363 | |
| 16.5-dpc testis | |||
| UI-M-BH0-ajs-d-08-0-UI.s1 NIHpBMAPpMpS1 | 398.4 | AI854280 | |
| Renin (Ren-1-d) | Red1 | 279.9 | M32352 |
| Cytochrome P450, family 17, subfamily a | Cyp17a1 | 239.9 | M64863 |
| Cysteine-rich secretory protein 1 | Crisp1 | 201.0 | M92849 |
| RNA binding motif protein, Y chromosome | RbmY1a1 | 132.9 | Y15131 |
| UI-M-BH2.2-aoq-f-11-0-UI.s1 NIHpBMAPpMpS3.2 | Cyp11a | 107.0 | AW121619 |
| RIKEN cDNA 2310045A20 gene | 92.6 | AI853896 | |
| Cystatin 9 | Cst9 | 70.4 | Y18243 |
| Steroidogenic acute regulatory protein | Star | 68.6 | AV224791 |
| Hydroxyacid oxidase (glycolate oxidase) 3 | Hao3 | 64.1 | AI648067 |
| 16.5-dpc ovary | |||
| RIKEN cDNA 4930532D21 gene | 190.6 | AV260742 | |
| Follistatin | Fst | 56.8 | Z29532 |
| RIKEN cDNA 1700026D08 gene | 48.5 | AW047638 | |
| Intersectin (SH3 domain protein 1A) | Itsn | 47.7 | AI643935 |
| RIKEN cDNA C030048H21 gene | 42.7 | AU017363 | |
| Mus musculus 0-day neonate thymus cDNA | Xist | 41.0 | L04961 |
| Mus musculus gene for spot35/calbindin-D28k | Calb1 | 39.7 | D26352 |
| Mus musculus transcribed sequences | 34.1 | AI180790 | |
| RIKEN cDNA 6330403A02 gene | 32.7 | AI852234 | |
| G protein-coupled receptor 49 | Gpr49 | 31.1 | AF110818 |
| 18.5-dpc testis | |||
| Renin (Ren-1-d); mouse renin (Ren-1-d) gene | Ren 1 | 1019.5 | M32352 |
| UI-M-BH0-ajs-d-08-0-UI.s1 NIHpBMAPpMpS1 | 190.1 | AI854280 | |
| Cytochrome P450, family 17, subfamily a | Cyp17a1 | 105.9 | M64863 |
| RNA binding motif protein, Y chromosome | RbmY1a1 | 100.9 | Y15131 |
| Mus musculus transcribed sequences | 99.6 | AI462234 | |
| UI-M-BH2.2-aoq-f-11-0-UI.s1 NIHpBMAPpMpS3.2 | Cyp11a | 86.6 | AW121619 |
| Testis-specific beta defensin-like | Tdl | 74.5 | AV207700 |
| Chemokine (C-C motif) ligand 17 | Ccl17 | 67.9 | AJ242587 |
| Selected mouse cDNA on the Y | Smcy | 65.6 | AI550379 |
| Serine (or cysteine) proteinase inhibitor, clade A | Serpina5 | 61.8 | AV283483 |
| 18.5-dpc ovary | |||
| LIM homeobox protein 8 | Lhx8 | 126.8 | D49658 |
| RIKEN cDNA 4930532D21 gene | 104.6 | AV260742 | |
| Expressed sequence C86987 | 91.2 | AW061168 | |
| Phosducin-like 2 | Pdc12 | 69.6 | AV204767 |
| Mus musculus gamma-B-crystallin gene | Crygb | 52.8 | Z22573 |
| G protein-coupled receptor 49 | Gpr49 | 51.8 | AF110818 |
| Developmental pluripotency-associated 3 | Dppa3 | 49.3 | AI606389 |
| RIKEN cDNA 4833435D08 gene | 42.1 | AI152472 | |
| vg85f05 × 1 Barstead mouse pooled organs MPLRB4 | 41.7 | AI481141 | |
| RIKEN cDNA 5730557K01 gene | 39.1 | AV342639 | |
Sox9, fold change 5.9; Sry, fold change 3.5.
The second method of analysis selectively identified differentially expressed genes. These genes exhibited a significant change in expression exclusively within the ovary or the testis at some point in the time course. After statistical analysis and filtering as described in Materials and Methods, 3693 unique transcripts were identified as being differentially expressed in either the testis or ovary time courses. Of these 3693 unique transcripts, 1936 were differentially expressed exclusively in the ovary while 1296 were differentially expressed exclusively in the testis. There were 461 transcripts differentially expressed in both the testis and ovary during the course of its development. The number of differentially expressed transcripts for each tissue at each time point is presented in Table 1. Hierarchical clustering analysis was performed on the 3963 unique transcripts, and the results are depicted in Figure 1. Based on the resultant dendogram of clustering experimental samples on the vertical axis, it is apparent that the transcripts expressed differentially in the male and female at 11.5 dpc are quite similar, and both sexes continue to share a similar expression profile through the completion of sex determination at 12.5 dpc. However, a significant divergence in expression patterns between the testis and ovary begins at 14.5 dpc and continues through 18.5 dpc, with 18.5-dpc testis and ovary predictably possessing the most distinctive expression profiles. During this period following sex determination, the expression profiles present in the testis at 14.5, 16.5, and 18.5 dpc and the corresponding ovarian time points share some similarity to each other as a group but very distinctive expression profiles are present when comparing the two tissues. Additionally, clustering individual genes on the horizontal axis produced distinct groups of genes demonstrating similar expression patterns. The clustering of gene expression profiles resulted in distinct groups of transcripts with up- and downregulation occurring at every experimental time point in the two time courses with the most drastic regulation of transcripts occurring in the testis and ovary at 16.5 and 18.5 dpc. Although many distinct patterns of regulation are present using this clustering technique, they are viewed more easily using alternative methods.
FIG. 1.
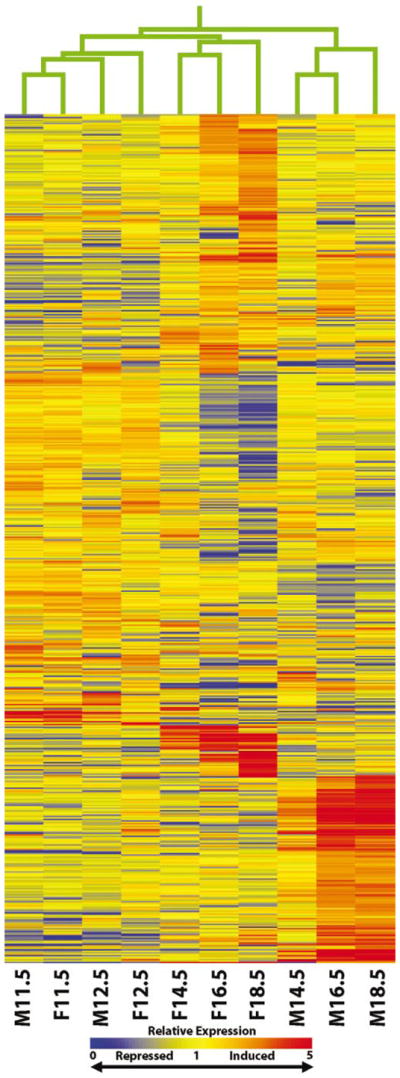
Hierarchical clustering of significantly expressed transcripts in the male and female embryonic gonad. A nonphylogenetic gene tree generated in GeneSpring depicting clustering of 3693 significantly expressed transcripts derived from the analysis of testis and ovary time courses. Genes are clustered on the horizontal axis and experimental samples on the vertical axis. The tree structure at the top of the diagram indicates the similarity of each experimental sample in the testis and ovary time course. Clustering was performed using a smooth correlation with the default separation ratio and minimum distance. Coloring is based on relative expression levels (normalized data): red is indicative of elevated expression compared with median level of each gene, blue is reduced compared with median level, while yellow is near the median level.
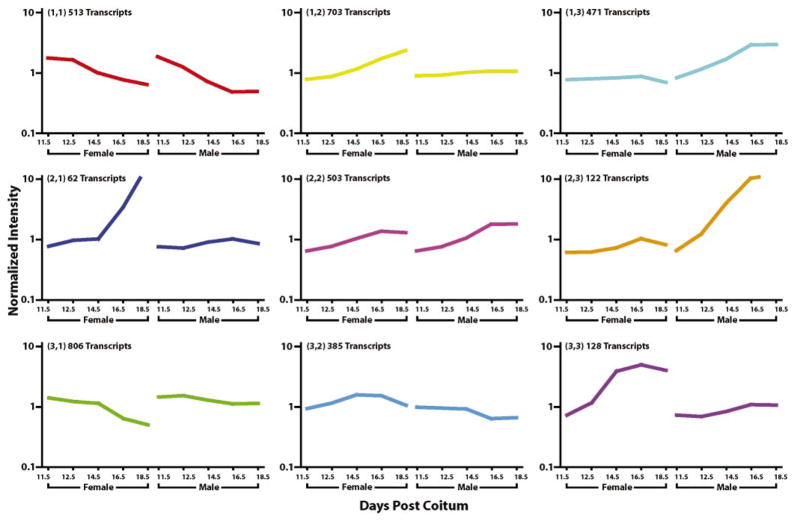
Analysis of the same group of statistically significant transcripts using K-means clustering was used to characterize the overall patterns of expression occurring during development of the embryonic testis and ovary. This analysis revealed a multitude of expression patterns occurring within and between the two time courses. The nine predominant patterns of expression are depicted in Figure 2. The majority of these transcripts exhibited differential expression either early or late in the time course, indicating that these two periods are the most transcriptionally active in the development of both the embryonic testis and ovary. Specifically, based on K-means clustering, 1319 transcripts (~36% of the total number of significant unique transcripts) demonstrated downregulation by 12.5 dpc in both the testis and ovary while only 503 transcripts (~14%) were upregulated at 16.5 or 18.5 dpc in both the testis and ovary. Conversely, 893 transcripts (~24%) were upregulated late in development in the ovary only and 593 transcripts (~16%) were upregulated in the testis only. Another group of 385 transcripts (~10%) exhibited an entirely unique expression pattern with slight upregulation late in ovarian development and gradual downregulation throughout testicular development. These groups of transcripts included differentially expressed transcripts in the testis and ovary as well as in only one tissue. Smaller but no less distinct patterns were not predominant enough to appear in the K-means clustering but are noteworthy. These are primarily groups of transcripts demonstrating regulation occurring only at 14.5 or 16.5 dpc but at no other time points. These are evident to some extent in the hierarchical clustering analysis by the red and blue areas in the male and female samples at 14.5 and 16.5 dpc but do not comprise a large set of the significant transcripts. It should be emphasized that the patterns of expression shown in Figure 2 are shown as averages and that transcripts in each cluster can exhibit patterns of expression that vary from the average when examined on an individual basis.
FIG. 2.
K-means clustering of significantly expressed transcripts. K-means clustering in GeneSpring was used to isolate distinct patterns of expression within and between the male and female embryonic gonad expression profiles. Clustering was performed using a smooth correlation and default settings. Graphs of each cluster are shown as an average of all genes in each cluster. Numerous patterns were identified during the development of the embryonic gonad with the predominate themes being a decrease in expression after determination of sex at 11.5 dpc and a significant upregulation trend occurring after 14.5 dpc, which occurs in both the testis and ovary time courses. Clustering is based on the 3963 unique and significantly expressed transcripts. The Y-axis is in logarithmic scale.
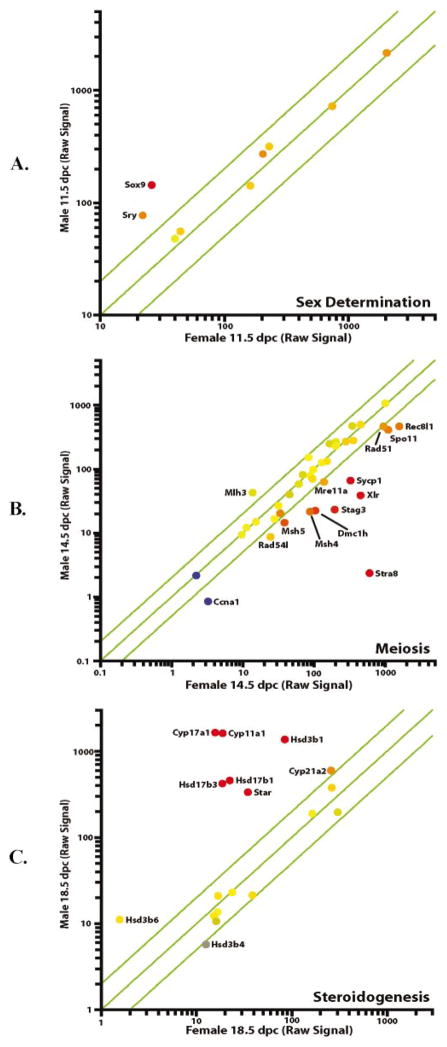
The importance and ultimately the usefulness of a time course of this nature is largely centered around the reliability of the data. Therefore, three distinct and well-documented events occurring during the course of embryonic gonad development were examined to evaluate the reliability of the data sets: sex determination, meiosis, and the synthesis of testosterone. Because it is well known that the process of sex determination is centered around 11.5 dpc, genes previously reported to be involved in sex determination were investigated. In the male gonad, expression of Sry, Sox9, Dax1, Sf1, and Wt1 among others were elevated at 11.5 dpc and then fell to low levels of expression by 14.5 dpc, consistent with their documented involvement with gonad differentiation [1, 2, 7–9]. At 11.5 dpc, many of these genes share similar expression levels in both the male and female, with the exception of Sry and Sox9, which are only expressed at significant levels in the testis (Fig. 3). By 14.5 dpc, Sry and Sox9 dropped to the same background level of expression in testis and ovary. DAX1 levels in the testis were 25% higher than the ovary at 11.5 dpc but, by 12.5 dpc, the level in the testis dropped dramatically whereas the level in the ovary remained high until a decrease was observed at 14.5 dpc. Related genes associated with further divergence between the two sexes include Amh and Dmrt1, both of which achieve high levels of expression in the testis at 16.5 dpc and remain elevated until 18.5 dpc.
FIG. 3.
Scatter plots of characteristic functional activities occurring during embryonic gonad development: (A) sex determination, (B) meiosis, and (C) steroidogenesis. Scatter plots were made within GeneSpring. Axes are in logarithmic scale of raw signal. Coloring is based on expression levels in sex where functional activity is most significant.
Once sex determination has been completed, the testis and ovary begin to proceed down drastically different paths of development. One of the most apparent differences is the synthesis of testosterone in the testis, resulting in the further development of the structures derived from the Wolffian ducts [20]. As described in previous work, Sox8 and AMH levels increased 3.5-fold and 4.5-fold, respectively, immediately after sex determination [21]. During this period, the genes encoding enzymes involved in biosynthesis of testosterone, including StAR, 3β-Hsd, Cyp17a1, and 17β-Hsd demonstrated male-specific increases in expression beginning at 14.5 dpc, with transcript levels typically reaching a plateau by 18.5 dpc. The levels reached by 18.5 dpc were greater than 100-fold higher than in the embryonic ovary and demonstrated comparable changes from earlier time points in the testis. Figure 3 depicts a number of testis-specific transcripts that are directly involved in testosterone biosynthesis in addition to a number of related transcripts that do not exhibit differential expression between the sexes. Other genes involved in this process include insulin-like growth factor I, which has been previously linked to the appearance of the steroidogenic transcripts [22] and was also elevated in the testis at this stage.
Meiosis is another major, sex-specific event occurring during this period, but occurs exclusively in the embryonic ovary and is highlighted by the 100-fold increase in expression of the female-specific meiosis marker Stra8 [23] at 14.5 dpc to reach a level more than 254-fold higher than in the testis (Table 2). Other genes involved in meiosis, such as Spo11, Dmc1h, Sycp1, Stag3, and Xlr, exhibited between a 3- and 25-fold higher level of expression in the ovary (Fig. 3) and a 3- to 146-fold higher level when compared with the ovary at 11.5 dpc.
Ontological analysis using EASE yielded insight into identifying important functional themes present within the transcripts expressed at specific time points in the male and female gonads. For example, at 11.5 dpc, there are no functional categories in either the male or female gonad with EASE scores of less than 0.1, which is similar to a P-value with a confidence interval in the 90th percentile. This could be due to a number of factors, the most likely being the high degree of similarity between the male and female gonad at this time, which, due to the methods employed in this analysis, results in a small number of unique transcripts in either sex at this time and thus does not provide a large enough group of transcripts to effectively analyze functional themes.
As the development of the embryonic testis and ovary progressed, various functional themes were apparent. Of significance is the predominance of signaling and cell-communication transcripts in the testis at 14.5 dpc and apoptosis-related transcripts at 16.5 dpc. Many more significant functional themes were present in the development of the ovary compared with the testis. The reason for this is not clear but may be directly related to the greater number of regulated transcripts in the ovary compared with the testis. Noteworthy functional themes during ovarian development include DNA binding, nucleic acid metabolism, and cyto-skeleton elements at 12.5 dpc, DNA packaging, chromatin-related transcripts, and lipid metabolism at 14.5 dpc (concurrent with the onset on meiosis), cell cycle and mitosis transcripts at 16.5 dpc, and immune-related transcripts at 18.5 dpc. Other functional groups were found during the analysis but these were the predominating themes.
DISCUSSION
The embryonic gonad has the potential to develop into a fully functional organ capable of producing the gametes necessary for sexual reproduction. The complete characterization of the transcriptional events occurring during the development of the embryonic testis and ovary is a necessary and important step in determining how the transcriptome of the embryonic gonad plays a role in achieving this result. To this end, two time courses tracking the changes in gene expression in the male and female embryonic gonad through five different time points between the periods of sex determination and birth were developed.
Other genome-scale studies have been undertaken in an attempt to further identify important genes involved in various stages of embryonic gonad development, specifically the sex determination process. These included two studies using high-throughput whole-mount in situ hybridization methods to attempt to identify novel genes involved in male and female gonad development [24, 25]. These studies used cDNA libraries derived from 9.5-dpc whole-mouse embryos and examined the genes present in gonad tissue from 11.5- and 14.5-dpc mice. A similar study by Grimmond et al. was employed using a cDNA library derived from urogenital ridge tissue and was able to identify novel, differentially expressed genes between the male and female at 11.5 and 12.5 dpc [26]. Other studies have used large-scale techniques such as suppressive-subtractive hybridization and alternative cDNA techniques and technologies to examine gene expression in various stages of embryonic gonad development [13, 27–30] but none have simultaneously examined the testis and ovary during the complete progression of gonad development beginning with sex determination at 11.5 dpc to just before birth at 18.5 dpc at the broad level covered in this study. The current study will hopefully alleviate this gap in available information on gene expression patterns in the embryonic gonad by having the advantage of complete time courses from sex determination to birth for both the embryonic testis and ovary.
Certain details of the study require emphasis. The exact stage of each embryo was confirmed and the sheer number of embryos required to complete a singlet at a given time point, especially at 11.5 dpc and 12.5 dpc, was challenging. This study used embryos with 18 ± 3 tail somites for the 11.5-dpc time point and 28 ± 3 tail somites for the 12.5-dpc time point. It was difficult to limit collection to embryos with an identical number of tail somites because the timing of ovulation, fertilization, and implantation within a litter is variable. Although each sample of RNA used for the array hybridization was generated from pooling the gonad from many embryos, the duplicates represent collection from unique embryos, often months apart. If the quality of the RNA prepared from the tissue was not sufficient for hybridization to the array, it was discarded and new tissue was collected. In contrast with a number of studies that used amplification techniques to generate enough RNA for array hybridization, no supplemental amplification steps were employed for this study. Furthermore, the data generated in this study, while trying to associate changes in gene expression to changes in functional state, are only measurements of steady-state mRNA levels and therefore do not correlate absolutely to observed changes in function. As an additional caveat, although the amount of the murine genome represented on the MGU74v2 array set is substantial, there is a noticeable group of known genes that are either represented inaccurately on the arrays or not at all.
When viewing the overall profile of gene expression from 11.5 dpc to 18.5 dpc, two major periods of transcriptional activity were apparent. While it is not plausible to attribute all this differential gene expression to specific events, such as sex determination, meiosis in the female, or steroid production in the male, it was possible to identify these events within the profile. The first period of activity occurs between 11.5 and 14.5 dpc, where the levels of significant gene expression are generally decreasing. This is the timeframe where the bipotential gonad develops into either a testis or an ovary dependent on the expression of Sry and subsequent downstream events such as the expression and action of Amh. A large number of genes have been previously implicated in this process and a number of them were identified in this study. Sry and Sox9, key components to the differentiation puzzle, were, as expected, only expressed at significant levels in the testis at 11.5 dpc, whereas other genes involved in sex determination shared similar levels of expression in the testis and ovary. This observation supports the critical role a relatively small number of factors have in sex determination. Interestingly, when an exhaustive analysis was performed that required a gene to be differentially expressed at only one time point in a tissue, have a signal of at least 50, and be present at least twofold higher in the testis than in the ovary at 11.5 dpc, Sry was the only gene that passed the requirements. The similarity of overall expression observed between the 11.5-dpc testis and ovary depicted in Figure 1 also supports the high degree of similarity between the tissues at this stage and the relatively small number of genes required to initiate sex determination.
While it’s accepted that the developing embryo is destined to be female until Sry is produced, it is important to consider that, once a sex-determining factor is produced and sex differentiation is triggered, gene repression may prove to be as critical in the developing embryo as gene activation. This study begins at embryonic Day 11.5, before which any number of factors, including the appearance of Sry, may be already influencing sex determination and affecting the relative number of unique transcripts. After differentiation, and evident at 14.5 dpc, there is a lull in the transcriptional activity observed in both sexes. This may reflect a quiescent, transitional period following the expression of genes involved in sex determination at or before 11.5 dpc and just before the induction of genes involved in sex-specific development and preparation for birth. Beginning at 16.5 dpc, the number of unique genes being expressed in the ovary and testis nearly triples.
Primordial germ cells migrate to the genital ridge by 11.5 dpc and proliferate rapidly during this period [31]. After sex determination has occurred, proliferation continues, but there is also a transient burst of apoptosis in the testis [32, 33]. By 16.5 dpc, the ovarian germ cells have entered and been arrested in meiosis and the male gonocytes are in mitotic arrest. Through EASE analysis, it was not surprising to see transcripts in the ovary at 14.5 and 16.5 dpc being assigned to gene categories related to meiosis. In the testis, however, EASE analysis reveals transcripts being assigned to gene categories involving apoptosis and cell death. Even though both sexes are undergoing cell death and apoptosis [34, 35], it is possible that it is much more distinguishable in the male because of the comparatively low level of transcriptional activity present.
Of overall interest is the observation that the number of genes expressed at 11.5 dpc in one tissue that satisfy the requirement of being at a signal of 50 or more and twofold higher than the other tissue (differentially expressed or not) is lower than all other time points. In addition, the level of expression (equivalent to raw average signal) of these genes is also lower than all other time points. This suggests that not only are fewer gene products necessary to drive all the cellular functions at this stage, including gonad differentiation, but also, the relative amount of those products is low. As development continues toward birth, the number of genes differentially expressed in the ovary and testis increase as does their average signal. Sry is present at a raw signal of 77 at 11.5 dpc. This is considered to be a relatively low abundance when array data are viewed and, yet, this level of expression is enough to single-handedly decide the sex of an organism.
As expected, after 11.5 dpc, the factors involved in development of testis-specific traits and suppression of a female gonadal phenotype are being produced. For example, the presence of comparatively high levels of Sox9 at 11.5 dpc triggers the expression of Sox8 and Amh after 12.5 dpc, acting to inhibit the formation of the female reproductive tract, further solidifying the maleness of the embryo [21]. In contrast with the male, the female embryonic gonad undergoes a meiotic event beginning at 14.5 dpc. This initiation of meiosis is clearly marked in the ovary by the dramatic, 100-fold increase in the meiotic marker Stra8, while expression of Stra8 is completely absent in the testis at this time. Other genes involved in meiosis are also elevated during this period and demonstrate a concerted drive toward meiosis. Although meiosis does occur in the embryonic ovary, it does become arrested in the diplotene stage of meiosis I. This allows for a unique opportunity to look at genes involved in the initiation of meiosis rather than in a convoluted fashion from other more mature meiotically active tissues. A distinct set of meiosis-related genes involved in DNA repair and the maintenance of DNA structure as well as the establishment of the synaptonemal complex such as synaptonemal complex protein (Sycp1), mutS homolog 4 (Msh4), mutS homolog 5 (Msh5), meiosis-specific sporulation protein SPO11 homolog (Spo11), and disrupted meiotic cDNA 1 (Dmc1) were present at elevated levels at 14.5 dpc and remained elevated until 18.5 dpc in the ovary while exhibiting significantly lesser levels of expression in the testis. All of these genes have previously been reported to be involved in the initiation of meiosis and their presence in this time course makes a strong argument that meiosis I is occurring by this point in the embryonic ovary (summarized by Svetlanov and Cohen [36]).
This study reports observations of global and temporal patterns of gene expression and provides a review of specific genes previously reported to be expressed within specific events of development, such as sex determination, meiosis, and steroidogenesis. It is not the authors’ intention to only identify a limited number of specific, novel genes associated with unique processes. In addition, this report validates the methodology applied to generate the dataset and provides a high level of confidence to researchers that wish to use this readily available wealth of information for their own purposes.
Acknowledgments
The authors would like to thank Derek Pouchnik in the Laboratory for Biotechnology and Bioanalysis I at Washington State University, Pullman, WA, for microarray hybridization and scanning, Lizhong Yang for GeneSpring analysis, and Chris Hostetler for technical assistance and critical review of the manuscript.
Footnotes
Supported by HD 10808 and U54 HD 42454 from NICHD.
References
- 1.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins JR, Koopman P, Berta P. Testis-determining factor and Y-linked sex reversal. Curr Opin Genet Dev. 1991;1:30–33. doi: 10.1016/0959-437x(91)80037-m. [DOI] [PubMed] [Google Scholar]
- 3.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 4.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 5.Capel B. The battle of the sexes. Mech Dev. 2000;92:89–103. doi: 10.1016/s0925-4773(99)00327-5. [DOI] [PubMed] [Google Scholar]
- 6.Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 7.Swain A, Zanaria E, Hacker A, Lovell-Badge R, Camerino G. Mouse Dax1 expression is consistent with a role in sex determination as well as in adrenal and hypothalamus function. Nat Genet. 1996;12:404–409. doi: 10.1038/ng0496-404. [DOI] [PubMed] [Google Scholar]
- 8.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 9.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 10.Morrish BC, Sinclair AH. Vertebrate sex determination: many means to an end. Reproduction. 2002;124:447–457. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- 11.Capel B, Albrecht KH, Washburn LL, Eicher EM. Migration of mesonephric cells into the mammalian gonad depends on Sry. Mech Dev. 1999;84:127–131. doi: 10.1016/s0925-4773(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 12.McLaren A, Southee D. Entry of mouse embryonic germ cells into meiosis. Dev Biol. 1997;187:107–113. doi: 10.1006/dbio.1997.8584. [DOI] [PubMed] [Google Scholar]
- 13.Bullejos M, Bowles J, Koopman P. Searching for missing pieces of the sex-determination puzzle. J Exp Zool. 2001;290:517–522. doi: 10.1002/jez.1095. [DOI] [PubMed] [Google Scholar]
- 14.Parker KL, Schimmer BP. Genes essential for early events in gonadal development. Ann Med. 2002;34:171–178. [PubMed] [Google Scholar]
- 15.Clarkson MJ, Harley VR. Sex with two SOX on: SRY and SOX9 in testis development. Trends Endocrinol Metab. 2002;13:106–111. doi: 10.1016/s1043-2760(01)00541-0. [DOI] [PubMed] [Google Scholar]
- 16.Peters H. Migration of gonocytes into the mammalian gonad and their differentiation. Philos Trans R Soc Lond B Biol Sci. 1970;259:91–101. doi: 10.1098/rstb.1970.0048. [DOI] [PubMed] [Google Scholar]
- 17.Greco TL, Payne AH. Ontogeny of expression of the genes for steroidogenic enzymes P450 side-chain cleavage, 3 beta-hydroxysteroid dehydrogenase, P450 17 alpha-hydroxylase/C17–20 lyase, and P450 aromatase in fetal mouse gonads. Endocrinology. 1994;135:262–268. doi: 10.1210/endo.135.1.8013361. [DOI] [PubMed] [Google Scholar]
- 18.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 19.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George FW, Wilson JD. Sex determination and differentiation. In: Neill EKAJ, editor. The Physiology of Reproduction. New York: Raven Press; 1994. [Google Scholar]
- 21.Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, Van De Kant HJ, Wegner M, De Rooij DG, Behringer RR, Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 22.Villalpando I, Lopez-Olmos V. Insulin-like growth factor I (IGF-I) regulates endocrine activity of the embryonic testis in the mouse. J Steroid Biochem Mol Biol. 2003;86:151–158. doi: 10.1016/s0960-0760(03)00265-6. [DOI] [PubMed] [Google Scholar]
- 23.Menke DB, Koubova J, Page DC. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev Biol. 2003;262:303–312. doi: 10.1016/s0012-1606(03)00391-9. [DOI] [PubMed] [Google Scholar]
- 24.Neidhardt L, Gasca S, Wertz K, Obermayr F, Worpenberg S, Lehrach H, Herrmann BG. Large-scale screen for genes controlling mammalian embryogenesis, using high-throughput gene expression analysis in mouse embryos. Mech Dev. 2000;98:77–94. doi: 10.1016/s0925-4773(00)00453-6. [DOI] [PubMed] [Google Scholar]
- 25.Wertz K, Herrmann BG. Large-scale screen for genes involved in gonad development. Mech Dev. 2000;98:51–70. doi: 10.1016/s0925-4773(00)00452-4. [DOI] [PubMed] [Google Scholar]
- 26.Grimmond S, Van Hateren N, Siggers P, Arkell R, Larder R, Soares MB, de Fatima Bonaldo M, Smith L, Tymowska-Lalanne Z, Wells C, Greenfield A. Sexually dimorphic expression of protease nexin-1 and vanin-1 in the developing mouse gonad before overt differentiation suggests a role in mammalian sexual development. Hum Mol Genet. 2000;9:1553–1560. doi: 10.1093/hmg/9.10.1553. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, 3rd, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci U S A. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowles J, Bullejos M, Koopman P. A subtractive gene expression screen suggests a role for vanin-1 in testis development in mice. Genesis. 2000;27:124–135. [PubMed] [Google Scholar]
- 29.McClive PJ, Hurley TM, Sarraj MA, van den Bergen JA, Sinclair AH. Subtractive hybridisation screen identifies sexually dimorphic gene expression in the embryonic mouse gonad. Genesis. 2003;37:84–90. doi: 10.1002/gene.10231. [DOI] [PubMed] [Google Scholar]
- 30.Menke DB, Page DC. Sexually dimorphic gene expression in the developing mouse gonad. Gene Expr Patterns. 2002;2:359–367. doi: 10.1016/s1567-133x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 31.Tam PP, Snow MH. Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J Embryol Exp Morphol. 1981;64:133–147. [PubMed] [Google Scholar]
- 32.Roosen-Runge EC, Leik J. Gonocyte degeneration in the postnatal male rat. Am J Anat. 1968;122:275–299. doi: 10.1002/aja.1001220208. [DOI] [PubMed] [Google Scholar]
- 33.De Rooij DG, Lok D. Regulation of the density of spermatogonia in the seminiferous epithelium of the Chinese hamster: II. Differentiating spermatogonia. Anat Rec. 1987;217:131–136. doi: 10.1002/ar.1092170204. [DOI] [PubMed] [Google Scholar]
- 34.Tilly JL. Apoptosis and ovarian function. Rev Reprod. 1996;1:162–172. doi: 10.1530/ror.0.0010162. [DOI] [PubMed] [Google Scholar]
- 35.Coucouvanis EC, Sherwood SW, Carswell-Crumpton C, Spack EG, Jones PP. Evidence that the mechanism of prenatal germ cell death in the mouse is apoptosis. Exp Cell Res. 1993;209:238–247. doi: 10.1006/excr.1993.1307. [DOI] [PubMed] [Google Scholar]
- 36.Svetlanov A, Cohen PE. Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Exp Cell Res. 2004;296:71–79. doi: 10.1016/j.yexcr.2004.03.020. [DOI] [PubMed] [Google Scholar]