Abstract
Parent-of-origin–dependent expression of imprinted genes is mostly associated with allele-specific DNA methylation of the CpG islands (CGIs) called germ line differentially methylated regions (gDMRs). Although the essential role of gDMRs for genomic imprinting has been well established, little is known about how they evolved. In several imprinted loci, the CGIs forming gDMRs may have emerged with the insertion of a retrotransposon or retrogene. To examine the generality of the hypothesis that the CGIs forming gDMRs were novel CGIs recently acquired during mammalian evolution, we reviewed the time of novel CGI emergence for all the maternal gDMR loci using the novel data analyzed in this study combined with the data from previous reports. The comparative sequence analyses using mouse, human, dog, cow, elephant, tammar, opossum, platypus, and chicken genomic sequences were carried out for Peg13, Meg1/Grb10, Plagl1/Zac1, Gnas, and Slc38a4 imprinted loci to obtain comprehensive results. The combined data showed that emergence of novel CGIs occurred universally in the maternal gDMR loci at various time points during mammalian evolution. Furthermore, the analysis of Meg1/Grb10 locus provided evidence that gradual base pair–wise sequence change was involved in the accumulation of CpG sequence, suggesting the mechanism of novel CGI emergence is more complex than the suggestion that CpG sequences originated solely by insertion of CpG-rich transposable elements. We propose that acquisition of novel CGIs was a key genomic change for the evolution of imprinting and that it usually occurred in the maternal gDMR loci.
Keywords: imprinted gene, CpG island, DMR, retrotransposon, marsupial, monotreme
Introduction
Genomic imprinting is a unique epigenetic regulation inducing monoallelic expression to subset of genes depending on the parental origin. It is known that plants and insects have genomic imprinting, but in higher vertebrates, interestingly, it has not been observed outside the therian mammals (the eutherians and marsupials). To date, nearly 100 imprinted genes have been identified in the mouse, and many genetic studies demonstrate their important roles to control fetal and placental development and growth, maternal behavior, and also carcinogenesis (Ferguson-Smith et al. 1991; Guillemot et al. 1995; Lefebvre et al. 1998; Li et al. 1999; Ono et al. 2006; Sekita et al. 2008; Monk 2010). Whereas most human orthologues are also imprinted, only 6 out of 13 genes so far examined are imprinted in marsupials (Renfree et al. 2009). No imprinted genes have been reported in monotremes (Pask et al. 2009; Renfree et al. 2009). All three groups of mammals have a placenta (albeit short lived), but after a short period of intrauterine development, monotreme young are delivered in an egg. Imprinting therefore may have coevolved with the evolution of mammalian viviparity (Renfree et al. 2009).
The distribution of imprinted genes on the mouse genome is not random. They are most often seen in clusters termed imprinted domains. Imprinted expression of multiple genes in an imprinted domain is coordinately regulated by a single genomic element called the germ line differentially methylated region (gDMR) or imprinting control region. gDMRs are CpG rich, and differential DNA methylation is observed between two parental alleles. The difference of DNA methylation on gDMRs is established during gametogenesis and maintained throughout development. Either genetic or epigenetic disruption of gDMR leads to a disruption of the expression pattern of surrounding imprinted genes and is associated with some human syndromes. Because differential methylation of gDMRs is one of the most essential processes of genomic imprinting, the acquisition of gDMR in the genome must be a pivotal event for the evolution of imprinting in mammals.
There are several reports suggesting that retrotransposition is involved in the acquisition of gDMR. We previously reported that the insertion of Peg10, a retrotransposon-derived imprinted gene essential for placental development in the mouse, must have occurred in therian ancestor after the divergence of marsupials and eutherians from monotremes (Suzuki et al. 2007). Although the DNA sequence of the DMR in 5′ region of PEG10 does not share significant homology with any known retrotransposon sequence, the evidence that the CpG island (CGI) forming DMR has also newly emerged in the therian ancestor and the several characteristic features of the methylation pattern and of the position of the putative transcriptional regulatory region in the marsupial PEG10 provide the possibility that the 5′ region of PEG10 corresponds to a long terminal repeat of the ancient retrotransposon from which PEG10 originated (Suzuki et al. 2007). Also some of the small imprinted genes that reside in an intron of other genes, such as Mcts2, Nap1l5, Inpp5f_v2, U2af1-rs1, and Nnat, are thought to be inserted into their present positions by retrotransposition. Mcts2 retrotransposition occurred in the ancient line of Euarchontoglires (synonymous with supraprimates) after the divergence of the Laurasiatheria, whereas retrotransposition for Nap1l5, Inpp5f_v2, and Nnat occurred in the eutherian ancestor after the divergence of marsupials, and U2af1-rs1 was retrotransposed in the common ancestor of rodents (Evans et al. 2005; Wood et al. 2007). Interestingly, in every case, the CGIs forming the gDMR likely emerged as novel CGIs at the same time as the retrotransposition of each gene occurred.
There are other gDMR loci that do not have obvious evidence suggesting retrotransposition. It is unclear whether they also suddenly emerged as novel CGIs at some time point in mammalian evolution or whether the evolutionary conserved CGIs somehow favored differential methylation. There are some data that address this question. The gDMRs for Peg1/Mest, Lit1/Kcnq1ot1, and Airn/Air emerged as novel CGIs in the common ancestor for eutherians after the divergence of marsupials (Killian et al. 2000; Suzuki et al. 2005; Ager et al. 2008). In rodents, novel CGIs associated with Impact gene occurred in the rodent ancestor (Okamura et al. 2000). The Snrpn gene was generated by a gene duplication event that occurred in the therian ancestor after the divergence of monotremes (Rapkins et al. 2006). A CGI also exists at 5′ region of the opossum SNRPN gene, suggesting that the origin of the CGI had the same or similar timing as the gene duplication event, although it is unlikely to be differentially methylated in marsupials because of the lack of an imprinting control transcript (IC transcript) and imprinting of SNRPN (Rapkins et al. 2006). The Peg3 region is probably eutherian specific because no orthologous region has been found in marsupial and monotreme genomes. Gene duplication events may be involved in the generation of Peg3 as there are numerous zinc finger protein genes around Peg3 in mouse and human.
How and why imprinted loci arose during mammalian evolution is not yet clear. In this study, we carried out comparative sequence analyses for the maternal gDMR loci with orthologous genomic regions of Peg13, Meg1/Grb10, Plagl1/Zac1, Gnas, and Slc38a4 of various mammalian species to test the generality of the hypothesis that the CGIs forming gDMRs have emerged as novel CGIs during mammalian evolution. We provide the first comprehensive view for the origins of gDMRs and discuss how they may have been acquired in the mammalian genome.
Materials and Methods
Comparative Sequence Analyses
Each genomic sequence was obtained from public database using the Ensembl genome browser (http://www.ensembl.org, the last accessed date; 21 October 2011). The graphs showing conserved genomic regions were created using mVISTA (http://genome.lbl.gov/vista, the last accessed date; 21 October 2011). The graphs showing CpG dinucleotide distribution were created using GENETYX-MAC, Version 13.1.6 (GENETYX Corporation), with the following parameters for calculation: 1/200 and 1/400 of the sequence length for the span and the step, respectively. To represent the position of each CpG sequence in the genomic sequences as a bar, we used the R statistical computing software (http://www.r-project.org, the last accessed date; 21 October 2011).
Results
Origin of the Peg13 CGI in Euarchontoglires
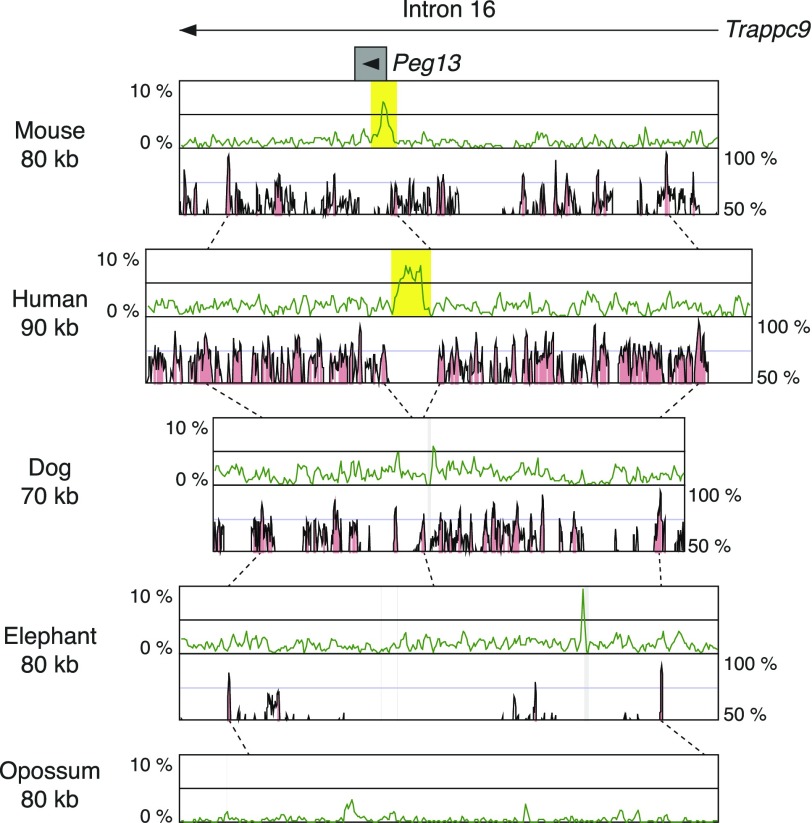
Peg13 is a single exon noncoding gene located in the intron 16 of Trappc9 on mouse chromosome 15. Peg13 has a CGI over the promoter region, and this CGI shows germ line–derived maternal methylation (Ruf et al. 2007). It is consistent with the paternal expression of Peg13, so this gDMR is thought to be essential for Peg13 imprinting. We obtained orthologous regions in human, dog, elephant, and opossum genomes using the evolutionary conserved regions in the corresponding intron of TRAPPC9 gene in each species. In the comparison between mouse and human, the CGI was conserved in the expected position, but Peg13 sequence was not highly conserved between mouse and human, consistent with the rapid sequence evolution of noncoding genes (fig. 1). In dog, elephant, and opossum, there were no CGIs conserved in the corresponding genomic region, suggesting that the CGI is not conserved in Metatheria, Afrotheria, and Laurasiatheria. These results indicate that the Peg13 CGI have emerged in the Euarchontoglires after the divergence of the Laurasiatheria. In the comparison between human and dog, there was constant high level of conservation throughout this genomic region, except for the small region surrounding the CGI (fig. 1). Interestingly, two conserved peaks seen at the both ends of this region were very close to the position in the dog sequence. This result strongly suggests that the genomic region corresponding to Peg13 was inserted into the genome of the Euarchontoglires ancestor. It remains unclear whether the inserted DNA itself was CpG rich or whether CpG sequences were accumulated after the insertion event. However, in both cases, the insertion of DNA was potentially the trigger for the emergence of novel CGI forming the gDMR in this locus.
FIG. 1.—
Comparison of CpG contents and conservation among the orthologous genomic regions around the Peg13 gDMR. The upper green graphs show CpG contents in the genomic sequences. The lower pink graphs show conserved regions in the genomic sequences between one species and the other species located just below (e.g., The pink graph seen in the mouse row is the comparison of mouse [base] and human and the graph in the human row is the comparison of human [base] and dog). The broken lines indicate where some conservation peaks in the upper row correspond in the next lower row. The arrowhead indicates the transcription start site with the direction, and the gray box shows exon. Gaps in the sequences are represented by the light gray shadows in graph regions. The CGI forming gDMR in mouse and the corresponding CGI in other species are yellow highlighted.
Eutherian Origin of the Meg1/Grb10 Downstream CGI
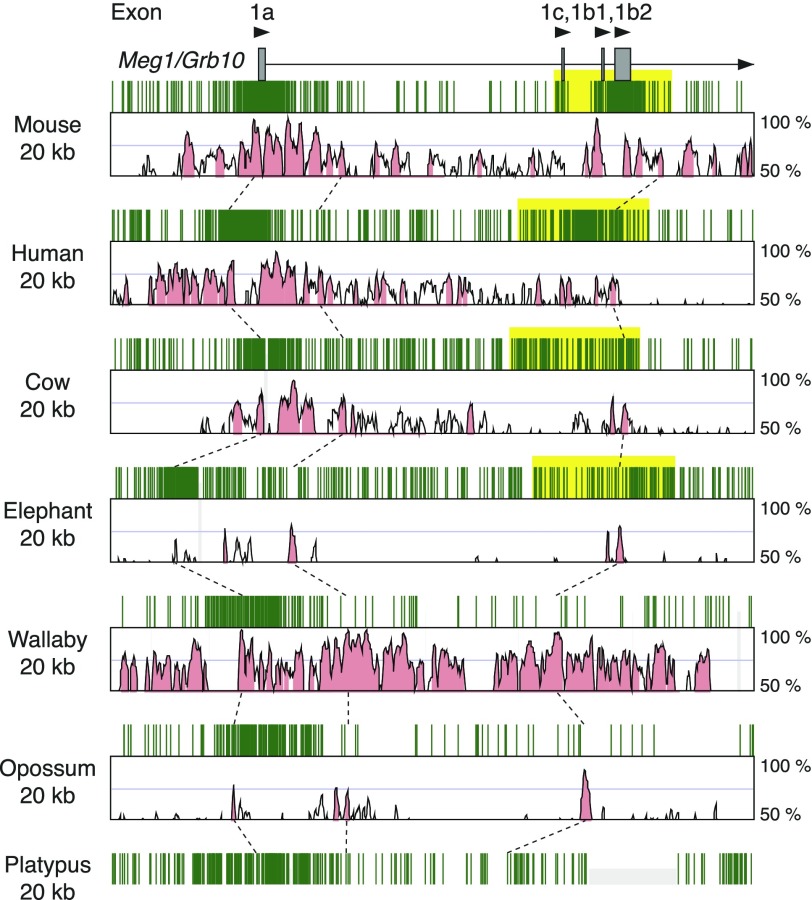
There are two CGIs in mouse Meg1/Grb10 promoter regions containing several transcription start sites (fig. 2). The major transcript produced from the upstream CGI is expressed in almost all tissues, whereas transcripts from the downstream CGI have a brain-specific expression pattern (Hikichi et al. 2003). Maternal methylation is observed only in the downstream CGI, and it is a gDMR. The brain-specific transcripts therefore show paternal expression, and the maternal expression of the major transcript is secondarily regulated by the differential methylation of the downstream CGI (Shiura et al. 2009). Orthologous genomic regions of platypus, opossum, tammar, elephant, cow, human, and mouse showed that the upstream CGI is conserved among all three mammalian subgroups, but only eutherian species have the downstream CGI (fig. 2). Therefore, the CGI forming gDMR in Meg1/Grb10 locus must have emerged in the eutherian ancestor after the divergence of marsupials but before the Afrotheria split.
FIG. 2.—
Comparison of CpG contents and conservation among the orthologous genomic regions around the Meg1/Grb10 gDMR. For this locus, each CpG site in the genomic sequences is represented by green bar because of the relatively shorter 20 kb genomic regions to compare. Explanations for other components are the same as the figure 1.
Eutherian Origin of the Plagl1/Zac1 Downstream CGI
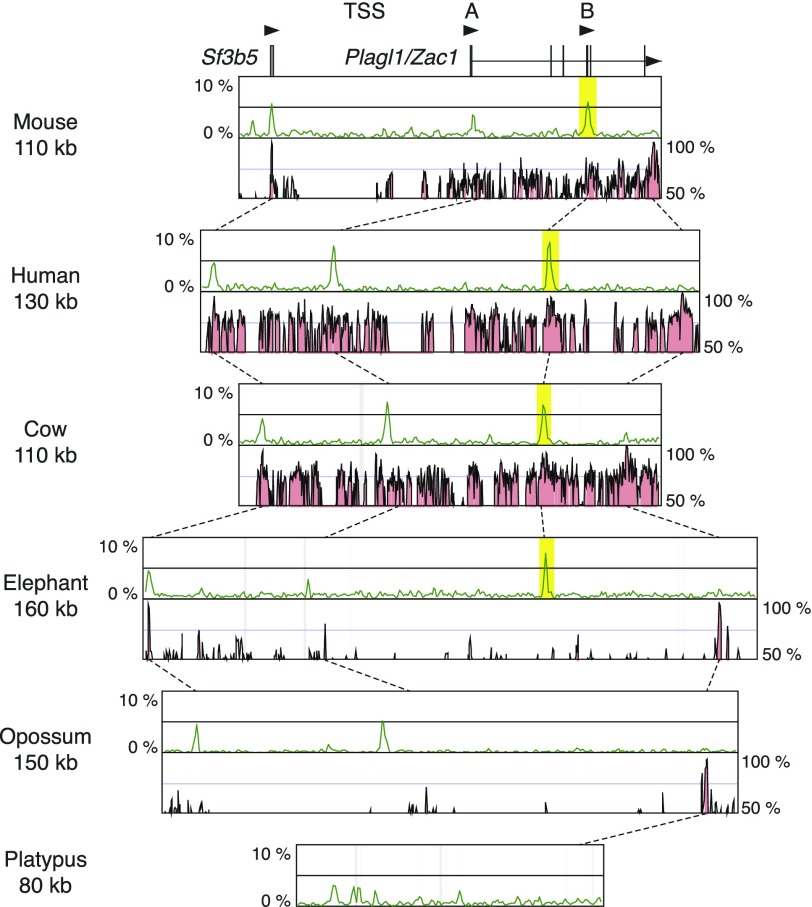
Plagl1/Zac1 has two different transcription start sites with CGIs for each (fig. 3). The transcript from the downstream promoter shows paternal expression, and the CGI is a maternally methylated gDMR (Smith et al. 2002). On the other hand, the transcript from the upstream promoter is expressed biallelically, and the upstream CGI is unmethylated in human (Valleley et al. 2007). Comparing the orthologous genomic regions among mouse, human, cow, elephant, opossum, and platypus, it was clear that the downstream CGI is seen only in the eutherian species similar to the Meg1/Grb10 locus (fig. 3). Therefore, the CGI forming gDMR in Plagl1/Zac1 locus also must have emerged as a novel CGI in the eutherian ancestor before the Afrotheria diverged.
FIG. 3.—
Comparison of CpG contents and conservation among the orthologous genomic regions around the Plagl1/Zac1 gDMR. Explanations for the each component are the same as the figure 1.
Eutherian Origin of the Nesp CGI in Gnas Imprinted Domain
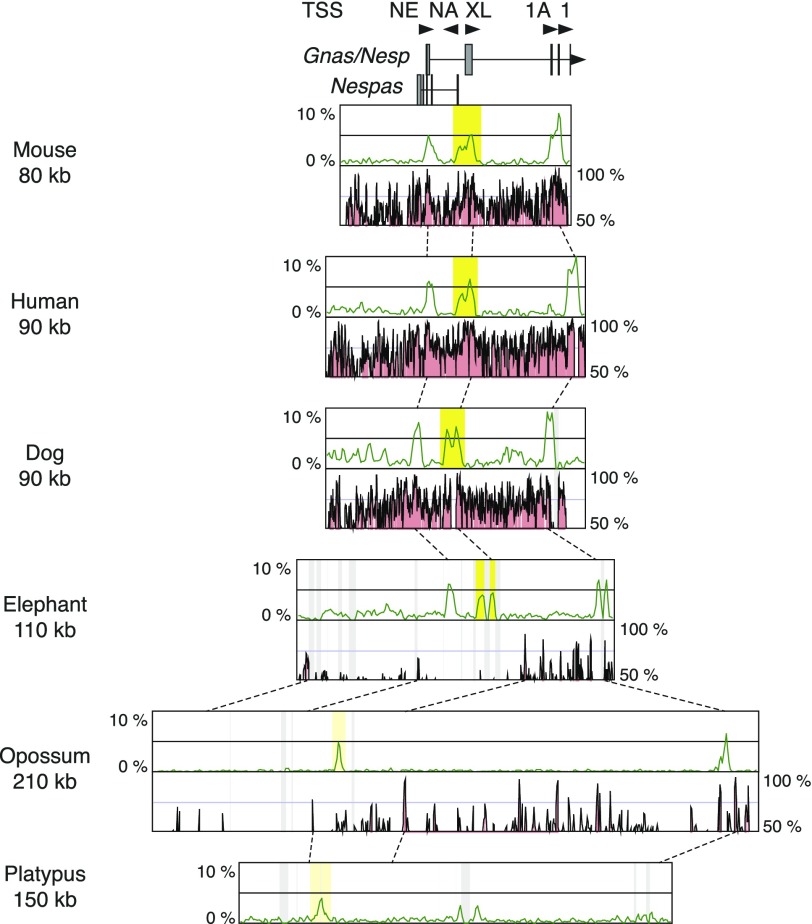
There are three CGIs containing total five transcription start sites in the mouse Gnas promoter region consisting of Nesp, Nespas, Gnasxl, Gnas, and Gnas1A (fig. 4). The upstream CGI on Nesp promoter is known to be a paternally methylated secondary DMR (Liu et al. 2000), whereas the middle CGI on Nespas and Gnasxl promoters and the exon 1A side of the downstream CGI are both maternally methylated gDMR (Liu et al. 2000; Coombes et al. 2003). There are two gDMRs in this domain. However, the downstream CGI methylation is dependent on the methylation of the middle CGI (Williamson et al. 2006). Therefore, the middle CGI is thought as the primary gDMR in this domain. Comparative analysis with the orthologous genomic regions in human, dog, elephant, opossum, and platypus showed that all three CGIs were conserved in eutherians but not in the marsupial and monotreme species (fig. 4). In opossum and platypus, there was only one CGI within the genomic region that eutherians have two CGIs. From our data, it was not possible to determine whether the CGI in noneutherian species corresponded to the upstream or the middle CGI in eutherians. However, unpublished data suggest that marsupial Gnasxl does have a CGI but has no differential methylation (Kelsey G, Ivanova E, personal communication). Therefore, unlike other loci, the upstream Nesp CGI, which is not a gDMR, has emerged as a novel CGI in this domain of the eutherian ancestor, but not Gnasxl CGI itself which forms the gDMR in the mouse.
FIG. 4.—
Comparison of CpG contents and conservation among the orthologous genomic regions around the Gnas gDMR. Explanations for the each component are the same as the figure 1. For the names of transcription start sites, “NE” represents Nesp, “NA” for Nespas, “XL” for Gnasxl, “1A” for Gnas1A, and “1” for Gnas exon 1.
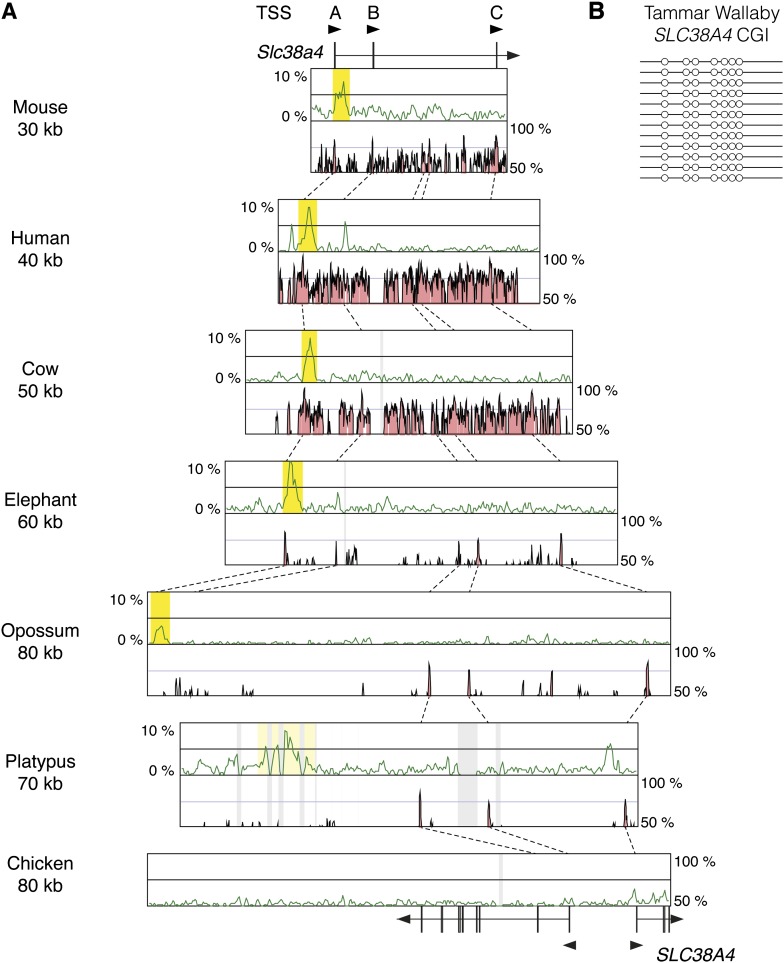
The Slc38a4 CGI Is Conserved at Least in Therian Mammals
Slc38a4 gene is highly expressed in placenta and liver of mouse, but the allelic pattern of expression is different between these tissues. Paternal expression is observed in the placenta, whereas the liver shows biallelic expression (Smith et al. 2003). The CGI of this gene is a maternally methylated gDMR (Chotalia et al. 2009). To determine when this CGI emerged, we compared orthologous genomic regions in human, cow, elephant, opossum, platypus, and chicken. Unexpectedly, the CGI was found in all the mammalian species including monotremes, unlike other gDMR loci (fig. 5A). However, in the comparison between opossum and platypus, there was no conserved region nearby the CGI. The CGI in the platypus sequence may not correspond to the CGIs in other species, although it is possible that the conserved regions are hidden in sequence gaps. Considering the possibility that the CGI in marsupials is also differentially methylated, we checked the methylation pattern of the orthologous CGI in a marsupial, the tammar wallaby, Macropus eugenii. The tammar CGI was clearly unmethylated, suggesting there is no imprinting of SLC38A4 in marsupials (fig. 5B). The Slc38a4 gene is located on mouse chromosome 15 and opossum chromosome 8. However, from about 90 kb upstream of opossum SLC38A4, the opossum sequence has synteny with mouse chromosome 2 (data not shown), suggesting that chromosome rearrangement occurred after the divergence of marsupials, as occurred in the Snrpn locus (Rapkins et al. 2006). Further knowledge about imprinting mechanism in this locus will be required to find what kind of genomic changes occurred with the acquisition of differential methylation. In the chicken, there was no orthologous CGI, and an expressed sequence tag database search revealed that the only transcription start site found in chicken SLC38A4 gene corresponds to the downstream alternative transcription start site in mouse, whereas another gene transcribed toward the opposite direction exists in the upstream of chicken SLC38A4 (fig. 5A). Thus, in this locus, the emergence of the CGI and the acquisition of differential methylation occurred at different times in mammalian evolution.
FIG. 5.—
Comparison of CpG contents and conservation among the orthologous genomic regions around the Slc38a4 gDMR. (A) Explanations for the each component are the same as the figure 1. (B) White circles indicate unmethylated CpGs.
Discussion
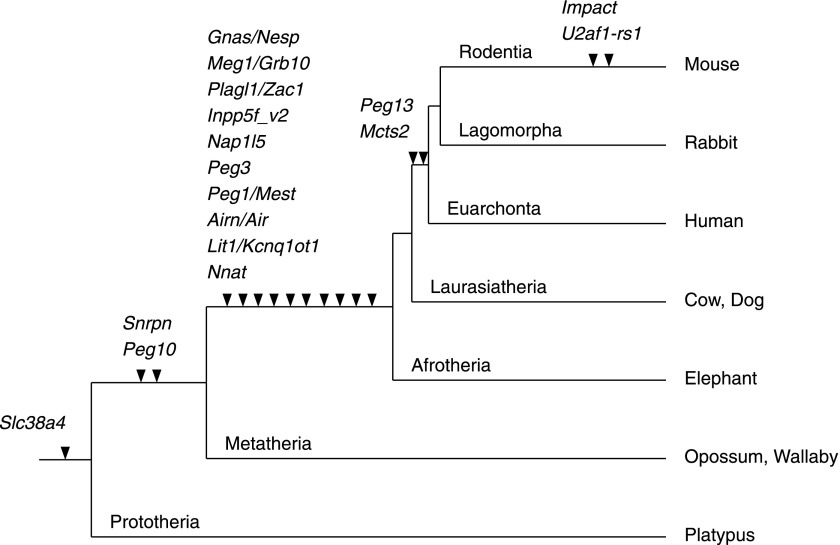
We analyzed the time of emergence of the CGIs forming maternal gDMR in mouse combining previously reported and our current data (fig. 6). Interestingly, all the maternal gDMRs, except mouse Gnas locus in which the Nesp CGI is a secondary DMR, were novel CGIs that emerged during mammalian evolution (fig. 6). Slc38a4, Snrpn, and Gnas loci suggest that differential methylation was acquired only in the eutherian lineage because the CGIs in Slc38a4 and Gnas loci of marsupials are unmethylated. Chotalia et al. (2009) hypothesized that an intronic location for the differential methylation of the maternal gDMRs is important, a hypothesis strikingly consistent with the outcome of this study. The novel CGIs of Slc38a4, Snrpn, and Gnas loci were unlikely to have emerged in introns in the ancestral mammal, whereas in most other loci, CGIs emerged in introns. Then in eutherian lineage, by the acquisition of the IC transcript and Nesp to Snrpn and Gnas loci, respectively, the location of the CGIs became internalized within the transcription unit. In Slc38a4 locus, we found that chromosome rearrangement occurred after the divergence of marsupials, just like that which occurred in the Snrpn locus (Rapkins et al. 2006). It would be interesting to know whether any upstream transcript over the Slc38a4 CGI was acquired by this chromosome rearrangement like the IC transcript in the Snrpn locus. Thus, the emergence of novel CGI in introns may be a condition for the acquisition of gDMRs, although a large-scale transgenic experiment that introduces novel CGIs will be required for the confirmation and further definition.
FIG. 6.—
The timing of the novel CGI emergence in each maternal gDMR locus during mammalian evolution. The arrowheads represent the acquisition of novel CGI to the each locus. The genes associated with novel CGI emergence are shown above the arrowheads.
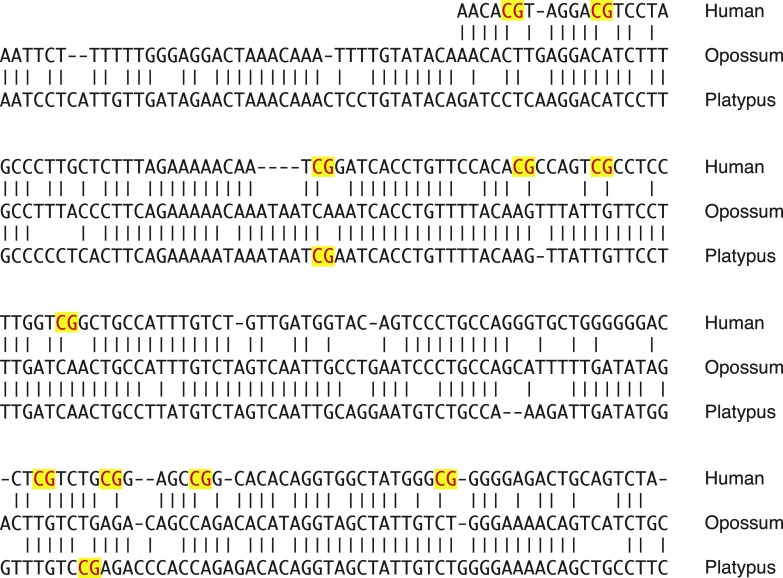
We next focused on how these novel CGIs emerged. Considering the multiple evidence of involvement of retrotransposition events for the acquisition of imprinting, the insertion of transposable elements or retrogenes are most likely associated with the emergence of these CGIs. The CpG sequences could be derived from the inserted sequences themselves or from gradual base pair–wise sequence changes that accumulate CpG sequences. Interestingly, we found evidence for the latter case in the analysis of Meg1/Grb10 locus. We showed that the Meg1/Grb10 downstream CGI only exists in eutherian species (fig. 2). However, a small conserved region inside the downstream CGI in the eutherian species was also detected in the marsupial and monotreme species despite the observation that the regions were not CpG rich in these species (fig. 2, the right broken line). We therefore aligned the genomic sequences of this region in human, opossum, and platypus to analyze the sequence changes in this small conserved region (fig. 7). The alignment data clearly showed overall sequence homology among the three species from different mammalian subgroups. The high conservation between the opossum and platypus sequences suggests that the original sequence was similar to their sequences. Whereas only 2 CpG sites were found in total of the opossum and platypus sequences, the human sequence has 10 CpG sites that still retain the overall sequence homology. The opossum and platypus sequences corresponding to the each CpG site in the human sequence are mostly conserved. Therefore, most CpG sites in the human sequence must have been acquired after the divergence of marsupial rather than the surprisingly similar sequence changes that occurred independently in the opossum and platypus genomes. This finding provides the evidence that the gradual base pair–wise sequence change was involved in the accumulation of CpG sequences. Schulz et al. (2010) have previously provided some data to show that maternal gDMRs tended to gain CpG sequences during eutherian evolution compared with nonimprinted CpG-rich promoters, consistent with our observation. Although it is unclear whether this phenomenon occurred consequent upon a nearby insertion of transposable element, the existence of this phenomenon supports the suggestion that inserted sequence is not the only source of CpG sequences and that inserted sequence itself is not necessarily CpG-rich. Although it is difficult to speculate how CpG sequences could be gained, GC-biased gene conversion may be one possible mechanism (Galtier et al. 2001; Duret and Galtier 2009). These results suggest the mechanism of novel CGI emergence is more complex than simply a result of the insertion of CpG-rich transposable elements. Under the supposition that there is a consensus mechanism to acquire these novel CGIs, we conclude that the most rational hypothesis is that the mammalian genome has enhanced CpG sequence density in the genomic region surrounding the insertion site of transposable elements, although some additional conditions must be considered. This is consistent with the observation that the CGIs are expanded upstream beyond the transcription start sites in some retrogene-associated imprinted loci and also with the difficulty to find common sequence among the gDMRs.
FIG. 7.—
Genomic sequence alignment of the small conserved region from Meg1/Grb10 gDMR corresponding region among three mammalian subgroups. The aligned genomic sequences are from the small conserved region in each species indicated by the right broken line in the figure 2. The vertical bars represent conserved nucleotides. Each CpG site is red colored and yellow highlighted.
In this study, we showed that many CGIs have emerged as novel CGIs in the maternal imprinted loci at various time points during mammalian evolution. Because protein coding genes tend to be highly conserved among mammalian species, changes of transcription regulation and gain/loss of genes rather than protein function evolution may have been the greater driving force for mammalian evolution. It is well known that CGIs are the crucial platform for epigenetic modifications to regulate transcription. We predict that novel CGIs have also emerged in other genomic loci than imprinted domains, and they have contributed to evolve the unique features in mammals. Of these, only when certain conditions were satisfied, the CGIs that had emerged became differentially methylated. This might explain why imprinted genes are often associated with fetal–maternal nutrient transfer, placental development, and some maternal behavior that must have evolved in mammals. Acquisition of novel CGIs is a key genomic change for the evolution of genomic imprinting that generally occurred in the maternal gDMR loci.
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (Postdoctoral Fellowship for Research Abroad to S.S.) and the Australian Research Council (Federation Fellowship to M.B.R.].
References
- Ager EI, Pask AJ, Gehring HM, Shaw G, Renfree MB. Evolution of the CDKN1C-KCNQ1 imprinted domain. BMC Evol Biol. 2008;8:163. doi: 10.1186/1471-2148-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotalia M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes C, et al. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol Cell Biol. 2003;23:5475–5488. doi: 10.1128/MCB.23.16.5475-5488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Evans HK, Weidman JR, Cowley DO, Jirtle RL. Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol Biol Evol. 2005;22:1740–1748. doi: 10.1093/molbev/msi165. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Cattanach BM, Barton SC, Beechey CV, Surani MA. Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature. 1991;351:667–670. doi: 10.1038/351667a0. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, et al. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet. 1995;9:235–242. doi: 10.1038/ng0395-235. [DOI] [PubMed] [Google Scholar]
- Hikichi T, Kohda T, Kaneko-Ishino T, Ishino F. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 2003;31:1398–1406. doi: 10.1093/nar/gkg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian JK, et al. M6P/IGF2R imprinting evolution in mammals. Mol Cell. 2000;5:707–716. doi: 10.1016/s1097-2765(00)80249-x. [DOI] [PubMed] [Google Scholar]
- Lefebvre L, et al. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. 1998;20:163–169. doi: 10.1038/2464. [DOI] [PubMed] [Google Scholar]
- Li L, et al. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284:330–333. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20:5808–5817. doi: 10.1128/mcb.20.16.5808-5817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D. Deciphering the cancer imprintome. Brief Funct Genomics. 2010;9:329–339. doi: 10.1093/bfgp/elq013. [DOI] [PubMed] [Google Scholar]
- Okamura K, et al. Comparative genome analysis of the mouse imprinted gene impact and its nonimprinted human homolog IMPACT: toward the structural basis for species-specific imprinting. Genome Res. 2000;10:1878–1889. doi: 10.1101/gr.139200. [DOI] [PubMed] [Google Scholar]
- Ono R, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- Pask AJ, et al. Analysis of the platypus genome suggests a transposon origin for mammalian imprinting. Genome Biol. 2009;10:R1. doi: 10.1186/gb-2009-10-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapkins RW, et al. Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet. 2006;2:e182. doi: 10.1371/journal.pgen.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu Rev Genomics Hum Genet. 2009;10:241–262. doi: 10.1146/annurev-genom-082908-150026. [DOI] [PubMed] [Google Scholar]
- Ruf N, et al. Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum Mol Genet. 2007;16:2591–2599. doi: 10.1093/hmg/ddm216. [DOI] [PubMed] [Google Scholar]
- Schulz R, et al. The parental non-equivalence of imprinting control regions during mammalian development and evolution. PLoS Genet. 2010;6:e1001214. doi: 10.1371/journal.pgen.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekita Y, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 2008;40:243–248. doi: 10.1038/ng.2007.51. [DOI] [PubMed] [Google Scholar]
- Shiura H, et al. Paternal deletion of Meg1/Grb10 DMR causes maternalization of the Meg1/Grb10 cluster in mouse proximal chromosome 11 leading to severe pre- and postnatal growth retardation. Hum Mol Genet. 2009;18:1424–1438. doi: 10.1093/hmg/ddp049. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Dean W, Konfortova G, Kelsey G. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res. 2003;13:558–569. doi: 10.1101/gr.781503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, et al. The mouse Zac1 locus: basis for imprinting and comparison with human ZAC. Gene. 2002;292:101–112. doi: 10.1016/s0378-1119(02)00666-2. [DOI] [PubMed] [Google Scholar]
- Suzuki S, et al. Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech Dev. 2005;122:213–222. doi: 10.1016/j.mod.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Suzuki S, et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007;3:e55. doi: 10.1371/journal.pgen.0030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valleley EM, Cordery SF, Bonthron DT. Tissue-specific imprinting of the ZAC/PLAGL1 tumour suppressor gene results from variable utilization of monoallelic and biallelic promoters. Hum Mol Genet. 2007;16:972–981. doi: 10.1093/hmg/ddm041. [DOI] [PubMed] [Google Scholar]
- Williamson CM, et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet. 2006;38:350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- Wood AJ, et al. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]