Abstract
Group B streptococci (GBS) (Streptococcus agalactiae) are a major cause of sepsis and meningitis in neonates and infants and of invasive disease in pregnant women, nonpregnant, presumably immunocompromised adults, and the elderly. Nine GBS serotypes based on capsular polysaccharide antigens have been described. The serotype distributions among invasive and colonizing isolates differ between pediatric and adult populations and have changed over time. Thus, periodic monitoring of GBS serotype distributions is necessary to ensure the proper formulation and application of an appropriate GBS vaccine for human use and to detect the emergence of novel serotypes. Since the mid-1990s, the proportion of GBS isolates that are nontypeable by standard serologic methods has increased, creating a need for more sensitive typing methods. We describe a typing method that uses DNA dot blot hybridization with probes generated by PCR from the GBS capsular genes for serotypes Ia, Ib, and II to VIII. PCR primers were designed to amplify type-specific GBS capsular gene sequences. Gene probes were constructed from the PCR products and used to classify isolates based on hybridization profiles. A total of 306 previously serotyped invasive and colonizing isolates were used to compare our dot blot capsular typing (DBCT) identification method with Lancefield serotyping (LS). A dot blot capsular type was assigned to 99% (303 of 306) of the isolates, whereas 273 of 306 isolates (89%) were assigned a Lancefield serotype. The overall agreement between the methods was 95% (256 of 270 isolates typeable by both methods). We conclude that the DBCT method is a specific and useful alternative to the commonly used LS method.
Group B streptococci (GBS) (Streptococcus agalactiae) cause invasive disease in neonates, young infants, and pregnant women and an increasingly greater proportion of disease in nonpregnant adults (27), especially those with underlying medical conditions, and the elderly (7). Widespread use of intrapartum chemoprophylaxis has resulted in a 70% decline in the number of early-onset neonatal GBS cases (26). A similar decline, however, has not been observed among older neonates, infants, or nonpregnant adults (16).
The polysaccharide capsule, which is the basis for serotyping, is the primary known GBS virulence factor (22, 25). To date, nine distinct serotypes (Ia, Ib, and II to VIII) have been described (11, 12). The distributions of these serotypes, however, vary by geographic location and study population. In the United States, serotypes III and Ia are most commonly associated with GBS disease in neonates and infants (11) and in pregnant women (7). Since its emergence in the early 1990s, serotype V has become the primary cause of GBS infection in nonpregnant adults and also causes significant morbidity in infants and pregnant women (3, 10, 11). In the future, vaccination may protect individuals from GBS disease (2), but to be effective, it must include the more virulent serotypes in circulation in the target population. Therefore, sensitive and specific methods for determining serotypes are required.
A myriad of GBS serotyping methods are available; these include the capillary precipitin (18), latex agglutination (24), coagglutination (8), double immunodiffusion (13), and enzyme immunoassay (1) methods. These tests often have a complicated interpretation and are expensive, and reagents are available for only a subset of serotypes (1). The Lancefield capillary precipitin method has been regarded as the “gold standard” for serotyping GBS isolates.
Since the mid-1990s, the proportion of isolates considered nontypeable by the Lancefield capillary precipitin method has increased (7) and ranges from 2 to 18% (2-4, 11). Isolates may be reported as nontypeable due to the expression of an uncharacterized polysaccharide for which antibodies are not yet available (29), an insertion or mutation in genes essential for capsule expression (28), or the presence of a reversible nonencapsulated phase variant (6). Furthermore, isolates from healthy persons may produce less capsule or type-specific antigen than those from patients with invasive GBS disease (23). Dot blot capsular typing (DBCT) is an attractive alternative to the Lancefield capillary precipitin method because it detects the presence of capsule genes rather than capsule expression, is more specific and reproducible, and is easier to perform than antibody-based serotyping (28).
Kong et al. (15) validated a typing method using the published sequences of the capsular polysaccharide synthesis (cps) gene clusters of GBS serotypes Ia and III (5, 31). Primers were designed to identify serotypes Ia, Ib, III, IV, V, and VI directly by PCR and all nine GBS serotypes by sequence heterogeneity or elimination. Investigators found an overall agreement of 100% between serotype identification by molecular typing and conventional antibody-based serotyping. Other groups have developed PCR-based assays to detect and genotype GBS isolates, but these analyses require further development and refinement (14, 28).
We describe a GBS typing method that uses DNA dot blot hybridization with capsular gene PCR products as probes for GBS strains initially serotyped by the Lancefield serotyping (LS) method. We provide compelling evidence for concordance between these methods.
MATERIALS AND METHODS
GBS strains.
We used the following GBS control strains (serotypes in parentheses): A909 (Ia), DK14 (Ib), DK23 (II), COH1 (III), CNCTC 1/82 (IV), CNCTC 10/84 (V), NT6 (VI), 87-603 (VII), and JM9 (VIII) (C. E. Rubens collection) (9). We tested 291 GBS isolates from four collections obtained from previous epidemiologic studies; these isolates were obtained from 76 symptomatic and asymptomatic pregnant women seen at a University of Michigan Medical Center clinic between August 1999 and March 2000 (19), 191 nonpregnant female and male students enrolled at the University of Michigan (3, 20, 21), and 24 neonates from Houston with early-onset GBS disease between January 1993 and December 1996 (32). Fifteen additional isolates obtained specifically for validating gene probes were used to detect serotype IV (N = 9) (1 isolate previously referenced in the literature) (30), VII (N = 1) (provided by C. J. Baker), or VIII (N = 5) (provided by L. C. Madoff) (17) isolates.
LS.
All 306 isolates had been serotyped previously by the Lancefield capillary precipitin test at the Streptococcal Immunology Laboratory, Baylor College of Medicine, Houston, Tex. (18).
PCR.
PCR amplification of probe sequences was performed by using a slight modification of a previously described protocol (33). DNA lysates were prepared from 1 ml of GBS overnight culture in Todd-Hewitt broth (BBL, Sparks, Md.). Each reaction mixture (25 μl) included 25 pmol of each primer, 0.4 mM deoxynucleoside triphosphate mixture, 4 mM MgCl2, 2 μl of DNA lysate, 2 U of Platinum Taq DNA polymerase, and PCR buffer (Gibco, BRL Life Technologies, Inc., Gaithersburg, Md.). The denaturation, annealing (specific temperatures are listed in Table 1), and elongation temperatures and times used were 94°C for 30 s, 45 to 55°C (depending on the primer melting temperatures) for 30 s, and 72°C for 20 to 50 s (depending on the lengths of the amplicons), respectively, for 35 cycles. A total of 10 μl of PCR product was separated by electrophoresis on a 2% agarose gel, stained with 0.02 μg of ethidium bromide ml−1, and visualized by UV transillumination.
TABLE 1.
Oligonucleotide primers used to construct gene probes for DBCT of GBS isolates
| Primera | Target gene | Size of PCR amplicon (bp) | GenBank accession no.b | Annealing temp (°C) | Forward (top) and reverse (bottom) primer sequences |
|---|---|---|---|---|---|
| cpsIaH | cpsH | 759 | AB028896.2 | 54 | 5′-AACTCCTGATTTTGATAGAA-3′ |
| 5′-AGCAGGCCACTTTTGTAGAA-3′ | |||||
| cpsIaK | cpsK | 195 | AB028896.2 | 51 | 5′-ATTTATTTGTGACACTATCA-3′ |
| 5′-AATAGATAGACTTGTGTATC-3′ | |||||
| cpsIbH | cpsH | 307 | AB050723.1 | 47 | 5′-CGTTATTTAGAAGTCCAG-3′ |
| 5′-GGTATTATAAGATTTGAA-3′ | |||||
| cpsIIK | cpsK | 526 | NA | 55 | 5′-CCAACGGCAATAAAATACAT-3′ |
| 5′-GCATTGAGATTAGAGTAGTC-3′ | |||||
| cpsIIIH | cpsH | 389 | AF163833.1 | 45 | 5′-CTTTGGAAGAGTGAGTTTAG-3′ |
| 5′-TGACCAATTAGTGTAGCATA-3′ | |||||
| cpsIVM | cpsM | 408 | AF355776.1 | 53 | 5′-AGATTTGATTGGTTTTGTTG-3′ |
| 5′-CTATATGAGGAAGTTGTTGT-3′ | |||||
| cpsVO | cpsO | 185 | AF349539.1 | 53 | 5′-TTTTCCACATAATACATCTT-3′ |
| 5′-TAACCTTCTCCTTCACACTA-3′ | |||||
| cpsVIH | cpsH | 586 | AF337958.1 | 46 | 5′-AATGGATTCCTTCTATGA-3′ |
| 5′-ATTATCCTGTTTTGTTTG-3′ | |||||
| cpsVIIM | cpsM | 293 | NA | 54 | 5′-GTGCAATTAGAGGACAAAAA-3′ |
| 5′-CATCGAATCAGGAAAAATAG-3′ | |||||
| cpsVIIIJ | cpsJ | 570 | NA | 54 | 5′-AAACAAAGATAATGGTGGTC-3′ |
| 5′-GAAATAGCTGCCTGATAAAT-3′ |
Designations indicate the serotype from which the probe was amplified and the target gene.
NA, not available (C. E. Rubens, personal communication).
DBCT.
Probe sequences were amplified by PCR and subsequently fluorescein labeled in accordance with the manufacturer's protocol (ECF random prime labeling; Amersham Biosciences, Piscataway, N.J.). We performed dot blot hybridization using the following protocol.
GBS isolates were inoculated into 600 μl of Todd-Hewitt broth, grown overnight at 37°C with 5% CO2, and then centrifuged at 3,000 rpm for 20 min (244 Microplate rotor, HN-SII centrifuge; International Equipment Co., Needham Heights, Mass.) to pellet the bacteria. Cells were lysed with 800 μl of lysis buffer (0.4 M NaOH, 10 mM EDTA) overnight at 68°C. Samples then were centrifuged at 3,000 rpm for 20 min to pellet cellular debris, and 50 μl of DNA in the crude extract was applied to a nylon transfer membrane (Hybond-N+; Amersham Biosciences). DNA hybridization was performed in accordance with the manufacturer's protocol (ECF amplification module; Amersham Biosciences), and visualization was done with a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) as described previously for Escherichia coli (34). The DNA hybridization data were quantified relative to positive and negative controls present on each membrane by using ImageQuant (Molecular Dynamics). Strains with a signal intensity approximately greater than 30% of the mean fluorescence of the positive controls (after subtraction of the mean negative control value for the positive controls and test strain) for duplicate membranes were considered positive.
All isolates were tested in duplicate with separate membranes. Isolates with discordant results were retested. If discordant results were observed upon repeat testing or if there were discrepancies between the dot blot capsular type and the Lancefield serotype, then the isolate was analyzed by PCR. An attempt was made to amplify each potential gene suggested by the LS and DBCT results, and the resulting PCR products were sequenced at the University of Michigan Medical School DNA Core Facility by using an Applied Biosystems (Foster City, Calif.) 3700 automated sequencer. BLAST homology searches, used for confirmation, were performed by using the National Center for Biotechnology Information Internet server. All isolates were assigned a dot blot type based on cps gene homology (Table 2). Each dot blot capsular type, with the exception of type III, was classified by using a unique probe. Probe cpsIaK displayed significant cross-reactivity with the cpsK genes of types Ia and V, whereas probe cpsIIIH cross-reacted with the type VI cpsH gene in addition to the type III cpsH gene.
TABLE 2.
Homology between GBS serotypes and gene probes used for DBCTa
| Gene probe | % Homology of the following serotype:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VI | VII | VIII | |
| cpsIaH | 100 | ||||||||
| cpsIaK | 100 | 100 | 95 | ||||||
| cpsIbH | 69 | 100 | |||||||
| cpsIIK | 100 | ||||||||
| cpsIIIH | 100 | 91 | |||||||
| cpsIVM | 100 | ||||||||
| cpsVO | 100 | ||||||||
| cpsVIH | 65 | 100 | |||||||
| cpsVIIM | 100 | ||||||||
| cpsVIIIJ | 100 | ||||||||
As indicated by a pairwise BLAST alignment (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html).
Validation.
The sensitivity, specificity, and predictive value estimates were calculated for the DBCT method relative to the LS method, which was considered the gold standard. Estimates were obtained for each serotype.
The sensitivity of the DBCT method for each serotype was calculated as the number of concordant positive results divided by the number of isolates found positive by the LS method for a given serotype. Specificity was calculated as the number of concordant negative results divided by the total number of isolates found negative by the LS method for a given serotype. The positive predictive value was calculated as the number of concordant positive results divided by the number of isolates that tested positive by the DBCT method. The negative predictive value was calculated as the number of concordant negative results divided by the number of isolates that tested negative by the DBCT method.
RESULTS
Comparison of DBCT and LS methods.
A total of 306 isolates previously serotyped by LS were typed by DBCT (Table 3 and Fig. 1). A dot blot capsular type was assigned to 99% (303 of 306) of the isolates by DNA dot blot hybridization or PCR and sequencing, whereas a serotype was assigned to only 89% (273 of 306) of the isolates by the LS method (P < 0.001). For the most part, the DBCT results correlated well with the LS results.
TABLE 3.
Concordance matrix comparing DBCT and LS results for 306 GBS isolates from various sources
| LS | No. of isolates with the following dot blot type:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VI | VII | VIII | Nontypeable | Total | |
| Ia | 60 | 1a | 1 | 62 | |||||||
| Ib | 1a | 32 | 1a | 1 | 35 | ||||||
| II | 39 | 1b | 1a | 41 | |||||||
| III | 2a | 31 | 4c | 2c | 39 | ||||||
| IV | 10 | 1a | 11 | ||||||||
| V | 70 | 1 | 71 | ||||||||
| VI | 6 | 6 | |||||||||
| VII | 2 | 2 | |||||||||
| VIII | 6 | 6 | |||||||||
| Nontypeable | 7 | 4 | 3 | 2 | 16 | 1 | 33 | ||||
| Total | 70 | 36 | 42 | 35 | 10 | 93 | 8 | 2 | 7 | 3 | 306 |
Discordant result has been confirmed by PCR and sequencing to match the DBCT result.
Confirmed to be serotype V by PCR and sequencing.
Discordant result has been confirmed by PCR and sequencing to match the LS result.
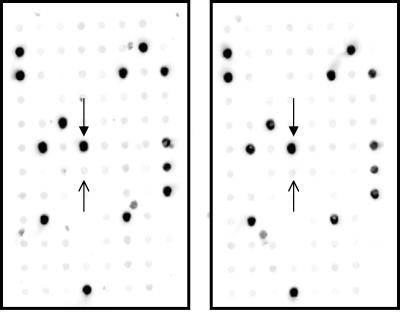
FIG. 1.
Representative DNA DBCT analysis of 96 GBS isolates with probe cpsIaH to identify serotype Ia isolates. Samples were analyzed in duplicate. Closed arrows (pointing down) indicate the positive control; open arrows (pointing up) indicate the negative control.
Among isolates typeable by both methods, DBCT and LS results were in agreement for 256 of 270 isolates (95%). All 14 isolates with discordant results were further tested by PCR and sequencing; retest results for 7 matched the DBCT result, retest results for 6 matched the LS result, and retest results for 1 matched neither (Table 3). We attempted to amplify each possible gene suggested from these isolates. Of the isolates whose retest results matched the DBCT result, three were found to be type Ia by DBCT but Ib (N = 1) and III (N = 2) by LS, one was found to be type III by DBCT but Ib by LS, and three were found to be type V by DBCT but Ia, II, and IV by LS. Of the isolates whose retest results matched the LS result, four were found to be type III by LS but V by DBCT, and two were found to be type III by LS but VI by DBCT. One isolate was found to be type III by DBCT and II by LS but was determined to be type V by PCR. Thirty-three isolates were nontypeable by LS but were assigned a dot blot capsular type, whereas 3 isolates were nontypeable by DBCT but were assigned a serotype by LS. The retest results for the three isolates that were nontypeable by DBCT were confirmed to match the LS result by PCR.
Validation.
Typing sensitivity results ranged from 79% (type III) to 100% (types VI, VII, and VIII), and specificity results ranged from 90% (type V) to 100% (types IV, VII, and VIII). The positive predictive value of the DBCT method ranged from 75% (types V and VI) to 100% (types IV and VII). The negative predictive value ranged from 97% (type III) to 100% (types IV to VIII) (Table 4). Positive and negative predictive values are sensitive to the prevalence of the serotype; therefore, these estimates are not generalizable to populations with different serotype distributions.
TABLE 4.
Sensitivity, specificity, and predictive values of LS (standard) and DBCT for 306 GBS isolates from various sources
| Serotype | %
|
|||
|---|---|---|---|---|
| Sensitivity | Specificity | Predictive value
|
||
| Positive | Negative | |||
| Ia | 97 | 96 | 86 | 99 |
| Ib | 91 | 99 | 89 | 99 |
| II | 95 | 99 | 93 | 99 |
| III | 79 | 99 | 89 | 97 |
| IV | 91 | 100 | 100 | 100 |
| V | 99 | 90 | 75 | 100 |
| VI | 100 | 99 | 75 | 100 |
| VII | 100 | 100 | 100 | 100 |
| VIII | 100 | 100 | 86 | 100 |
These data suggest that if DBCT were used as the gold standard, the sensitivity and specificity results would be equivalent to the positive and negative predictive values calculated previously with LS as the standard, respectively. Similarly, the new positive and negative predictive values would be equivalent to the sensitivity and specificity results calculated previously with LS as the standard, respectively. Because DBCT had fewer nontypeable results, LS is a less sensitive method overall.
DISCUSSION
The capillary precipitin serotyping method, developed by Rebecca Lancefield, has been applied broadly to the serotyping of GBS isolates since its introduction in the 1930s (18). However, the proportion of isolates found nontypeable by this method was significant and has increased over time (29), potentially skewing the serotype distribution and having an adverse impact on vaccine formulation. DBCT is an attractive alternative to LS. Kong et al. reported 100% typeability for their molecular typing method based on PCR and sequencing (15). Similarly, 99% of the isolates in our test population could be typed by DBCT, compared to 89% that could be typed by LS. Among isolates that were typeable by both methods, the agreement on serotype was relatively high (95%).
Aside from typeability, DBCT possesses other attributes that make it preferable to LS. DBCT is straightforward and does not require preparation of antisera. DNA dot blot hybridization is amenable to high-throughput GBS typing and is adaptable to phenotypic analysis of libraries on a glass slide by microarray technology. Because DBCT detects the presence of capsule genes rather than capsule expression, isolates with capsule production below the threshold of detection by LS or that lack capsule expression may still be typed by DBCT. Additionally, dot blot hybridization is less susceptible to the contamination inherent with PCR-based methods. The DBCT method has a cost and time advantage over the method of Kong et al. (15) because it does not require any DNA sequencing.
The sensitivity, specificity, and predictive values for the DBCT method were reasonably high, although variability across serotypes was apparent. Variability can be attributed to isolates that were nontypeable by LS but typeable by DBCT or to discordant serotype results. Thirty-three isolates were nontypeable by LS but were assigned a dot blot capsular type, and 14 results were discordant between the methods. Discordant results, as determined by PCR and sequencing, matched the LS result in six instances, the DBCT result in seven instances, or neither in one instance. These findings suggests that the majority of variability can be attributed to increased typeability by the DBCT method and discordant results that match the DBCT result. However, because this method identifies the serotype indirectly by characterizing the cps genotype, isolates that have defective cps operons and thus are acapsular or potentially produce an aberrant capsule would be misclassified. Further research is needed to distinguish among these possibilities.
Six discordant results were found to agree with the LS result; each was serotype III. For each of these results, the gene probe homology was incomplete, meaning that one of the two probes did not hybridize, and the results were extrapolated based on a result obtained with a single probe. Four results were interpreted as type V but were confirmed to be type III by PCR, based on hybridization with probe cpsIaK but not with probe cpsVO. These isolates did not, however, hybridize to probe cpsIIIH, suggesting that they lack the type III cps polymerase and therefore that the production of type III cps would not be expected. Similarly, two results were interpreted as type VI but were confirmed to be type III by PCR, based on hybridization with probe cpsIIIH but not with probe cpsIaK or cpsVIH (Table 2). Discordant LS and DBCT results could be explained by the presence of a defective gene that is detectable by the gene probe due to the addition of a novel gene that is different from but encodes the same activity as the gene expected from LS. Another possibility is that isolates that were nontypeable by DBCT may have a novel serotype yet to be identified. To avoid these discrepancies in the future, results with incomplete hybridization should be analyzed by PCR and sequencing.
In conclusion, we developed a GBS DBCT method as an alternative to capillary precipitin serotyping. DBCT is sensitive and specific and reduces misclassification of nontypeable isolates. It is suitable for analyzing various populations and can be used to monitor isolates causing disease as well as those colonizing otherwise healthy individuals. Application of DBCT in future epidemiologic studies and surveillance activities will facilitate the appropriate formulation of candidate GBS vaccines.
Acknowledgments
We thank Melissa Hickman and Kristen Taylor for LS of isolates and L. C. Madoff for providing strains.
This work was supported by grants R21AI44868 (B. Foxman), T32AI49816 (B. Foxman), and AI22498 (C. E. Rubens). Lancefield serotyping was paid for in part by grant 334-SAP/99 (S. D. Manning) from the Blue Cross Blue Shield of Michigan Foundation and the University of Michigan Medical School's Advisory Council on Clinical Research (M. D. Pearlman). Collection and serotyping of isolates obtained from C. J. Baker were supported in part by NIH-NIAID contract AI-75325.
REFERENCES
- 1.Arakere, G., A. E. Flores, P. Ferrieri, and C. E. Frasch. 1999. Inhibition enzyme-linked immunosorbent assay for serotyping of group B streptococcal isolates. J. Clin. Microbiol. 37:2564-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, S., B. Trollfors, T. Lagergard, G. Zackrisson, and B. A. Claesson. 2000. Serotypes and clinical manifestations of group B streptococcal infections in western Sweden. Clin. Microbiol. Infect. 6:9-13. [DOI] [PubMed] [Google Scholar]
- 3.Bliss, S. J., S. D. Manning, P. Tallman, C. J. Baker, M. D. Pearlman, C. F. Marrs, and B. Foxman. 2002. Group B streptococcus colonization in male and nonpregnant female university students: a cross-sectional prevalence study. Clin. Infect. Dis. 34:184-190. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173:365-373. [DOI] [PubMed] [Google Scholar]
- 5.Chaffin, D. O., S. B. Beres, H. H. Yim, and C. E. Rubens. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 7.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 8.Hakansson, S., L. G. Burman, J. Henrichsen, and S. E. Holm. 1992. Novel coagglutination method for serotyping group B streptococci. J. Clin. Microbiol. 30:3268-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, T. O., D. W. Shelver, J. F. Bohnsack, and C. E. Rubens. 2003. A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J. Clin. Investig. 111:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison, L. H., D. M. Dwyer, and J. A. Johnson. 1995. Emergence of serotype V group B streptococcal infection among infants and adults. J. Infect. Dis. 171:513. [DOI] [PubMed] [Google Scholar]
- 11.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, and A. Schuchat. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infections Program. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 12.Hickman, M. E., M. A. Rench, P. Ferrieri, and C. J. Baker. 1999. Changing epidemiology of group B streptococcal colonization. Pediatrics 104:203-209. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, D. R., and P. Ferrieri. 1984. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J. Clin. Microbiol. 19:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke, D., C. Menard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 15.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachenauer, C. S., C. J. Baker, M. J. Baron, D. L. Kasper, C. Gravekamp, and L. C. Madoff. 2002. Quantitative determination of immunoglobulin G specific for group B streptococcal beta C protein in human maternal serum. J. Infect. Dis. 185:368-374. [DOI] [PubMed] [Google Scholar]
- 17.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 18.Lancefield, R. C. 1934. Serological differentiation of specific types of bovine haemolytic streptococci (group B). J. Exp. Med. 59:441-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning, S. D., B. Foxman, C. L. Pierson, P. Tallman, C. J. Baker, and M. D. Pearlman. 2003. Correlates of antibiotic-resistant group B streptococcus isolated from pregnant women. Obstet. Gynecol. 101:74-79. [DOI] [PubMed] [Google Scholar]
- 20.Manning, S. D., M. D. Pearlman, P. Tallman, C. L. Pierson, and B. Foxman. 2001. Frequency of antibiotic resistance among group B streptococcus isolated from healthy college students. Clin. Infect. Dis. 33:e137-e139. [DOI] [PubMed] [Google Scholar]
- 21.Manning, S. D., P. Tallman, C. J. Baker, B. Gillespie, C. F. Marrs, and B. Foxman. 2002. Determinants of co-colonization with group B streptococcus among heterosexual college couples. Epidemiology 13:533-539. [DOI] [PubMed] [Google Scholar]
- 22.Nizet, V., P. Ferrieri, and C. E. Rubens. 2000. Molecular pathogenesis of group B streptococcal disease in newborns, p. 180. In D. Stevens and E. Kaplan (ed.), Streptococcal infections: clinical aspects, microbiology and molecular pathogenesis. Oxford University Press, New York, N.Y.
- 23.Palacios, G. C., E. K. Eskew, F. Solorzano, and S. J. Mattingly. 1997. Decreased capacity for type-specific-antigen synthesis accounts for high prevalence of nontypeable strains of group B streptococci in Mexico. J. Clin. Microbiol. 35:2923-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, C. J., N. M. Vandel, D. K. Ruprai, E. A. Martin, K. M. Gates, and D. Coker. 2001. Detection of group B streptococcal colonization in pregnant women using direct latex agglutination testing of selective broth. J. Clin. Microbiol. 39:408-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubens, C. E., M. R. Wessels, J. M. Kuypers, D. L. Kasper, and J. N. Weiser. 1990. Molecular analysis of two group B streptococcal virulence factors. Semin. Perinatol. 14:22-29. [PubMed] [Google Scholar]
- 26.Schrag, S. J., E. R. Zell, R. Lynfield, A. Roome, K. E. Arnold, A. S. Craig, L. H. Harrison, A. Reingold, K. Stefonek, G. Smith, M. Gamble, A. Schuchat, and T. Active Bacterial Core Surveillance. 2002. A population-based comparison of strategies to prevent early-onset group B streptococcal disease in neonates. N. Engl. J. Med. 347:233-239. [DOI] [PubMed] [Google Scholar]
- 27.Schuchat, A. 1999. Group B streptococcus. Lancet 353:51-56. [DOI] [PubMed] [Google Scholar]
- 28.Sellin, M., C. Olofsson, S. Hakansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slotved, H. C., S. Sauer, and H. B. Konradsen. 2002. False-negative results in typing of group B streptococci by the standard Lancefield antigen extraction method. J. Clin. Microbiol. 40:1882-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suara, R. O., R. A. Adegbola, C. J. Baker, O. Secka, E. K. Mulholland, and B. M. Greenwood. 1994. Carriage of group B streptococci in pregnant Gambian mothers and their infants. J. Infect. Dis. 170:1316-1319. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto, S., K. Miyake, Y. Koike, M. Watanabe, Y. Machida, M. Ohta, and S. Iijima. 1999. Molecular characterization of type-specific capsular polysaccharide biosynthesis genes of Streptococcus agalactiae type Ia. J. Bacteriol. 181:5176-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaleznik, D. F., M. A. Rench, S. Hillier, M. A. Krohn, R. Platt, M. L. Lee, A. E. Flores, P. Ferrieri, and C. J. Baker. 2000. Invasive disease due to group B streptococcus in pregnant women and neonates from diverse population groups. Clin. Infect. Dis. 30:276-281. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, L., B. Foxman, and C. Marrs. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40:3951-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, L., B. W. Gillespie, C. F. Marrs, and B. Foxman. 2001. Optimization of a fluorescent-based phosphorimaging dot blot DNA hybridization assay to assess E. coli virulence gene profiles. J. Microbiol. Methods 44:225-233. [DOI] [PubMed] [Google Scholar]