Abstract
Rotavirus strains detected as part of ongoing strain surveillance in Cameroon, and whose first-round reverse transcription-PCR product could not be genotyped by using conventional genotyping primers, were subjected to sequence analysis for strain characterization. We detected for the first time in Africa a human rotavirus with G5 specificity. The Cameroonian G5 strain had a short electrophoretic pattern and was of VP6 subgroup I specificity and a VP4 P[8] type. The VP7 gene shared a higher nucleic acid and amino acid homology with the porcine G5 strain CC117 (90 and 96%, respectively) than with human G5 strain IAL-28 (86 and 92%, respectively). Phylogenetic analysis showed Cameroonian strain MRC3105 clustered together in the same lineage as two other reported porcine G5 strains. The Cameroonian G5 strain, the first to be reported in humans outside of Latin America, may be a natural reassortant between animal and human rotavirus strains.
Rotaviruses are associated with approximately 500,000 to 600,000 deaths of infants and young children every year (18). Most of these deaths occur in sub-Saharan Africa and Asia (16, 18). Because of this high burden of disease and mortality, rotavirus vaccines remain a priority for the World Health Organization and other public-sector organizations, such as the Global Alliance for Vaccines and Immunization.
Molecular epidemiological studies of rotavirus use various viral markers for strain characterization (8). These consist of the electrophoretic migration pattern of the 11 segments of double-stranded RNA (dsRNA) of the rotavirus genome when separated by polyacrylamide gel electrophoresis (PAGE), the VP6 subgroup antigen specificity and the “genotypes” of the important outer capsid neutralization antigens (9, 10, 12). The VP4 and VP7 proteins are both involved in virus neutralization and protective immunity as they elicit the production of neutralizing antibodies in the host. A dual-nomenclature system exists for rotavirus strains based on the protease-sensitive VP4 (P-types) and the VP7 glycoprotein (G-types) (8).
Ten G types and 10 P types have been detected in human rotaviruses, although 4 G and 2 P types are most commonly found (10, 15). Rotavirus strains with G1 to G4 VP7 serotypes have been the target of the reassortant vaccines because of their common occurrence globally. However, recent reports from several studies have shown the increased incidence of rotavirus strains with previously unusual G and/or P genotypes (1, 5, 10, 11, 19, 21, 22). So, G9 strains have been seen to emerge globally over the past few years (22). VP7 serotype G5 strains, originally detected only in pigs, have been identified commonly in Brazilian children with diarrheal disease (11). They have also been described in children with acute diarrhea in Argentina (3) and Paraguay (4). Here we report the detection of a human rotavirus with G5 specificity in Cameroon. To our knowledge, this is the first report of human G5 rotavirus strains outside of Latin America and highlights the potential for strain diversity in different regions of the world.
From January to October 2000, a total of 890 fecal specimens were collected from young infants and children between 1 month and 5 years of age who presented with acute diarrheal illness at two hospitals in the South West and Western provinces of Cameroon (7). Ten percent stool suspensions in phosphate-buffered saline were initially screened by enzyme immunoassay as previously described for the detection of rotavirus antigens (7). The tests were read both visually and spectroscopically at a wavelength of 450 nm. Each plate included a negative and a positive control, and all tests were performed in duplicate.
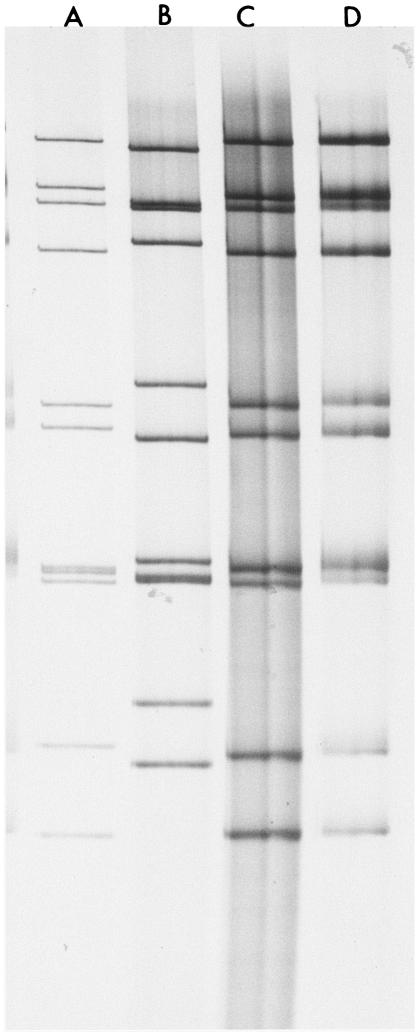
All of the rotavirus positive specimens were analyzed for VP6 subgroup specificity by monoclonal antibodies (kind donation from H. B. Greenberg) as previously described (14, 20). Furthermore, PAGE was used to determine the RNA electropherotypes of the strains and to select samples with adequate intact dsRNA for genotyping. Briefly, the dsRNA genome was extracted from all stool suspensions by the phenol-chloroform method and electrophoresed overnight at 100 V for 16 to 18 h in a 10% polyacrylamide vertical slab gel (7). Figure 1 illustrates the PAGE profiles of some rotavirus strains identified in Cameroon and a G5 South African porcine isolate.
FIG. 1.
RNA electrophoretic patterns of the Cameroonian G5 strain and some of the strains isolated in this study. Lanes A and C are human G9 genotypes with long RNA profiles. Lane B represents the electrophoretic profile of Cameroonian human G5 strain MRC3105, and lane D represents a porcine G5 strain with a long RNA profile from South Africa.
All PAGE-positive stool samples were subjected to molecular genotyping of the VP4 and VP7 genes by reverse transcription-PCR. In brief, the viral dsRNA was extracted with phenol-chloroform and purified with the RNAid kit (Bio 101). This purified dsRNA was specifically primed with VP7 and VP4 consensus primer pairs (9, 12) and subjected to reverse transcription-PCR as described in detail elsewhere (1, 9, 12, 21).
Rotavirus antigens were detected in 195 (21.9%) of the 890 stool samples. The epidemiological results are presented elsewhere (7). Twelve of the PAGE-positive samples could not be VP7 genotyped, although the full-length VP7 gene could be transcribed in the first-round PCR. Ten of the 12 VP7 untypeable strains (two samples did not have enough stool for further assays) were cloned and sequenced for determination of their VP7 specificity.
These full-length VP7 gene products were reamplified with the same consensus primer, recovered from the 2% agarose gel, and purified with the QIAquick gel extraction kit (QIAGEN, Chatsworth, Calif.). The purified gene product was cloned into the pGEM-T Easy Vector cloning system (Promega, Southampton, United Kingdom) as recommended by the manufacturer. The plasmids were recovered from the desired selected clones and digested with restriction enzymes to ascertain the size of the insert. Two independent clones for each sample with the desired inserts were submitted to the DNA sequencing laboratory at the Department of Microbiology, University of Cape Town, for sequencing on an ALFexpress automated sequencer with M13 forward and reverse primers.
Sequence analysis (National Center for Biotechnology Information BLASTIN program [http://www.ncbi.nlm.gov/BLAST/]) showed four rotavirus G9 strains, four rotavirus G1 strains, a single G4 strain, and the unusual G5 strain.
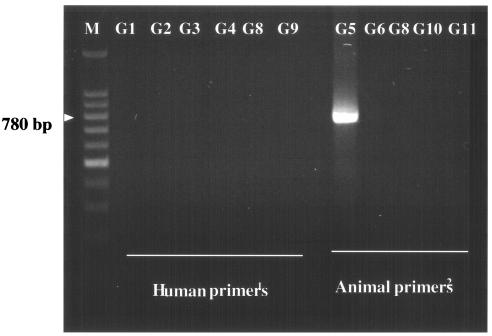
The G5 rotavirus strain (designated MRC3105) was detected in the stool of a 12-month-old girl who had been brought in for medical attention for diarrhea and vomiting at the Presbyterian Hospital in Kumba in the South West province of Cameroon. This G5 rotavirus strain was further characterized as having a short RNA electropherotype, a subgroup I VP6, and a P[8] VP4 specificity. To confirm the G5 VP7 specificity, we subjected this single strain to further typing by PCR with a pool of animal rotavirus primers (13). In this confirmation assay, the first-round VP7 RT product was subjected to a seminested PCR typing assay with the sense pool of animal typing primers consisting of FT5 (for G5), DT6 (for G6), HT8 (G8), ET10 (G10), and BT11 (G11) (11) and the conserved antisense primer sBeg9. Agarose gel analysis of the amplified gene product revealed a PCR product of 780 bp, the expected size for G5 rotaviruses (Fig. 2).
FIG. 2.
Agarose gel electrophoresis of the amplified gene product of the Cameroonian MRC3105 rotavirus specimen with a pool of human (12) and animal (13) primers. Lane M is a 100-bp marker. Highlighted is the expected 780-bp G5 PCR product.
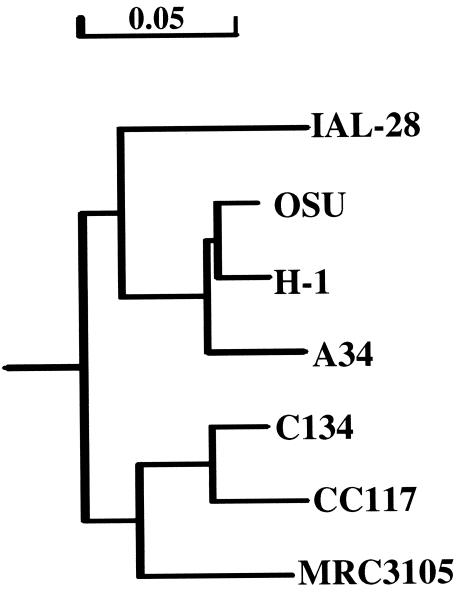
A comparison of the sequences of strain MRC3105 and other G5 strains recorded in the literature showed a relatively high similarity (90%) to the nucleotide sequence of porcine strain CC117 (accession number L35056) and slightly less to that of human strain IAL-28 from Brazil (86%) (accession number L79916). Similarly, the amino acid homology with porcine G5 strain CC117 (96%) was higher than that to human G5 strain IAL-28 (92%) (Table 1). A comparison of the amino acid compositions of the major VP7 neutralization domains (6) also showed a high amino acid homology between MRC3105 and porcine G5 strains C134 (accession number L35058) and CC117 at antigenic site B, two amino acid substitutions in antigenic region A (amino acids 96 and 92), and a diverse number of amino acid changes in region C (Table 2). From these data, a phylogenetic tree was developed linking MRC3105 to representative published animal G5 VP7 sequences, as well as IAL-28, a human G5 strain. The phylogenetic analysis showed three distinct lineages. This dendrogram indicates a common progenitor strain linking all of these G5 strains. The Cameroonian G5 strain (MRC3105) forms a cluster with closely related porcine strains CC117 and C134, which were isolated in Latin America (Fig. 3). Another cluster is formed by three other animal G5 strains, H-1 (equine), OSU (porcine, isolated in the United States), and A34 (porcine, isolated in Venezuela and Argentina). Having a progenitor in common with the latter animal strain cluster but clustering separately is Brazilian human G5 strain IAL-28 (11).
TABLE 1.
Percentages of deduced amino acid (bottom left) and nucleotide (upper right) homology of Cameroonian G5 and selected human and animal rotavirus G5 strains from other countriesa
| Strain | % Homology with strain:
|
||||||
|---|---|---|---|---|---|---|---|
| MRC3105 | IAL28 | A34 | OSU | H-1 | C134 | CC117 | |
| MRC3105 | 86 | 86 | 87 | 87 | 90 | 90 | |
| IAL28 | 92 | 87 | 89 | 90 | 86 | 85 | |
| A34 | 93 | 92 | 95 | 94 | 88 | 86 | |
| OSU | 94 | 92 | 97 | 97 | 89 | 86 | |
| H-1 | 94 | 94 | 97 | 99 | 88 | 86 | |
| C134 | 96 | 92 | 92 | 93 | 93 | 95 | |
| CC117 | 96 | 92 | 92 | 92 | 93 | 98 | |
TABLE 2.
Comparison of amino acid sequence of the VP7 antigenic sites of MRC3105 and selected G5 strainsa
| Strain | Sequenceb at antigenic site:
|
||
|---|---|---|---|
| A | B | C | |
| IAL 28 | NEAATEIADDKWTDT | MKYDINLQLDM | LTTDTNSFETVAST |
| H-1 | ---------t---e- | ----g------ | s---i--------- |
| C134 | -----q---n---e- | ----a------ | -------------a |
| CC117 | ---------n---e- | ----a------ | -------------a |
| OSU | ---------t---e- | ----g------ | s---i--------na |
| A34 | ---------t---e- | ----g------ | s---i--------na |
| MRC3105 | ---------s---e- | ----a------ | s----s-------xa |
The amino acid sequences were from IAL 28, H-1, C134, CC117, OSU, and A34. Dash indicates identical amino acids.
The first and last (left to right) residues are numbered as follows: site A, 87 and 101; site B, 143 and 152; site C, 208 and 221.
FIG. 3.
Phylogenetic relatedness of Cameroonian rotavirus G5 strain to other selected human and animal G5 strains. GenBank accession numbers of selected G5 strains: CC117, L35056; IAL-28, L79916; A34, L35059; H-1, AF242393; C134, L35058; OSU, X04613; MRC3105, AY327107.
Immunity to primary natural rotavirus infection is believed to be predominantly homotypic, broadening after subsequent infections (2, 15) and eventually leading to protective immunity to all antigenic types. The reassortant rotavirus vaccine formulations have been targeted at the most commonly circulating human rotavirus strains (G1 to G4) with the ultimate aim of providing protection and minimizing severe rotavirus disease. However, recent epidemiological studies in developing countries have shown increasing diversity of human rotaviruses. Rotavirus G5, G6, G8, and G10 strains are more commonly found in animals, particularly cattle and pigs (11). However, over the past decade, reports have shown increased detection of uncommon strains and high regional diversity among circulating rotaviruses. In Africa, G8 and G9 strains are common (1, 5, 21); in Asia, G9 strains have also been reported commonly (22); and G5 rotavirus strains have been commonly associated with diarrhea in Brazilian children (11). Because most of the Brazilian G5 strains have been reported to possess human rotavirus VP4 P types, the suggestion is that these unusual strains could be the result of reassortment between porcine and human rotaviruses. Cross-species transmission of rotaviruses has been documented (17), indicating the importance of this potential mechanism of rotavirus genetic diversity.
The Cameroonian rotavirus strain was isolated from a child with community-acquired rotavirus infection in rural Cameroon. The Cameroonian G5 strain exhibited a short electropherotype and VP6 subgroup I specificity. This is an unusual combination of these epidemiological markers for animal rotaviruses, which tend to have long RNA electropherotypes. In addition, the VP4 genotype, P[8], possessed by strain MRC3105 is the most prevalent human rotavirus P type worldwide and is usually associated with VP7 type G1, G3, G4, or G9 (10, 15). These factors, together with the relatively high nucleotide and amino acid sequence similarity of the VP7 gene of strain MRC3105 to that of porcine G5 strains and the marked similarity at the neutralization antigenic sites, encourages speculation that Cameroonian G5 strain MRC3105 is also likely to be the result of reassortment between a porcine G5 strain and a human strain. In the South West province of Cameroon, as in many rural African villages, people and animals, especially domestic farm animals, live in close proximity and usually use the same water source, providing favorable conditions for dual infection and potential reassortment. Studies on circulating animal strains in Cameroon will throw more light on this speculation.
The finding of G5 rotavirus in Africa adds to the global distribution of this strain and strengthens the need to continue strain surveillance in developing countries to further understand the extent of strain distribution and diversity. While surveillance has mostly been focused on infection in humans, there is a need to include domestic animals in these surveillance programs because of (i) their close contact with humans, especially in developing countries; (ii) increased reports of the detection of strains common to animals in human populations; and (iii) the increasing spread and detection of strains with unusual G and P types and untypeable strains. The availability and affordability of sequencing methods now provide an alternative tool for the characterization of these unusual strains to provide more information on circulating types and the natural reservoirs for these strains.
Nucleotide sequence accession number.
The nucleotide sequences of the G5 strains have been submitted to the GenBank database and assigned accession number AY327107.
Acknowledgments
Research grants from the Department of Vaccines and Biologicals, World Health Organization, and the Medical Research Council, South Africa, supported this study. We extend our sincere gratitude to the Deutscher Akademischer Austausch Dienst (German Academic Exchange Service) and the Claude Harris Foundation, Cape Town, South Africa, for providing research fellowships to M.D.E. and G.E.A., respectively.
We acknowledge our colleagues at the Presbyterian Hospital in Kumba and the Cabinet des Soins Clinic in Bafoussam, Cameroon, for support and collaboration in the collection of stool specimens and the staff of the MRC/MEDUNSA Diarrheal Pathogens Research Unit for technical assistance in the typing and sequencing analysis.
REFERENCES
- 1.Armah, G. E., C. T. Pager, R. H. Asmah, F. R. Anto, A. R. Oduro, F. Binka, and A. D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 2.Bishop, R. F. 1996. Natural history of human rotavirus infection. Arch. Virol. Suppl. 12:119-128. [DOI] [PubMed] [Google Scholar]
- 3.Bok, K., N. Castagnaro, A. Borsa, S. Nates, C. Espul, O. Fay, A. Fabri, S. Grinstein, I. Miceli, D. O. Matson, and J. A. Gomez. 2001. Surveillance for rotavirus in Argentina. J. Med. Virol. 65:190-198. [PubMed] [Google Scholar]
- 4.Coluchi, N., V. Munford, J. Manzur, C. Vazquez, M. Escobar, E. Weber, P. Marmol, and M. L. Racz. 2001. Detection, subgroup specificity, and genotype diversity of rotavirus strains in children with acute diarrhea in Paraguay. J. Clin. Microbiol. 40:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyall-Smith, M. L., I. Lazdins, G. W. Tregear, and I. H. Holmes. 1986. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc. Natl. Acad. Sci. USA 83:3465-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esona, M. D., G. E. Armah, and A. D. Steele. 2003. Molecular epidemiology of rotavirus infection in western Cameroon. J. Trop. Pediatr. 48:14-17. [DOI] [PubMed] [Google Scholar]
- 8.Estes, M. 1996. Rotavirus and their replication, p. 1625-1655. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 9.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 11.Gouvea, V., L. de Castro, M. Timenetsky, H. Greenberg, and N. Santos. 1994. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J. Clin. Microbiol. 32:1408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gouvea, V., M. C. Timenetsky, and N. Santos. 1994. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 32:1438-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg, H., V. McAuliffe, J. Valdesuso, R. Wyatt, J. Flores, A. Kalica, Y. Hoshino, and N. Singh. 1983. Serological analysis of the subgroup protein of rotavirus, using monoclonal antibodies. Infect. Immun. 39:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshino, Y., and A. Z. Kapikian. 1996. Classification of rotavirus VP4 and VP7 serotypes. Arch. Virol. Suppl. 12:99-111. [DOI] [PubMed] [Google Scholar]
- 16.Molbak, K., T. K. Fischer, and C. S. Mikkelsen. 2000. The estimation of mortality due to rotavirus infections in sub-Saharan Africa. Vaccine 19:393-395. [DOI] [PubMed] [Google Scholar]
- 17.Nakagomi, O., and T. Nakagomi. 1993. Interspecies transmission of rotaviruses studied from the perspective of genogroup. Microbiol. Immunol. 37:337-348. [DOI] [PubMed] [Google Scholar]
- 18.Parashar, U. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos, N., R. C. Lima, C. F. Pereira, and V. Gouvea. 1998. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J. Clin. Microbiol. 36:2727-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steele, A. D., and J. J. Alexander. 1987. Molecular epidemiology of rotavirus infection on black infants in South Africa. J. Clin. Microbiol. 25:2384-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steele, A. D., M. C. van Niekerk, and M. J. Mphahlele. 1995. Geographic distribution of human rotavirus VP4 genotypes and VP7 serotypes in five South African regions. J. Clin. Microbiol. 33:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. G. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]