Abstract
Owing to the potential role of the tick Amblyomma cooperi in the enzootic cycle of Rickettsia rickettsii, the etiologic agent of Brazilian spotted fever (BSF), this study evaluated infection by Rickettsia species in A. cooperi ticks collected from an area in Brazil where BSF is endemic. Among a total of 40 A. cooperi adult ticks collected in an area of BSF endemicity in the state of São Paulo, PCR analysis detected DNA of Rickettsia bellii in 16 ticks (40%), and 3 other ticks (7.5%) were positive for a previously unidentified spotted-fever-group (SFG) rickettsia. Cultivation in Vero cell cultures by the shell vial technique with individual A. cooperi ticks resulted in two isolates of R. bellii and one isolate genotypically characterized as an SFG rickettsia. The two R. bellii isolates were established in Vero cell cultures in the laboratory and were confirmed to be R. bellii by molecular analysis of the gltA and 17-kDa protein-encoding genes and by electron microscopic analysis. The SFG rickettsial isolate could not be stably passaged in cell culture in the laboratory, but molecular analysis of early passages suggested that it was closely related to Rickettsia parkeri, Rickettsia africae, and Rickettsia sibirica. These results do not support the role of A. cooperi in the ecology of R. rickettsii in the area studied, but they add two more species of rickettsiae to the poorly developed list of species occurring in ticks in South America.
The genus Rickettsia comprises obligately intracellular bacteria, of which many cause zoonotic diseases in all continents inhabited by humans in the world. Due to their strictly intracellular survival inside host cells, rickettsiae are classically transmitted to humans by arthropod vectors, which include ticks, mites, fleas, or lice. Even though the study of rickettsiae has been conducted in an anthropocentric way, many Rickettsia species of no or still unknown pathogenicity to humans have been described. These findings have generated several speculations on the ecology of pathogenic rickettsiae, since it has been proven that the presence of a nonpathogenic rickettsia within a tick population can minimize the transmission of a pathogenic rickettsia (3, 17).
In Brazil, lethal human cases of Brazilian spotted fever (BSF), caused by Rickettsia rickettsii, have been reported since the first half of the 20th century (5, 6, 18, 19). In this country, R. rickettsii is transmitted to humans primarily by the tick species Amblyomma cajennense and Amblyomma aureolatum (6, 9). A. cajennense has been incriminated as the main vector of BSF to humans in Brazil. Horses, tapirs (Tapirus terrestris), and capybaras (Hydrochaeris hydrochaeris) are its primary hosts (14).
Observations on the ecology of BSF in the 1990s, in one area of endemicity in the state of São Paulo, suggested the potential role of capybaras as natural reservoirs of R. rickettsii. Even though this statement has never been proven, the following findings are in its favor. (i) Earlier studies in the 1940s showed that when experimentally infected with R. rickettsii, capybaras retain circulating rickettsiae for several days, with no clinical signs, but with a level of rickettsemia sufficient to infect feeding A. cajennense ticks (30, 31). (ii) The reemergence of BSF cases in many areas of the state of São Paulo since the early 1990s was coincidental with an explosive increase in the populations of free-living capybaras in these areas. (iii) Capybaras have been suggested to act as primary hosts for the tick A. cajennense. (iv) A spotted-fever-group (SFG) rickettsia (species identification was not performed) was isolated from the capybara tick Amblyomma cooperi, which was collected from an R. rickettsii-seropositive capybara from an area in the state of São Paulo where BSF is endemic (16).
All parasitic stages of A. cooperi feed primarily on capybaras, although larvae and nymphs can be found in several other host species (M. B. Labruna, A. Pinter, and R. H. F. Teixeira, submitted for publication), but there is no indication that A. cooperi is an aggressive tick to humans. Large populations of A. cooperi have been found in areas of the state of São Paulo where BSF is endemic. Due to the potential role of A. cooperi as an enzootic vector of R. rickettsii, this study evaluated the infection with Rickettsia species of A. cooperi ticks collected from an area of BSF endemicity in the state of São Paulo, Brazil.
MATERIALS AND METHODS
Study site and collection of ticks.
A total of 40 adult A. cooperi ticks (22 males and 18 females) were collected in three areas of Pedreira Municipality, State of São Paulo, where recent cases of BSF have been reported in humans. Two of these areas, farm 1 (22°44′19′′S, 46°55′27′′W) and farm 2 (22°47′03′′S, 46°54′10′′W), were both located on the banks of the Jaguari River, and the third area, farm 3 (22°41′14′′S, 46°53′17′′W), was located on the banks of the Camanducaia River. A total of 5, 15, and 20 ticks were collected from farms 1, 2, and 3, respectively. These three areas had horses grazing on mixed-overgrowth pastures, interspersed with remote forest areas, which were inhabited by large populations of free-living capybaras. Free-living A. cooperi ticks were collected from vegetation by dragging in January 2001. Collected ticks were brought alive to the laboratory, where they were immediately incubated at 35°C for 48 h before being subjected to the hemolymph test.
Hemolymph test.
Ticks were individually processed by the hemolymph test as described previously (2). Briefly, a drop of hemolymph from each tick was dried on a glass slide and stained by the Gimenez method (8). Ticks were then frozen at −80°C until processing for isolation of DNA or rickettsial organisms.
Isolation of rickettsiae.
Isolation of rickettsiae was attempted on five of the hemolymph-positive A. cooperi ticks by the shell vial technique as described previously (13). Briefly, individual ticks were thawed in a water bath at 37°C and disinfected for 10 min in iodine alcohol, followed by several washes in sterile water. Then each tick was triturated in 700 μl of brain heart infusion broth, the resultant tick homogenate was divided into three 200-μl aliquots, and each aliquot was inoculated into one shell vial containing a monolayer of confluent Vero cells. The remaining 100 μl of tick homogenate was used for DNA isolation as described below. After inoculation, the shell vials were centrifuged for 1 h at 700 × g and 22°C. Then the monolayer was washed once with minimal essential medium containing 5% bovine calf serum and subsequently incubated at 28, 32, or 37°C (one shell vial at each temperature) with a medium containing antibiotics (1% [each] penicillin, streptomycin, and gentamicin, and 1.5 g of amphotericin B/ml). After 3 days, the medium was changed to antibiotic-free medium, and the aspirated medium was examined by Gimenez staining for the presence of Rickettsia-like organisms (8). If the result was positive, the monolayer of the shell vial was harvested and inoculated into a 25-cm2 flask containing a monolayer of confluent uninfected Vero cells. Cells of the 25-cm2 flask were observed by Gimenez staining until more than 90% of the cells were infected, when they were harvested and inoculated into 150-cm2 flasks of Vero cells. A rickettsial isolate was considered established in the laboratory after at least 3 passages through 150-cm2 flasks, each reaching an infected-cell level of >90%. Cell passages of isolates were genotypically identified by sequencing the PCR product of the original infected tick and the resultant infected cells, as described below.
DNA isolation.
Frozen A. cooperi ticks were thawed in a water bath at 37°C, sterilized by immersion in iodine alcohol for 10 min followed by washing in sterile phosphate-buffered saline (PBS) solution, and then put individually into a sterile 1.5-ml microtube with 200 μl of PBS. Each tick was cut into small pieces with a sterile scissors and homogenized with a sterile micropestle. A 200-μl volume of tick homogenate was aspirated through a 21-gauge needle attached to a 1-ml syringe, and DNA was extracted by using the Dneasy tissue kit (Qiagen, Chatsworth, Calif.) according to the manufacturer's protocol for isolation of DNA from animal blood samples. Purified DNA was quantified in a digital spectrophotometer at a wavelength of 260 nm (MBA 2000l; Perkin-Elmer, Norwalk, Conn.) and stored at 4°C until use as a template for PCR amplifications. Five microliters of the template (approximately 300 ng of tick DNA) was used for each PCR. The remaining 100 μl from each tick homogenate, initially processed for isolation of rickettsiae in cell culture (described above), was added to 100 μl of PBS, and DNA was extracted by using the Dneasy tissue kit as described above.
DNAs of rickettsiae from cell cultures were isolated by using the IsoQuick nucleic acid extraction kit (Orca Research Inc., Bothell, Wash.). For this purpose, infected Vero cells were centrifuged at 4,000 × g for 5 min, and the resulting pellet was processed according to the manufacturer's protocol.
PCR amplification.
All tick samples were individually processed by a real-time PCR assay with primers CS-5 (forward) and CS-6 (reverse) (Table 1), designed to amplify a 147-bp fragment of the citrate synthase gene (gltA) of Rickettsia spp. A fluorogenic probe [5′ 6-FAM d(CATTGTGCCATCCAGCCTACGGT) BHQ-1 3′] (BioSearch Technologies, Novato, Calif.) positioned 76 bp downstream of the forward primer and 3 bp upstream of the reverse primer was used in the reactions. Real-time PCRs were performed in a Bio-Rad i-cycler apparatus with 25 μl per reaction, which contained 12.5 μl of the PCR iQSupermix (Bio-Rad, Hercules, Calif.), 0.75 μl of each primer at 15 μM, 0.25 μl of the probe at 15 μM, and 5.25 μl of molecular-grade water. Primers and probe concentrations were optimized in previous assays by spanning different initial concentrations of oligonucleotides. Real-time PCR cycling conditions were as follows: 1 cycle at 95°C for 2 min, followed by 50 cycles of 15 s at 95°C, 30 s at 50°C, and 30 s at 60°C. This real-time assay has successfully yielded fluorogenic signals from all Rickettsia species tested, which included R. rickettsii, Rickettsia prowazekii, Rickettsia canadensis, Rickettsia akari, Rickettsia felis, Rickettsia montanensis, and Rickettsia sibirica, and its sensitivity was determined to be 1 DNA copy of R. rickettsii and 100 DNA copies of Rickettsia bellii (M. B. Labruna and J. W. McBride, unpublished data). For each reaction, a negative control (5 μl of the same molecular-grade water mentioned above) and a positive control (300 ng of DNA of R. sibirica-infected Vero cells) were included.
TABLE 1.
Primer pairs used for amplification of rickettsial genes
| Gene and primer pair | Primers | Primer sequence (5′ → 3′) | Reference or source | Position on gene relative to the open reading frame |
|---|---|---|---|---|
| gltA | ||||
| 1 | CS-78 | GCAAGTATCGGTGAGGATGTAAT | This study | −78 to −56 |
| CS-323 | GCTTCCTTAAAATTCAATAAATCAGGAT | This study | 323 to 296 | |
| 2 | CS-239 | GCTCTTCTCATCCTATGGCTATTAT | Labruna et al., submitted | 239 to 263 |
| CS-1069 | CAGGGTCTTCGTGCATTTCTT | Labruna et al., submitted | 1069 to 1049 | |
| 3 | CS-5 | GAGAGAAAATTATATATCCAAATGTTGAT | This study | 922 to 948 |
| CS-1273 | CATAACCAGTGTAAAGCTG | 25 | 1098 to 1080 | |
| 4 | CS-5 | GAGAGAAAATTATATATCCAAATGTTGAT | This study | 922 to 948 |
| CS-6 | AGGGTCTTCGTGCATTTCTT | This study | 1068 to 1049 | |
| 17-kDa | ||||
| 5 | 17kD1 | GCTCTTGCAACTTCTATGTT | 32 | 31 to 50 |
| 17kD2 | CATTGTTCGTCAGGTTGGCG | 32 | 464 to 445 | |
| 6 | 17k-5 | GCTTTACAAAATTCTAAAAACCATATA | Labruna et al., submitted | −62 to −34 |
| 17k-3 | TGTCTATCAATTCACAACTTGCC | Labruna et al., submitted | +6 to 464 | |
| ompA | ||||
| 7 | Rr190.70p | ATGGCGAATATTTCTCCAAAA | 24 | 478 to 499 |
| Rr190.602n | AGTGCAGCATTCGCTCCCCCT | 24 | 990 to 969 |
The gltA gene was targeted in the real-time PCR because it has been detected in all rickettsial species (25). Once a tick was demonstrated by real-time PCR to contain rickettsial DNA, amplification of a larger fragment of the gltA gene was attempted by routine PCR using primers CS-78 (forward) and CS-323 (reverse) (Table 1). In this case, PCRs (25 μl) were performed in an Applied Biosystems Thermocycler (Gene Amp PCR System 2700) by adding 5 μl of the DNA template to 12.5 μl of the PCR iQSupermix, 1.0 μl of each primer at 20 μM, and 5.5 μl of molecular-grade water. PCR cycling conditions were as follows: 1 initial cycle at 95°C for 3 min; 40 cycles of 15 s at 95°C, 30 s at 48°C, and 30 s at 72°C; and 1 final cycle at 72°C for 7 min. For each reaction, a negative control (water) and a positive control (R. sibirica-infected Vero cells) were included as described for the real-time PCR. Ten microliters of the PCR product was separated by electrophoresis in a 1.5% agarose gel, stained with ethidium bromide, and examined by UV transillumination. The nucleotide sequence of the resulting PCR product was determined as described below.
For proper molecular characterization of rickettsiae isolated in cell culture, DNA of infected Vero cells was tested by routine PCRs using all the primers described in Table 1, targeting the following rickettsial genes: gltA, the 17-kDa protein-encoding gene, and ompA (a major outer membrane protein). These three genes have been characterized at the molecular level in most of the rickettsial species and are important molecular targets for taxonomy (1, 7, 24, 25, 32). PCRs were performed as described above for routine PCR.
Cloning and sequencing.
PCR products of the expected sizes were cloned by using the TOPO TA Cloning kit (Invitrogen, Carlsbad, Calif.) as described elsewhere (1). Plasmids containing the DNA inserts of the expected sizes were sequenced at least four times by using an ABI automated sequencer with M13 forward and M13 reverse sequencing primers (Invitrogen).
Phylogenetic analysis.
The sequences obtained were aligned for each gene (gltA, 17-kDa, and ompA) with the corresponding sequences of other Rickettsia species available in GenBank by using the CLUSTAL algorithm of the MegAlign program (Lasergene; DNAstar, Madison, Wis.). Phylogenetic relationships were inferred by using PAUP 4.0 β1 (29). For each gene analyzed, a phylogram was constructed by the neighbor-joining method, using Kimura's two-parameter model. Confidence values for individual branches of the resulting tree were determined by bootstrap analysis with 1,000 replicates. For the gltA and 17-kDa genes, R. bellii was designated as the outgroup, as shown in previous phylogenetic analyses (25; M. B. Labruna, D. H. Bouyer, J. McBride, L. M. A. Camargo, E. P. Camargo, and D. H. Walker, submitted for publication). For the ompA analysis, Rickettsia australis was used as the outgroup (27).
EM.
Infected Vero cell monolayers were fixed in Ito's fixative, a mixture of 1.25% formaldehyde, 2.5% glutaraldehyde, 0.03% trinitrophenol, 0.03% CaCl2, and 0.05 M cacodylate buffer at pH 7.3 (12); postfixed in 1% osmium tetroxide for 1 h; and stained en bloc in 1% uranyl acetate-0.1 M maleate buffer (pH 5.2). Pellets were dehydrated in ethanol, embedded in epoxy resin (Poly/Bed 812), and polymerized at 60°C overnight. Ultrathin sections (thickness, 70 nm) were prepared by using a Reichert Ultracut S ultramicrotome, placed on copper grids, stained with uranyl acetate and lead citrate, and examined in a Philips CM 100 electron microscope (EM) and a Philips 201 EM at 60 kV.
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers for the partial sequences of strain COOPERI generated in this study are AY362704 for the gltA gene, AY362705 for the 17-kDa gene, and AY362706 for the ompA gene. The accession numbers for the partial sequences of R. bellii isolate Ac25 are AY362702 for the 17-kDa gene and AY362703 for the gltA gene.
RESULTS
Infection of ticks by rickettsiae.
By the hemolymph test, a total of 12 A. cooperi ticks contained typical Rickettsia-like organisms inside hemocytes. Twenty ticks were hemolymph negative, and the other eight ticks yielded inconclusive results because hemolymph cells were completely lost during the washing procedures of the hemolymph test. A total of 19 ticks contained DNA of the rickettsial gltA gene by real-time PCR, including the 12 hemolymph-positive ticks, 3 hemolymph-negative ticks, and 4 hemolymph-inconclusive ticks. If we take the PCR assay as the valid result, the hemolymph test showed 80% sensitivity and 100% specificity. These calculations were performed on the 32 ticks that yielded conclusive results (positive or negative) in the hemolymph test.
Of the 19 ticks testing positive by real-time PCR, a total of 16 were also positive by routine PCR using primers CS-78 and CS-323, which amplify a 401-bp fragment of the gltA gene. The nucleotide sequences of the PCR products of 14 out of these 16 positive ticks were 100% identical to that of R. bellii (U59716), and the remaining 2 ticks contained DNA with nucleotide sequences 100% identical to both R. sibirica (U59734) and Rickettsia parkeri (U59732). The other three ticks were positive by real-time PCR but negative by routine PCR. These ticks' DNA showed a high (>40 cycles) critical threshold by real-time PCR, which suggests a low rickettsial concentration. Samples from these three ticks were subjected to a second real-time PCR, and the DNA sequence of the resultant product (100 nucleotides excluding the region corresponding to the primers) was 100% identical to the sequence of R. bellii for two ticks, while that for the third tick was 100% identical to that of several SFG rickettsiae, including R. sibirica (U59734) and R. parkeri (U59732). Overall, the PCR targeting the gltA gene showed that among 40 A. cooperii ticks evaluated in this study, 16 (40%) contained a rickettsia genotypically identified as R. bellii and 3 (7.5%) contained a rickettsia genotypically classified in the core of the SFG. Two out of 5 ticks (40%) from farm 1, 6 out of 15 ticks (40%) from farm 2, and 8 out of 20 ticks (40%) from farm 3 were positive for R. bellii. On the other hand, only two ticks (13%) from farm 2 and one tick (5%) from farm 3 were positive for the SFG rickettsia. Of the 16 ticks positive for R. bellii, 10 were male and 6 were female. There was no statistical difference in the incidence of infection by R. bellii between the two sexes (χ2 = 0.21; P = 0.65).
Isolation of rickettsiae.
Isolation assays by the shell vial technique were attempted with five of the hemolymph-positive ticks. Previous PCR with a sample of the tick homogenates showed four of these ticks to be positive for R. bellii and one to be positive for an SFG rickettsia. Rickettsiae were successfully isolated from two of the R. bellii-positive ticks and from one SFG rickettsia-positive tick. R. bellii was successfully isolated only from shell vials incubated at 28 or 32°C. Shell vials kept at 37°C were negative. R. bellii-infected cells from both 28 and 32°C shell vials were inoculated into 25-cm2 flasks, which showed 100% infected cells after 5 days, at both temperatures. Then infected cells from these flasks were inoculated into 150-cm2 flasks, which also showed 100% infected cells after 5 days. The R. bellii isolates displayed a strong cytopathic effect when incubated at 28°C, destroying the monolayer completely in a few days. However, when cells were incubated at 32°C, there was no visible effect, and the appearance of the monolayer was indistinguishable from that of normal uninfected control cells, even when they were 100% infected. Further attempts to propagate the R. bellii isolates in cells incubated at 37°C never resulted in more than 10% infected cells.
DNA of R. bellii-infected cells at the 4th passage was subjected to PCR targeting the gltA, 17-kDa, and ompA genes. PCR products of the expected sizes were obtained with the gltA and 17-kDa primers listed in Table 1, but no product was obtained with the ompA primers. We sequenced 1,153 and 499 nucleotides of the gltA and 17-kDa genes, respectively, of the two R. bellii isolates. The corresponding gene sequences of the two isolates were 100% identical to each other, the gltA sequences were 99.9% (1,152 of 1,153) similar to the R. bellii gltA sequence (U59716), and the 17-kDa sequences were 99.4% (496 of 499) similar to the R. bellii 17-kDa gene sequence (AF445380). We also sequenced the 1,153-bp fragment of the gltA gene from three PCR-positive A. cooperi ticks. One of them was 100% (1,153 of 1,153) similar to R. bellii gltA (U59716), and the other two were 99.9% similar (1,152 of 1,153). Thus, the two Rickettsia isolates from two A. cooperi ticks in this study, designated isolates Ac25 and Ac29, could be genetically identified as R. bellii. These isolates were successfully established in the laboratory and have been deposited as reference strains in the Rickettsial and Ehrlichial Diseases Laboratory at the University of Texas Medical Branch, Galveston, Tex.
A second Rickettsia species, genotypically classified in the SFG, was isolated in the shell vial incubated at 32°C but did not grow massively in the cells and was lost after 2 cell passages. However, PCR performed with DNA extracted from the 1st and 2nd cell passages resulted in expected product sizes for the rickettsial genes gltA, 17-kDa, and ompA. The gltA sequence was 99.3% (1,142 of 1,150) similar to that of R. sibirica (U59734). The 17-kDa sequence was 99.4% (392 of 394) similar to those of R. rickettsii (AY281069), Rickettsia peacockii (AF260571), and R. parkeri (U17008). The ompA sequence was 98.4% (483 of 491) similar to that of Rickettsia africae (U43790) and 98.0% (481 of 491) similar to that of R. parkeri (U43802). The Brazilian SFG isolate was designated strain COOPERI.
Phylogenetic analysis.
Due to the limited partial sequences of many rickettsiae available in GenBank, phylogenetic analyses were performed using 976, 393, and 415 bp of the gltA, 17-kDa, and ompA genes, respectively. Strain COOPERI was assigned to the SFG because it was positive for the ompA gene (so far found only in SFG species) and clustered with other SFG species in all phylogenetic trees (Fig. 1-3). Phylogenetic analysis inferred from the ompA gene placed strain COOPERI in a subgroup with R. parkeri (86% bootstrap support) within a clade composed of R. africae, R. parkeri, and R. sibirica (>60% bootstrap support) (Fig. 1).
FIG. 1.
Neighbor-joining phylogram based on partial ompA sequences, showing the phylogenetic placement of strain COOPERI among SFG rickettsial species. Levels of bootstrap support (>50%) for phylogenetic groupings are given. Percentages of difference between taxa are indicated by the scale of the drawing. Bar, 1% difference.
FIG. 3.
Neighbor-joining phylogram based on partial gltA sequences showing the phylogenetic placement of strain COOPERI among validated rickettsial species. Levels of bootstrap support (>50%) for phylogenetic groupings are given. Percentages of difference between taxa are indicated by the scale of the drawing. Bar, 0.5% difference.
By phylogenetic analysis based on the 17-kDa gene, strain COOPERI was placed in a clade with R. peacockii, R. rickettsii, R. parkeri, Rickettsia conorii, and R. sibirica (64% bootstrap support) (Fig. 2). A similar arrangement was observed by the gltA gene analysis, except for the additional inclusion of R. africae, Rickettsia honei, and Rickettsia slovaca in this clade (only 53% bootstrap support) (Fig. 3).
FIG. 2.
Neighbor-joining phylogram based on partial 17-kDa sequences showing the phylogenetic placement of strain COOPERI and R. bellii isolate Ac25 among rickettsial species. Levels of bootstrap support (>50%) for phylogenetic groupings are given. Percentages of difference between taxa are indicated by the scale of the drawing. Bar, 1% difference.
Ultrastructural observations.
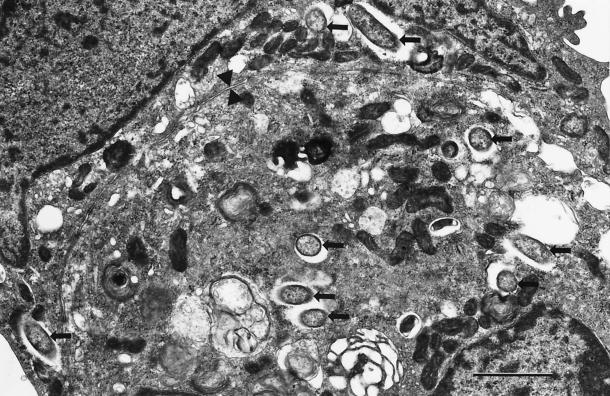
Ultrastructurally, the R. bellii strain isolated in the present study was morphologically identified within heavily infected Vero cells. The rickettsiae possessed typical bacillary morphology, and the majority of rickettsiae were observed free in the cytosol surrounded by electron lucent “halos,” corresponding to a slime layer up to 140 nm thick (Fig. 4). Several of the longer rickettsiae were in the process of binary division (Fig. 5). Most rickettsiae ranged from 0.28 to 0.38 μm in width and from 1.0 to 1.5 μm in length. Some rickettsiae had a “filamentous” appearance and could be as long as 3 μm (Fig. 6). A higher magnification of the cell wall revealed typical gram-negative morphology consistent with rickettsial species including a cytoplasmic membrane, periplasmic space, and an outer membrane with an inner leaflet slightly thicker than the outer leaflet (Fig. 7). The inner leaflets ranged in thickness from 5.0 to 9.5 nm. EM also demonstrated the presence of a thin electron-dense layer, up to 5 nm thick, immediately adjacent to the outer leaflet of the cell wall membrane, consistent with a microcapsular layer (Fig. 7).
FIG. 4.
Electron photomicrograph of two adjacent cells containing several intracytosolic rickettsiae (arrows). Note the prominent electron-lucent “halos” surrounding each rickettsia. Arrowheads mark the borders between the cells. Bar, 1.0 μm.
FIG. 5.
Electron photomicrograph of an intracytosolic rickettsia in the process of binary fission. Rickettsiae possess a characteristic gram-negative morphology, with an electron-lucent “halo” or slime layer (arrowheads) adjacent to the cell wall (small arrow) and a cytoplasmic membrane (large arrow) separated from the cell wall by the periplasmic space. Bar, 0.5 μm.
FIG. 6.
A filamentous rickettsia (marked by an asterisk), with associated slime layer (arrowheads), measuring 3.0 μm before extending out of the plane of section (arrow). Bar, 1.0 μm.
FIG. 7.
Rickettsial cytoplasmic membrane (large solid arrow) A higher magnification of the cell wall revealed a cell wall membrane with an inner leaflet (arrowheads) that is slightly thicker than the outer leaflet (small solid arrow), and an associated electron-lucent slime layer (asterisk) adjacent to the outer cell wall. A thin, electron-dense layer on the outer leaflet of the cell wall are morphologically consistent with a microcapsular layer (open arrows). Bar, 100 nm.
DISCUSSION
The present study showed that A. cooperi ticks from an area of BSF endemicity were infected by R. bellii at high infection rates (40%) and by an SFG rickettsia (strain COOPERI) at lower infection rates (7.5%). Even though we have tested only a small sample of ticks, our results offer some ecological considerations about the role of A. cooperi in the ecology of R. rickettsii, the agent of BSF in Brazil. As it is known that the presence of nonpathogenic rickettsiae within a tick population can minimize the transmission of pathogenic rickettsiae (3, 17), one might speculate that A. cooperi would not be an efficient enzootic vector of R. rickettsii in these areas, since almost 50% of the ticks were infected by rickettsial species other than R. rickettsii. In a parallel study, more than 600 free-living adult A. cajennense ticks were collected from the same pastures in the three farms of the present study. Interestingly, all of these ticks were negative for rickettsiae by the hemolymph test and by PCR targeting the gltA gene (11). These findings indicate that in this area, A. cajennense ticks are likely to be much more susceptible than A. cooperi ticks to infection by R. rickettsii. On the other hand, studies with large numbers of ticks in the United States demonstrated that some populations of Dermacentor andersoni (the primary vector of R. rickettsii in the western United States) were infected by as many as four rickettsial species: R. rickettsii, Rickettsia rhipicephali, R. bellii, and R. montanensis (3, 20). Thus, we cannot be sure that the A. cooperi populations of the present study were not infected by R. rickettsii, since we tested only a small number of ticks.
Strains of R. rickettsii from Brazil have been isolated and properly characterized (7, 21). However, no R. rickettsii strain has been isolated from the area of the present study, and the human BSF cases in this area were confirmed only by serological assays using R. rickettsii antigens or antisera and supported by clinical and epidemiological investigations (15). Since strain COOPERI was shown to be an SFG rickettsia genetically closely related to R. rickettsii, one would expect these rickettsiae to cross-react by serologic assays. In a previous study conducted in the same area as the present study, an SFG rickettsia was isolated from an A. cooperi tick collected on an R. rickettsii-seropositive capybara (16). This isolate was assigned to the SFG due to positive serological reactions with an anti-R. rickettsii antiserum. Although the species of this rickettsial isolate was not confirmed, there is a good chance that it might be strain COOPERI. Thus, it would be useful for future studies in this area of BSF endemicity to employ more-specific methods for diagnosing human cases of the disease. To date, at least three rickettsial species have been reported from this area of BSF endemicity: R. bellii and strain COOPERI in A. cooperi ticks (present study) and R. felis in fleas (Ctenocephalides felis felis) (11). Additionally, clinical, epidemiological, and serological evaluations support the presence of R. rickettsii (15).
In the phylogenetic analyses based on the 17-kDa and gltA genes, strain COOPERI was placed in groups with several SFG species, but always with low bootstrap values (Fig. 2 and 3). Previous phylogenetic studies of rickettsiae based on the 17-kDa and gltA genes have shown that these genes are very similar among most SFG species, which has resulted in low bootstrap support for the allocation of many SFG species in the phylogenetic trees (25; Labruna, Bouyer, et al., submitted for publication). For these closely related SFG species, analyses of the ompA and ompB genes are more informative (25). In fact, our phylogenetic analysis based on the ompA gene showed strain COOPERI to be closely related to R. parkeri, since these two rickettsiae clustered together 86% of the time (Fig. 1). However, the Kimura distance value between the partial ompA sequences of strain COOPERI and R. parkeri (0.014) was higher than that between strain COOPERI and R. africae (0.012). Further studies must establish strain COOPERI in cell culture in order to define its phenotype and perform other genotypic analyses.
From 1974 to 1983, a total of 263 isolates of R. bellii, infecting at least eight tick species of the genera Dermacentor, Haemaphysalis, Argas, and Ornithodoros, were identified in the United States (22). The present report is the first outside the United States and is also the first to identify R. bellii in an Amblyomma tick species. The presence of R. bellii in Brazil highlights the possibility that this Rickettsia species might have a much broader geographic range than previously thought. In fact, some other Rickettsia species formerly thought to be restricted to a few geographic areas are now realized to have a much broader distribution. This realization has resulted primarily from advances in molecular techniques, which have facilitated the study of rickettsiae around the world (10).
Recent phylogenetic analyses of rickettsiae have determined that R. bellii belongs to an ancestral group which diverged prior to the split of the typhus group (TG) and SFG (25). The presence of R. bellii infecting different tick species in different geographic areas may be an indication of recent horizontal transmission among ticks. However, this bacterium has never been reported to infect vertebrate hosts. Another possible mechanism of horizontal transmission of intracellular bacteria between arthropods is via parasitoid arthropods (28). As a matter of fact, parasitoids of the genus Ixodiphagus have been reported in ticks from North America (26) and South America (M. B. Labruna, C. D. Paula, and A. P. Prado, abstract from the 13th Annual Meeting of the Instituto Biológico 2000, abstr. 67, Arq. Inst. Biol. 67(Suppl.), 2000). Unfortunately, no study concerning infection by rickettsiae in tick parasitoids has been reported.
R. bellii has been described as a delicate, hemocyte-associated, rod-shaped bacterium, differing from the classical lanceolate, coccobacillary SFG rickettsiae (22). The intracellular bacteria observed inside hemocytes of hemolymph-postive A. cooperi ticks were delicate and rod-shaped, as were those stained by Gimenez stain in Vero cells (data not shown). Vero cell culture has been reported as a usually satisfactory system for primary recovery of R. bellii from hemolymph-positive ticks (22). Our results confirmed this assessment, in that isolates Ac25 and Ac29 grew very rapidly even in the 1st passage, infecting nearly 100% of the cells by 5 days. However, the procedures used by Philip et al. to isolate R. bellii in Vero cell cultures (22), referred to as the method of Cory et al. (4), consisted of incubating cells at 35°C. Using this procedure, Philip et al. (22) observed variations in the susceptibility of Vero cells to primary infection, shown by differences in the nature of cytopathogenicity according to locality and arthropod host (i.e., isolates from one area or tick species gave rise to a distinct cytopathic effect, whereas little or no cytopathic effect could be demonstrated for isolates from another area or tick species). In the present study, we isolated two strains from the same geographic area and the same tick species. These two strains behaved similarly to one another and were shown to cause a cytopathic effect only at 28°C. At 32°C, no cytopathic effect was observed, and the strains did not grow satisfactorily at 37°C. We also inoculated Ac25 onto Vero cell monolayers incubated at 35°C (data not shown), with the purpose of comparing the results with those of previous reports (22). In this case, Ac25 grew very slowly, never infected more than 50% of the cells, and caused no cytopathic effect. Thus, it seems that the capacity of isolates to induce a cytopathic effect in cell cultures is linked not only to the origin of the isolate but also to the temperature of incubation of the cells. Comparisons of cytopathogenicity of a Rickettsia species in one cell line should be done only when cultures are incubated at the same temperature.
Ultrastructurally, R. bellii possesses a morphology and dimensions consistent with rickettsiae except for its length. R. bellii is an unusually long rickettsia. While the longest R. bellii organism found by EM in the present study was 3.0 μm long (Fig. 6), this rickettsia had one end out of the plane of section. Therefore, it is difficult to characterize the length of this rickettsial species by transmission EM. A previous light microscopic study of an infected cell culture demonstrated that R. bellii ranges in size from 2.0 to 3.0 μm during the log phase of growth and can develop into filamentous forms as long as 10 to 15 μm in cases of nutrient exhaustion (22).
In the present study, R. bellii possessed a cell wall with an inner leaflet thicker than the outer leaflet and an adjacent slime layer separating the cell wall from the surrounding host cell cytoplasm. While the only other ultrastructural study of R. bellii reported the lack of a “discernible” microcapsular layer adjacent to the outer cell wall (22), the present study demonstrated the presence of a thin electron-dense layer on the outer leaflet of the cell wall membrane that could correspond to a microcapsular layer, giving the outer cell wall a characteristic “fuzzy” appearance. In some areas of the outer cell wall, the electron-dense layer was nonexistent, and in other areas this layer extended as far as 5 nm off the surface. The biological importance of these projections remains undefined. While R. bellii is thought not to possess the surface proteins conserved among TG or SFG rickettsial species, a previous study demonstrated that sera obtained from patients previously infected with R. conorii did react with lipopolysaccharide from R. bellii (23). In agreement with those of other rickettsial species, the processes of cellular entry into a phagosome, vacuolar escape, and subsequent intracytosolic existence appear to be conserved in R. bellii, as most of the rickettsiae were found free in the cytosol. A few rickettsiae were visualized in vacuoles undergoing apparent digestion, but this was a rare event.
Finally, our study adds two more rickettsiae to the list of species occurring in ticks from South America, a topic of little previous investigation. The only other Rickettsia species reported in ticks on this continent are R. rickettsii (9, 21) and strain ARANHA, reported recently in the tick Amblyomma longirostre from northwestern Brazil (Labruna, Bouyer, et al., submitted for publication). Our phylogenetic trees showed strain COOPERI to be distinct from strain ARANHA (Fig. 1 and 2). The low number of rickettsial species in South America is a result of the few studies conducted on this continent and will certainly increase as more areas are investigated.
Acknowledgments
This work was supported by the Fogarty International Center (grant D43TW00903 to D.H.W. and M.B.L.), National Institute of Allergy and Infectious Diseases (grant AI21242 to D.H.W. and T.W.), and Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (grant 02/00644-0 to M.B.L.; grant 00/02711-1 to S.M.G.).
REFERENCES
- 1.Bouyer, D. H., J. Stenos, P. C. Valdes, C. G. Moron, V. L. Popov, J. E. Zavala-Velazquez, L. D. Foil, D. R. Stothard, A. F. Azad, and D. H. Walker. 2001. Rickettsia felis: molecular characterization of a new member of the spotted fever group. Int. J. Syst. Evol. Microbiol. 51:339-347. [DOI] [PubMed] [Google Scholar]
- 2.Burgdorfer, W. 1970. The hemolymph test. Am. J. Trop. Med. Hyg. 19:1010-1014. [PubMed] [Google Scholar]
- 3.Burgdorfer, W. 1988. Ecological and epidemiological considerations of Rocky Mountain spotted fever and scrub typhus, p. 33-50. In D. H. Walker (ed.), Biology of rickettsial diseases, vol. 1. CRC, Inc., Boca Raton, Fla.
- 4.Cory, J., C. E. Yunker, J. A. Howarth, Y. Hokama, L. E. Hughes, L. A. Thomas, and C. M. Clifford. 1975. Isolation of spotted fever group and Wolbachia-like agents from field-collected materials by means of plaque formation in mammalian and mosquito cells. Acta Virol. 19:443-445. [PubMed] [Google Scholar]
- 5.Davis, G. E., and R. R. Parker. 1933. Additional studies on the relationship of the viruses of Rocky Mountain spotted fever and Sao Paulo exanthematic typhus. Pub. Health Rep. 48:1006-1011. [Google Scholar]
- 6.Dias, E., and A. V. Martins. 1939. Spotted fever in Brazil. Am. J. Trop. Med. 19:103-108. [Google Scholar]
- 7.Eremeeva, M. E., R. M. Klemt, L. A. Santucci-Domotor, D. J. Silverman, and G. A. Dasch. 2003. Genetic analysis of isolates of Rickettsia rickettsii that differ in virulence. Ann. N. Y. Acad. Sci. 990:717-722. [DOI] [PubMed] [Google Scholar]
- 8.Giménez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 9.Gomes, L. S. 1933. Typho exanthematico de São Paulo. Brasil-Medico 17:919-921. [Google Scholar]
- 10.Hechemy, K. E., T. Avsic-Zupanc, J. E. Childs, and D. A. Raoult. 2003. Rickettsiology: present and future directions: preface. Ann. N. Y. Acad. Sci. 990:xvii-xx. [DOI] [PubMed] [Google Scholar]
- 11.Horta, M. C. 2002. Pesquisa de infecção por riquétsias do grupo da febre maculosa em humanos, eqüídeos, caninos e em diferentes estádios de vida do Amblyomma cajennense, provenientes de uma área endêmica do Estado de São Paulo. M.S. dissertation. University of São Paulo, São Paulo, Brazil.
- 12.Ito, S., and Y. Rikihisa. 1981. Techniques for electron microscopy of rickettsiae, p. 213-227. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, New York, N.Y.
- 13.Kelly, J. P., D. Raoult, and P. R. Mason. 1991. Isolation of spotted fever group rickettsiae from triturated ticks using a modification of the centrifugation vial technique. Trans. R. Soc. Trop. Med. Hyg. 85:397-398. [DOI] [PubMed] [Google Scholar]
- 14.Labruna, M. B., C. E. Kerber, F. Ferreira, J. L. H. Faccini, D. T. De Waal, and S. M. Gennari. 2001. Risk factors to tick infestations and their occurrence on horses in the State of São Paulo, Brazil. Vet. Parasitol. 97:1-14. [DOI] [PubMed] [Google Scholar]
- 15.Lemos, E. R., F. B. Alvarenga, M. L. Cintra, M. C. Ramos, C. D. Paddock, T. Ferebee, S. R. Zaki, F. C. Ferreira, R. C. Ravagnani, R. D. Machado, M. A. Guimarães, and J. R. Coura. 2001. Spotted fever in Brazil: a seroepidemiological study and description of clinical cases in an endemic area in the state of São Paulo. Am. J. Trop. Med. Hyg. 65:329-334. [DOI] [PubMed] [Google Scholar]
- 16.Lemos, E. R. S., H. H. B. Melles, S. Colombo, R. D. Machado, J. R. Coura, M. A. A. Guimarães, S. R. Sanseverino, and A. Moura. 1996. Primary isolation of spotted fever group rickettsiae from Amblyomma cooperi collected from Hydrochaeris hydrochaeris in Brazil. Mem. Inst. Oswaldo Cruz 91:273-275. [DOI] [PubMed] [Google Scholar]
- 17.Macaluso, K. R., D. E. Sonenshine, S. M. Ceraul, and A. F. Azad. 2002. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second rickettsia. J. Med. Entomol. 39:808-813. [DOI] [PubMed] [Google Scholar]
- 18.Monteiro, J. L. 1933. Vacina contra o typho exantematico de S. Paulo. Mem. Inst. Butantãn 8:11-20. [Google Scholar]
- 19.Monteiro, J. L. 1933. Comportamento experimental do virus do typho exantematico de S. Paulo apos passagem pelo carrapato (Amblyomma cajennense). Mem. Inst. Butantãn 8:23-37. [Google Scholar]
- 20.Philip, R. N., and E. A. Casper. 1981. Serotypes of spotted fever group rickettsiae isolated from Dermacentor andersoni (Stiles) ticks in Western Montana. Am. J. Trop. Med. Hyg. 30:230-238. [DOI] [PubMed] [Google Scholar]
- 21.Philip, R. N., E. A. Casper, W. Burgdorfer, R. K. Gerloff, L. E. Hughes, and E. J. Bell. 1978. Serologic typing of rickettsiae of the spotted fever group by microimmunofluorescence. J. Immunol. 121:1961-1968. [PubMed] [Google Scholar]
- 22.Philip, R. N., E. A. Casper, R. L. Anacker, J. Cory, S. F. Hayes, W. Burgdorfer, and E. Yunker. 1983. Rickettsia bellii sp. nov.: a tick-borne rickettsia, widely distributed in the USA, that is distinct from the spotted fever and typhus biogroups. Int. J. Syst. Bacteriol. 33:94-106. [Google Scholar]
- 23.Raoult, D., and G. A. Dasch. 1989. Line blot and Western blot immunoassays for diagnosis of Mediterranean spotted fever. J. Clin. Microbiol. 27:2073-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regnery, R. L., C. L. Spruill, and B. D. Plikaytis. 1991. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173:1576-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roux, V., E. Rydkina, M. Eremeeva, and D. Raoult. 1997. Citrate synthase gene comparison, a new tool for phylogenetic analysis and its application for the rickettsiae. Int. J. Syst. Bacteriol. 47:252-261. [DOI] [PubMed] [Google Scholar]
- 26.Stafford, K. C., III, A. J. Denicola, and L. A. Magnarelli. 1996. Presence of Ixodiphagus hookeri (Hymenoptera: Encyrtidae) in two Connecticut populations of Ixodes scapularis (Acari: Ixodidae). J. Med. Entomol. 33:183-188. [DOI] [PubMed] [Google Scholar]
- 27.Stenos, J., and D. H. Walker. 2000. The rickettsial outer-membrane protein A and B genes of Rickettsia australis, the most divergent rickettsia of the spotted fever group. Int. J. Syst. Evol. Microbiol. 50:1775-1779. [DOI] [PubMed] [Google Scholar]
- 28.Stevens, L., R. Giordano, and R. F. Fialho. 2001. Male-killing, nematode infections, bacteriophage infection, and virulence of cytoplasmic bacteria in the genus Wolbachia. Annu. Rev. Ecol. Syst. 32:519-545. [Google Scholar]
- 29.Swofford, D. L. 1999. PAUP: phylogenetic analysis using parsimony, version 4.0 βl. Center for Agriculture and Bioscience International, Champaign, Ill.
- 30.Travassos, J., and A. Vallejo. 1942. Comportamento de alguns cavídeos (Cavia aperea e Hydrochoerus capybara) às inoculações experimentais do vírus da febre maculosa. Possibilidade desses cavídeos representarem o papel de depositários transitórios do vírus na natureza. Mem. Inst. Butantãn 15:73-86. [Google Scholar]
- 31.Travassos, J. and A. Vallejo. 1942. Possibilidade de Amblyomma cajennense se infectar em Hydrochoerus capybara experimentalmente inoculado com o vírus da febre maculosa. Mem. Inst. Butantãn 15:87-90. [Google Scholar]
- 32.Webb, L., C. Mitchell, D. C. Malloy, G. A. Dasch, and A. F. Azad. 1990. Detection of murine typhus infection in fleas by using the polymerase chain reaction. J. Clin. Microbiol. 28:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]