Abstract
Redox regulation of EGFR signaling helps protect cells against oxidative stress. In this study, we investigated whether the cytotoxicity of an EGFR tyrosine kinase inhibitor, erlotinib (ERL), was mediated by induction of oxidative stress in human head and neck cancer (HNSCC) cells. ERL elicited cytotoxicity in vitro and in vivo while increasing a panel of oxidative stress parameters which were all reversible by the antioxidant N-acetyl cysteine. Knockdown of EGFR using siRNA similarly increased these oxidative stress parameters. Overexpression of mitochondrial targeted catalase but not superoxide dismutase reversed ERL-induced cytotoxicity. Consistent with a general role for NADPH oxidase (NOX) enzymes in ERL-induced oxidative stress, ERL-induced cytotoxicity was reversed by diphenylene iodonium, a NOX complex inhibitor. ERL reduced the expression of NOX1, NOX2 and NOX5 but induced the expression of NOX4. Knockdown of NOX4 using siRNA protected HNSCC cells from ERL-induced cytotoxicity and oxidative stress. Our findings support the concept that ERL-induced cytotoxicity is based upon a specific mechanism of oxidative stress mediated by hydrogen peroxide production through NOX4 signaling.
Keywords: Erlotinib, EGFR, NAC, oxidative stress, NOX4
Introduction
Cancer cells are hypothesized to exist in a metabolic condition of increased intrinsic oxidative stress when compared to normal untransformed cells (1–3). Metabolic oxidative stress is thought to increase tumorigenesis by activating redox regulated signaling pathways involved in cell proliferation and survival (4–8). Previous studies have shown that the redox regulation of epidermal growth factor receptor (EGFR) signaling plays a major role in the protection of cancer cells against oxidative stress (8–10).
EGFR is a receptor tyrosine kinase that activates pro-survival and pro-growth signaling pathways including PI3K/Akt, MAPK and JNK (11). EGFR is an important molecular target for antineoplastic therapy as EGFR is found to be upregulated in the majority of lung cancers, glioblastoma and head and neck cancers (HNSCC) and is associated with a poor clinical prognosis (11–13).
Inhibitors of EGFR such as gefitinib, cetuximab and erlotinib, are available clinically and have successfully been used to treat colorectal, non small cell lung cancer (NSCLC), and HNSCC both as mono-therapy and combined with chemotherapy or radiation therapy although their mechanism of action remains unclear (14–16). Studies have suggested that inhibition of DNA repair may be involved in the radiosensitizing effect of EGFR inhibition since DNA protein kinases, which are important regulators of DNA repair, are down regulated with EGFR inhibition (16). In addition, EGFR inhibition has been shown to cause cell cycle arrest in G1 and change the cell cycle distribution of cancer cells by decreasing the percent of cells in the radio-resistant S phase (16). Finally, modulation of downstream signaling pathways may play an important role in the mechanism of EGFR inhibition as inhibition of downstream signaling pathways such as Akt have been shown to induce cytotoxicity in cancer cells (8, 17).
Previous studies have shown that oxidative stress led to EGFR phosphorylation, which conferred protection against oxidative stress-induced apoptosis (8, 9, 18). Since EGFR signaling is both up regulated in the majority of HNSCC and important for cell survival, and cancer cells are under increased metabolic oxidative stress (compared to normal cells), we propose that cancer cells may increase EGFR signaling to protect against metabolic oxidative stress-induced cell killing. Furthermore, if EGFR signaling is involved in the protection of cancer cells from oxidative stress, then inhibition of EGFR activation would be expected to increase endogenous metabolic oxidative stress . The aim of this study was to determine if inhibition of EGFR signaling induced cell killing due to oxidative stress in HNSCC cells in vitro and in vivo.
Materials and Methods
Cells and culture conditions
FaDu and Cal-27 HNSCC cells were obtained from the American Type Culture Collection ([ATCC] Manassas, VA). SQ20B HNSCC cells were a gift from Dr. Anjali Gupta (Department of Radiation Oncology, The University of Iowa). All cell lines were authenticated by the ATCC for viability (before freezing and after thawing), growth, morphology and isoenzymology. Cells were stored according to the supplier's instructions and used over a course of no more than 3 months after resuscitation of frozen aliquots. All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 4 mM L-glutamine, 1 mM sodium pyruvate, 1.5 g/L sodium bicarbonate and 4.5 g/L glucose with 10% fetal bovine serum (FBS; Hyclone, Logan, UT). Cultures were maintained in 5% CO2 and air humidified in a 37°C incubator.
Drug Treatment
N-acetyl cysteine (NAC), diphenylene iodonium (DPI), pegylated catalase (CAT) and pegylated superoxide dismutase (SOD) were obtained from Sigma (St. Louis, MO). NAC was dissolved in 1 M sodium bicarbonate (pH 7.0), and DPI was dissolved in DMSO. Erlotinib (Tarceva, ERL) and Acetadote (Cumberland Pharmaceuticals, Nashville TN) were obtained from the inpatient pharmacy at the University of Iowa Hospitals and Clinics. ERL was provided as a solid tablet which was ground into a fine powder and subsequently dissolved in 100% DMSO. All drugs were used without further purification. Drugs were added to cells at final concentrations of 20 mM NAC, 50 nM DPI, 100 U/mL CAT, 100 U/mL SOD and 10 µM ERL. The required volume of each drug was added directly to complete cell culture media on cells to achieve the desired final concentrations. All cells were placed in a 37°C incubator and harvested at the time points indicated.
Western blot analysis
Cell lysates were standardized for protein content, resolved on 4–12% SDS polyacrylamide gels, and blotted onto nitrocellulose membranes. Membranes were probed with rabbit anti-EGFR, anti-phospho-EGFR, anti-β-actin (Cell Signaling Technologies, Beverly, MA), anti-NOX1, anti-NOX2 (Abcam, Cambridge MA), goat anti-NOX4 and anti-NOX5 (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies. Antibody binding was detected using an ECL chemiluminescence kit (Amersham, Arlington Heights, IL).
PI staining for DNA content
Fixed cells were washed with PBS and incubated with RNAase A (0.1 mg/mL) for 30 min followed by incubation with propidium iodide (PI) for 1 h. DNA content of PI-stained cells was analyzed by FACScan (Becton–Dickinson) and the percentage of cells with G1, S, and G2/M DNA content was calculated using MODFIT software (Verity Software House).
Clonogenic cell survival experiments
Attached cells from experimental dishes were analyzed for clonogenic survival as previously described (19).
Glutathione assay
Cell pellets were thawed and homogenized in 50 mM phosphate buffer (pH 7.8) containing 1.34 mM diethylenetriaminepentaacetic acid (DETAPAC) buffer. Total glutathione content was determined by the method of Anderson (20). Glutathione disulfide (GSSG) was analyzed as described previously (21). All glutathione determinations were normalized to the protein content using the Lowry method. (22).
siRNA Transfection
EGFR, NOX4 and control siRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). HNSCC cells were transfected with 20 nM siRNA at 80% confluence in reduced-serum Eagle’s Minimum Essential Medium (EMEM, Santa Cruz, CA) for 24 hours. Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used for transfections following protocols provided by the manufacturer. Biochemical analyses were performed 48–72 h after transfection.
Tumor cell implantation
Twenty four female 4–5 week old athymic-nu/nu nude mice were purchased from Harlan Laboratories (Indianapolis, IN). Mice were housed in a pathogen-free barrier room in the Animal Care Facility at the University of Iowa and handled using aseptic procedures. All procedures were approved by the IACUC committee of the University of Iowa and conformed to the guidelines established by NIH. Mice were allowed at least 3 days to acclimate prior to beginning experimentation, and food and water were made freely available. Tumor cells were inoculated into nude mice by subcutaneous injection of 0.1 mL aliquots of saline containing 4 × 106 FaDu cells into the right flank using 26-gauge needles.
Tumor measurements
All mice started treatment 1 week after tumor inoculation. At this point, tumors sizes ranged from 0.06 – 0.08 cm3. Mice were evaluated daily and tumor measurements taken three times per week using Vernier calipers. Tumor volumes were calculated using the formula: tumor volume = (length × width2) /2 where the length was the longest dimension, and width was the dimension perpendicular to length.
In vivo drugs administration
Mice were divided into 4 groups (n = 6 mice/group). ERL group: Erlotinib was suspended in water and administered orally 12.5 mg/kg every other day for 6 total doses. NAC group: N-acetyl cysteine (Acetadote, Cumberland Pharmaceuticals, Nashville, TN) was administered i.p. 325 mg/kg every day for 10 total doses. NAC+ERL group: mice were administered 325 mg/kg NAC every day (i.p.) plus 12.5 mg/kg ERL orally every other day for a total of 10 NAC and 6 ERL doses. Control group: mice were administered i.p. saline every other day and water orally every day. Mice were euthanized via CO2 gas asphyxiation or lethal overdose of sodium pentobarbital (100 mg/kg) when tumor diameter exceeded 1.5 cm in any dimension.
Immunofluorescence Staining
Slides were blocked for 30 min with normal goat serum and incubated overnight at 4°C with rabbit anti-human pEGFR (Santa Cruz Biotechnology, Santa Cruz, CA, 1: 450 dilution). Secondary detection was performed with AlexaFluor488 anti-rabbit (Invitrogen). Counterstain was performed with ToPro3 (far red). Negative control slides were obtained by omitting the primary or secondary antibody. The images were acquired using a Bio-Rad Radience 2100MP confocal microscope at 60x magnification with Zen2009 software. Images were analyzed by quantification of the fluorescence intensity using image analysis and recognition software, Image J (National Institutes of Health, Bethseda MA) and averaged for 3 animals/group for each treatment group.
Transduction of Antioxidant Enzymes
AdCMV Bgl II (AdEMPTY), AdCMVCAT (AdCAT) and AdCMVMCAT (AdMCAT) were purchased from Viraquest (North Liberty, Iowa). Each gene was inserted into the E1 region of an Ad5 E1/particle E3 deleted replication deficient adenoviral vector. The adenovirus constructs were originally prepared by Dr. John Engelhardt (AdEMPTY and AdCAT) and Dr. Andre Melendez (AdMCAT) (23,24). Viral particles (100 MOI) were added to cells for 24 h, and the media was changed to complete media prior to each experiment. Catalase activity was measured on cell homogenates by monitoring the disappearance of 10 mM H2O2 in 50 mM potassium phosphate (pH 7.0) spectrophotometrically at 240 nm. Activities were expressed in mk units/mg protein as described (25).
Measurement of intracellular prooxidant levels
Attached cells were labeled with 5-(and-6)-carboxy-2’,7’-dichlorodihydrofluorescein diacetate, (DCFH, 10 µg/mL) dissolved in 0.1% DMSO for 15 min at 37°C or dihydroethidium (DHE, 10 µM) dissolved in 0.1% DMSO for 40 min at 37°C. Culture plates were placed on ice, trypsinized, re-suspended in ice cold PBS, and analyzed using a FACScan flow cytometer (excitation 488 nm, emission 530 nm band-pass filter [DCFH]; excitation 488 nm, emission 585 nm band-pass filter [DHE]). The mean fluorescence intensity (MFI) of 10,000 cells was analyzed in each sample and corrected for auto-fluorescence from unlabeled cells.
Statistical analysis
Statistical analysis was done using GraphPad Prism version 5 for Windows (GraphPad Software, San Diego, CA). Differences between 3 or more means were determined by one-way ANOVA with Tukey post-tests. Linear mixed effects regression models were used to estimate and compare the group-specific change in tumor growth curves. All tests were two-sided and carried out at the 5% level of significance. All statistical analysis was performed at the p<0.05 level of significance.
Results
Effect of Erlotinib on EGFR expression
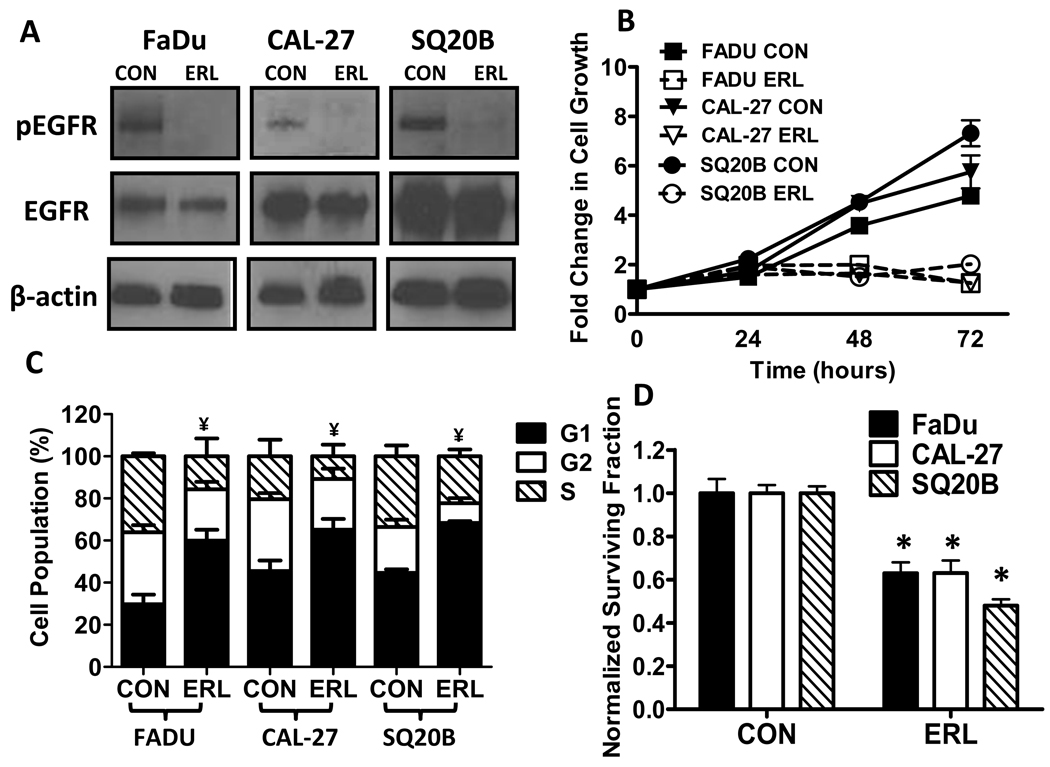
Erlotinib (ERL), an FDA approved EGFR tyrosine kinase inhibitor, was used to determine the effect of inhibiting EGFR signaling in HNSCC cells. Figure 1A shows that in FaDu, Cal-27 and SQ20B cells treated with 10 µM ERL for 24 h, phosphorylated EGFR (pEGFR, active form) was decreased without causing significant effects on total EGFR (tEGFR). The dose of 10 µM was for our studies because this dose for 24 hrs was effective at inducing moderate but significant toxicity in our HNSCC cell model. ERL at 5 µM is also capable of inhibiting EGFR expression after 24 hrs but a significant cytotoxic effect is not observed until 48 hrs after treatment in our HNSCC cell model (data not shown).
Figure 1. Effect of Erlotinib (ERL) on EGFR expression, cell growth and cytotoxicity in HNSCC cells.
(A) EGFR expression in FaDu, Cal-27 and SQ20B cells was assayed by Western blot for EGFR and phospho-EGFR Tyr1068 (pEGFR) in the presence or absence of 10 µM ERL for 24 hours. β-actin was used as a loading control. (B) FaDu (squares), Cal-27 (triangles), and SQ20B (circles) cells were treated with 10 µM ERL then grown and counted at 0, 24, 48 and 72 hours. (C–D) FaDu, Cal-27 and SQ20B cells were treated with 10 µM ERL for 24 hours then analyzed by flow cytometry for cell cycle distribution (C) and clonogenic survival (D). Error bars represent ± 1SD of N = 3 experiments. *, P < .05 versus respective control (CON); ¥ refers to differences (p<0.05) of cells in G1 compared to respective control.
Effect of Erlotinib on HNSCC cell growth and cell cycle
To examine the effect of ERL on HNSCC cell growth and cell cycle distribution, FaDu, Cal 27, and SQ20B cells were treated with ERL, then counted and analyzed for PI staining at 24, 48 and 72 h after treatment. A near complete inhibition of cellular growth was observed with 10 µM ERL over a course of 72 h in all cell lines (Fig. 1B). We also observed an increased accumulation of cells in the G1 phase of the cell cycle in all 3 cell lines after 24 h of ERL treatment (Fig. 1C). These results support the conclusion that ERL inhibited cell growth in HNSCC cells by blocking the cells in the G1 phase of the cell cycle.
Effect of Erlotinib on Clonogenic Cell Survival and Oxidative Stress
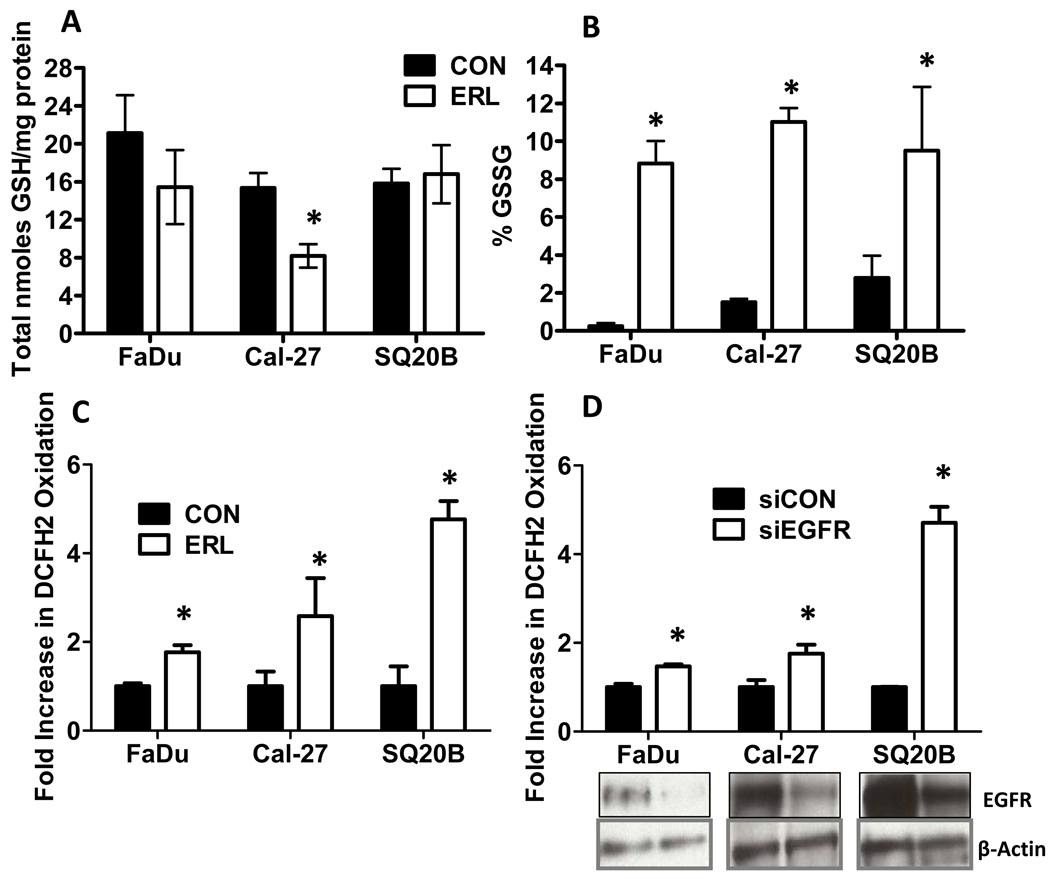
The cytotoxic effect of ERL was determined by analyzing clonogenic survival in HNSCCs after 24 h treatment with ERL. ERL (10 µM) treated cells showed a significant reduction (40–60%) in cell survival compared to control (Fig. 1D). We then determined if inhibition of EGFR with ERL would induce oxidative stress by analyzing disruptions in glutathione (GSH/GSSG) metabolism. The GSH/GSSG redox couple is the major thiol-disulfide antioxidant system in cells (26). The percentage of GSH that was oxidized (GSSG) suggests a shift toward an oxidizing environment and was used as an indicator of oxidative stress (26). ERL induced a significant decrease in total glutathione (GSH) levels (calculated as GSH+GSSG) in only Cal-27 cells (Fig. 2A), while inducing an increase in %GSSG (calculated as GSSG/[GSH+GSSG])×100) in all 3 cell lines (Fig. 2B). Intracellular prooxidant production (presumably hydroperoxides) in all cell lines was determined by measuring DCFH oxidation in the presence and absence of ERL. ERL significantly increased DCFH oxidation in all 3 cell lines (Fig. 2C). These data suggest that 10 µM ERL induces cytotoxicity and oxidative stress in HNSCC cells as characterized by disruptions in thiol metabolism and hydroperoxide production.
Figure 2. Effect of EGFR inhibition on oxidative stress parameters.
(A–C) FaDu, Cal-27 and SQ20B cells were treated with 10 µM ERL for 24 hours then analyzed for total glutathione (GSH, A), percentage glutathione disulfide (%GSSG, B) and DCFH oxidation (C). (D) FaDu, Cal-27 and SQ20B cells were transfected with siRNA against EGFR (siEGFR) or control (siCON) and assayed for EGFR expression. β-Actin was used as a loading control. Error bars represent ± 1SD of N = 3 experiments. *, P < .05 versus control (CON) or CON siRNA (siCON).
EGFR knockdown Induced Oxidative Stress in SQ20B cells
To confirm that ERL-induced oxidative stress was due to inhibition of EGFR signaling, EGFR expression was knocked down with siRNA in FaDu, Cal-27 and SQ20B cells. When the cells were analyzed for DCFH oxidation, EGFR knockdown resulted in a significant increase in DCFH oxidation (Fig. 2D). These results provide further support for the hypothesis that inhibition of EGFR signaling induces oxidative stress in HNSCC cells.
The effect of NAC and ERL on Clonogenic Cell Killing and Oxidative stress
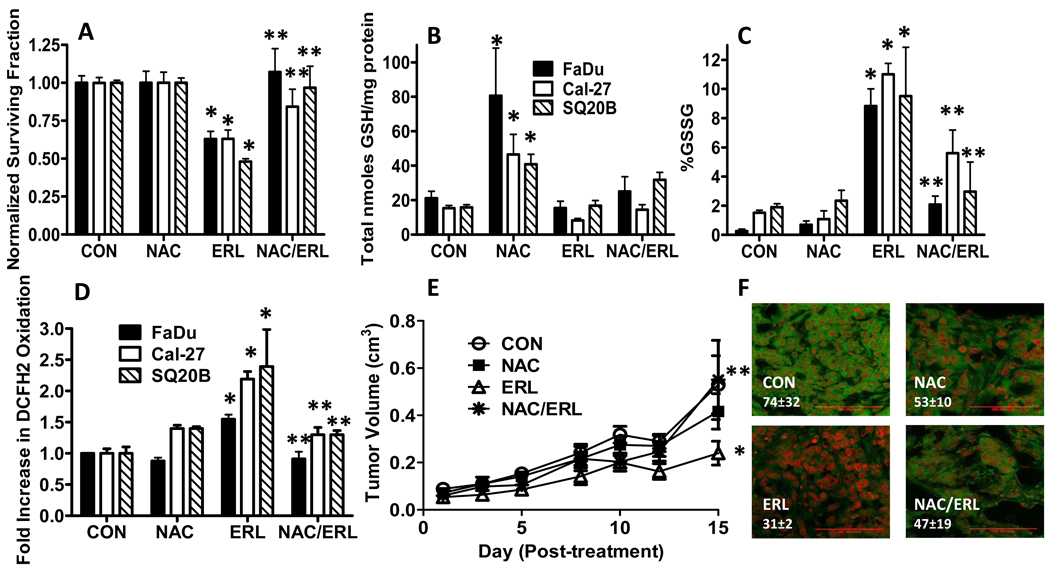
To further analyze the role of oxidative stress in ERL-induced cell killing, FaDu, Cal-27 and SQ20B cells were pretreated with NAC (a thiol antioxidant) before ERL treatment, then analyzed for clonogenic survival and GSH/GSSG levels. NAC was able to completely reverse the cytotoxicty induced by ERL in FaDu and SQ20B cells, and partially (but significantly) reverse ERL induced cytotoxicty in Cal-27 cells (Fig. 3A). NAC significantly increased total GSH levels in all cell lines but only modestly increased total GSH levels in the presence of ERL (Fig. 3B). Additionally, NAC suppressed the increase in %GSSG (Fig. 3C) and DCFH oxidation (Fig. 3D) that was induced by ERL in all cell lines. These results show that inhibition of EGFR signaling with ERL induces disruptions in thiol metabolism and cytotoxicity consistent with oxidative stress, which was reversed by NAC, supporting the hypothesis that ERL-induced cytotoxicity in HNSCC cells may be related to increased oxidative stress.
Figure 3. Effect of N-acetyl-cysteine (NAC) on ERL-induced cytotoxicity and oxidative stress.
(A–D) FaDu, Cal-27 and SQ20B cells were treated with 10 µM ERL for 24 hours with or without treatment with 20 mM NAC for 1 hour before and during ERL exposure, then analyzed for clonogenic survival (A), total glutathione (GSH, B), percentage glutathione disulfide (%GSSG, C) and DCFH oxidation (D). Error bars represent ± 1SD of N = 3–4 experiments. (E) Athymic (nu/nu) mice bearing FaDu xenograft tumors were treated with 0.3 g/kg NAC i.p. and/or 12.5 mg/kg ERL p.o. daily for 2 weeks. Control mice received water p.o. daily for 2 weeks. Data points represent the average values for 6 mice. (F) Immunohistochemical analysis of pEGFR expression (green) in tumors. Tumors were counterstained with ToPro3 (red). Red line shown (in the bottom right corner of each image) represents a magnification scale bar of 100 microns. Values shown (in the bottom left corner of each image) represent quantification of pEGFR fluorescence intensity using image analysis and recognition software, Image J and averaged for 3 animals/group for each treatment group. *, p< 0.05 versus control. **, p<0.05 versus ERL.
Effect of NAC on ERL-induced cytotoxicity in vivo
The in vivo activity of NAC and ERL in FaDu tumor bearing athymic nude mice was examined (Fig. 3E,F). The control and NAC treatment groups demonstrated no difference in tumor growth (Fig. 3E), whereas ERL induced a significant inhibition of growth in FaDu tumors when compared to control or NAC alone (Fig. 3E). The combination of NAC and ERL resulted in tumor growth that was not significantly different from control or NAC treated tumors (Fig. 3E). These results support the conclusion that NAC is able to reverse the tumor growth inhibition induced by ERL in FaDu cells in vivo, which confirms the results seen in FaDu cells in vitro (Fig. 3A). When we analyzed and quantified pEGFR levels using immunohistochemistry in FaDu tumors treated with NAC and/or ERL (3 mice from each group), decreased pEGFR expression was observed in ERL treated tumors, which was reversed with NAC (Fig. 3F) again confirming the results seen in vitro. Because of the small number of tumors (n=3) that were evaluated from each group, the differences in the mean fluorescence intensity values for pEGFR expression (shown in figure 3F) between treatment groups did not reach statistical significance.
Role of Hydrogen peroxide and Superoxide in ERL-induced cytotoxicity
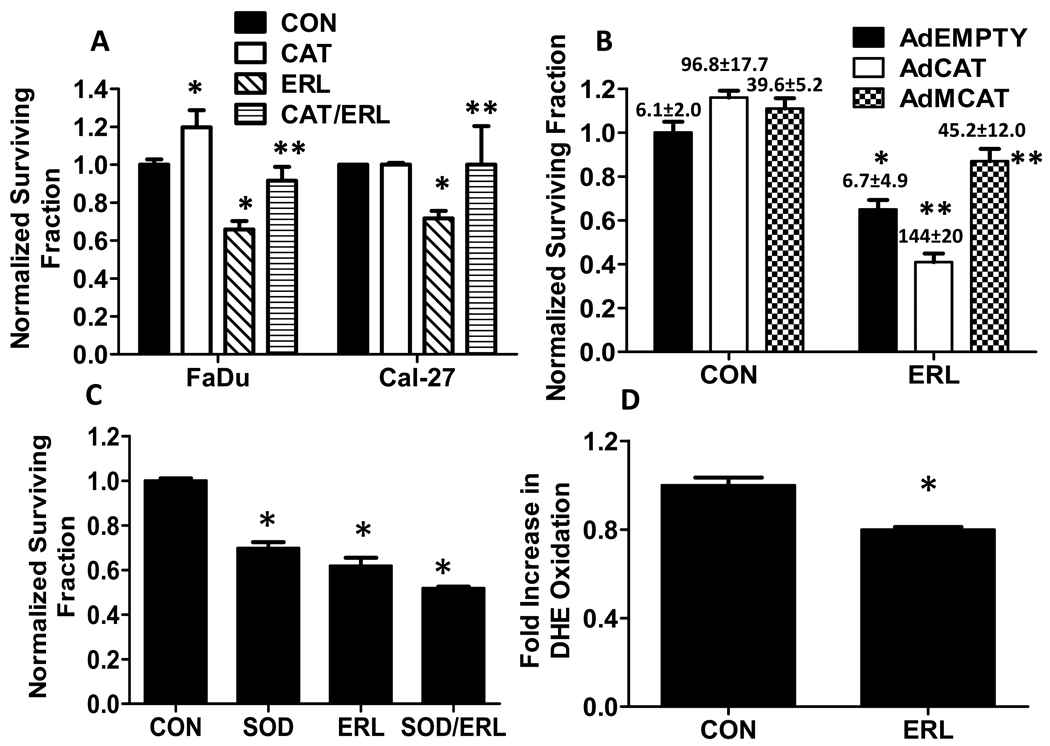
To identify if H2O2 or O2.− was involved in ERL-induced cytotoxicity, FaDu and Cal-27 cells were pretreated for 1 h with 100 U/mL pegylated catalase (CAT) before treatment with ERL for 24 h. CAT significantly reversed ERL-induced cytotoxicity in FaDu and Cal-27 cells (Fig. 4A). Additionally, AdMCAT but not AdCAT significantly increased intracellular CAT activity (values above bars in Fig. 4B) and reversed ERL-induced cytotoxicity (Fig. 4B) in FaDu cells, suggesting that mitochondrial H2O2 may be involved. In contrast, pegylated superoxide dismutase (SOD) was unable to reverse ERL-induced cytotoxicity and increased cell killing when used alone and in combination with ERL (Fig. 4C). DHE oxidation, which is believed to be indicative of O2.− production, was not increased but decreased in the presence of ERL (Fig. 4D). Altogether, the results in figure 4 suggest that H2O2 and not O2.− is involved in ERL-induced cytotoxicity and oxidative stress.
Figure 4. Role of hydrogen peroxide and superoxide in ERL-induced cytotoxicity in HNSCC cells.
(A) FaDu and Cal-27 cells were treated with 10 µM ERL for 24 h with or without treatment with 100 U/mL pegylated catalase (CAT) for 1 h before and during ERL exposure, then analyzed for clonogenic survival. (B) FaDu cells were transiently transduced with adenoviral vectors encoding catalase targeted to the cytosol (AdCAT), catalase targeted to mitochondria (AdMCAT) or empty vector (AdEmpty) then treated with 10 µM ERL for 24 hours before clonogenic survival analysis. Values above bars represent catalase activity measurements in Units/mg protein (U/mg protein). (C) FaDu cells were treated for 24 hours with 10 µM ERL, then analyzed for DHE oxidation. Error bars represent ± 1SD of 3–5 experiments. *, p< 0.05 versus control. **, p<0.05 versus ERL.
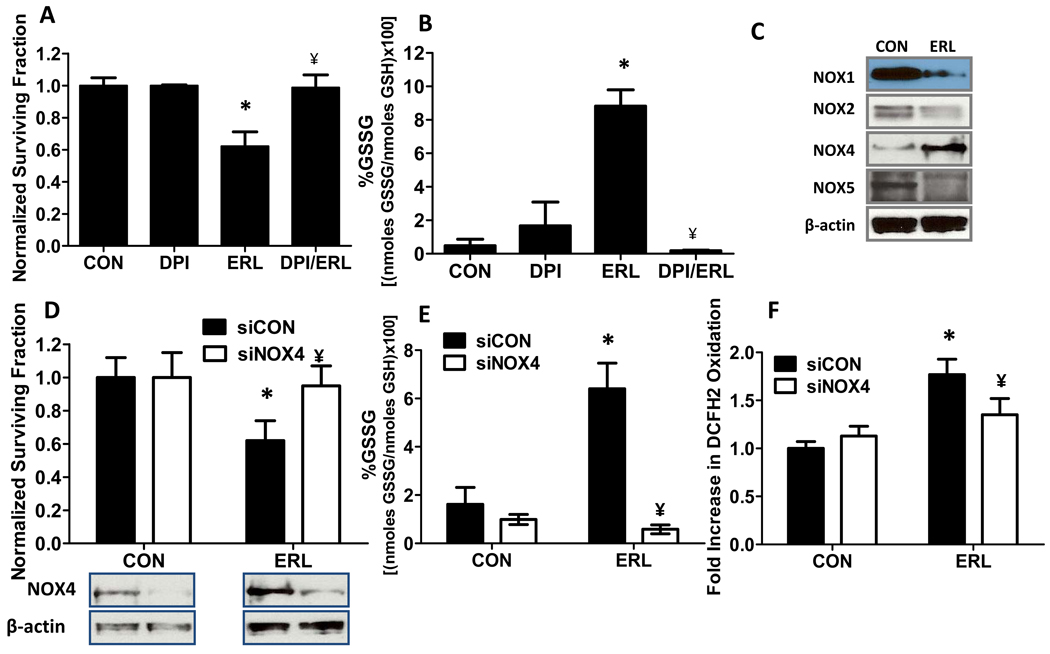
Role of NOX enzymes in Erlotinib-induced cytotoxicity and oxidative stress in FaDu cells
To determine if NOX enzymes were playing a role in ERL-induced cytotoxicity and oxidative stress, FaDu cells were treated with the NOX enzyme inhibitor diphenylene iodonium (DPI) in combination with ERL for 24 h. DPI significantly and completely protected FaDu cells from ERL-induced cytotoxicity (Fig. 5A) and oxidative stress (Fig. 5B), suggesting that ERL-induced oxidative stress may be mediated by NOX enzymes. In efforts to identify which NOX enzymes were involved in ERL-induced oxidative stress, levels of immunoreactive NOX1, NOX2, NOX4 and NOX5 were analyzed by Western blot. NOX3 is not expressed in our HNSCC cell model. Treatment of FaDu cells with ERL for 24 h decreased NOX1, NOX2 and NOX5 expression, however, NOX4 expression was increased (Fig. 5C). Knockdown of NOX4 expression using siRNA protected the cells against ERL-induced cytotoxicity (Fig. 5D) and oxidative stress parameters such as %GSSG (Fig. 5E) and DCFH oxidation (Fig. 5F). Overall, these results support the hypothesis that inhibition of EGFR signaling by ERL leads to down regulation of NOX1, NOX2 and NOX5, and activation of NOX4-mediated ROS production (believed to be H2O2), which causes oxidative stress and cell killing.
Figure 5. Role of NADPH Oxidase (NOX) enzymes in Erlotinib (ERL) induced toxicity and oxidative stress in FaDu cells.
FaDu cells were treated with 10 µM ERL with or without 50 nM diphenylene iodonium (DPI) for 24 h then analyzed for clonogenic survival (A) and percentage glutathione disulfide (%GSSG, B). (C) FaDu cells were assayed by Western blot for Rac1, NOX1, NOX2, NOX4 and NOX5 in the presence or absence of 10 µM ERL for 24 h. (D–F) FaDu cells were transfected with siRNA against NOX4 (siNOX4) or control (siCON), treated with 10 µM ERL for 24 hours then analyzed for clonogenic survival (D), NOX4 expression (D), percentage glutathione disulfide (%GSSG, E) and DCFH oxidation (F). Error bars represent ± 1SD of N = 3 experiments. *, P < .05 versus CON; ¥, P < .05 versus ERL.
Discussion
Metabolic oxidative stress, which is observed in cancer cells, is thought to enhance tumor progression by activating redox regulated signaling pathways involved in cell proliferation, survival and metastasis (3–10). In particular, oxidative stress stimuli have been shown to induce autophosphorylation of EGFR, which confers protection against oxidative stress-induced apoptosis (8–9). Exposure to anticancer agents, has been shown to induce a stress response, which includes activation of EGFR signaling, that may be responsible for the development of resistance to many anticancer agents in tumor cells (27). Therefore, inhibition of EGFR signaling is a logical strategy for enhancing oxidative stress in cancer cells and may prevent development of resistance to conventional chemotherapeutic agents. Here, we have provided several lines of evidence demonstrating that inhibition of EGFR signaling with the tyrosine kinase inhibitor ERL, induces cancer cell killing via enhancing oxidative stress.
EGFR over expression is observed in the majority of HNSCC cells, with the truncated mutant form of EGFR, EGFR variant III (EGFRvIII), being detected in a fraction of HNSCC cases (28). The HNSCC cell lines used in this study, FaDu, Cal-27, and SQ20B all constitutively express the activated form of EGFR (pEGFR, Figure 1A), with SQ20B over expressing EGFRvIII, which also causes its constitutive activation. We observed that ERL inhibited cell growth in all 3 cell lines resulting in G1 arrest (Fig. 1B,C) and clonogenic cell killing (Fig. 1D) which supports previous data with ERL and other EGFR tyrosine kinase inhibitors (16, 29–31).
When the effects of ERL on oxidative stress were analyzed by monitoring glutathione redox states (GSH/GSSG) and DCFH oxidation, ERL was found to induce significant increases in %GSSG (Fig. 2B) and DCFH oxidation (Fig. 2C) in all 3 cell lines compared to control cells. Confirmation that inhibition of EGFR was responsible for ERL-induced oxidative stress was obtained when it was shown that EGFR knockdown using siRNA induced a significant increase in DCFH oxidation in all 3 cell lines (Fig. 2D). We were unable to completely knockdown EGFR expression in SQ20B cells with siRNA. We believe this is due to the high basal levels of EGFR present in SQ20B and/or the presence of mutated EGFR in this cell line. To further support the hypothesis that ERL can induce oxidative stress, the non specific thiol antioxidant NAC was shown to significantly suppress the increase in %GSSG and DCFH oxidation in all 3 cell lines (Fig. 3C,D). Additionally, NAC was able to significantly reverse the cytotoxicity induced by ERL in all the cell lines in vitro (Fig. 3A) and in FaDu cells in vivo (Fig. 3E) suggesting that increased oxidative stress was causally involved in ERL-induced cytotoxicity.
Interestingly, EGFR remained phosphorylated with NAC treatment despite the presence of ERL in FaDu cells in vitro (data not shown) and in FaDu xenografts (Fig. 3F). Possible explanations for this result may be that NAC bound in the active site of EGFR, preventing ERL from binding to the ATP binding site or NAC may be binding to ERL directly. We found that there was no statistically significant difference in the ability of NAC to rescue cells from the toxicity seen when NAC was added 1 h after ERL, relative to when NAC was added 1 h before ERL (data not shown). These results suggest that the direct reaction of ERL with NAC does not seem to completely account for the protective effects of NAC and that some other mechanism (which could include direct binding to EGFR or inhibition of thiol mediated redox signaling) seems to play a role in the toxicity seen with ERL. Although, the role of oxidative stress in ERL-induced cytotoxicity is unclear based solely on the results observed with NAC, we have shown that CAT and overexpression of mitochondrial targeted CAT were able to significantly reverse the effect of ERL (Fig. 4A,B) confirming the role if H2O2-mediated oxidative stress in ERL-induced cytotoxicity and oxidative stress in our HNSCC cell model. Investigating the precise interactions between NAC, EGFR and ERL is beyond the scope of this report, but is currently being investigated.
We next determined if NADPH oxidase (NOX) enzymes could be involved in ERL-induced oxidative stress in FaDu cells. We chose to pursue these experiments in FaDu cells because of our success with this cell line in vivo, and because of the high siRNA transfection efficiency observed in this cell line compared to Cal-27 and SQ20B. NOX enzymes are a family of transmembrane enzymes (NOX1–5, Duox1 and 2) that produce ROS in response to stimuli including growth factors (32). Pretreatment of FaDu cells with the non-specific NOX inhibitor DPI, significantly protected cells from ERL toxicity and oxidative stress (Fig. 5A,B) confirming the general role of NOX enzymes in ERL-induced cytotoxicity and oxidative stress. However, when NOX1, NOX2, NOX4 and NOX5 expression were analyzed in the presence of ERL, NOX1, NOX2 and NOX5 were found to be down regulated while NOX4 expression was increased in FaDu cells (Fig. 5C) suggesting that NOX4 may be mediating the oxidative stress induced by ERL.
To determine if ROS production via NOX4 was responsible for ERL-induced cytotoxicity and oxidative stress, we knocked down NOX4 using siRNA in FaDu cells and found that NOX4 siRNA significantly reversed the cytotoxicity, %GSSG and DCFH oxidation induced by ERL (Fig. 5D–F). These results support the hypothesis that NOX4 plays a major role in the oxidative stress induced by ERL. NOX4 has been found to differ from the other NOX enzymes in that it is the only NOX enzyme that increases fluxes of H2O2 to a greater extent than O2.− (33). Additionally, NOX4 siRNA has been found to decrease H2O2 production but not O2.− production (34) and prevent oxidative stress and apoptosis caused by TNF-α in cerebral microvascular endothelial cells (35).
In our studies, we provide 3 lines of evidence for the involvement of NOX4-mediated H2O2 production in ERL-induced oxidative stress: [1] ERL was able to induce DCFH (Fig. 2C) but not DHE oxidation (Fig. 4D); [2] CAT but not SOD was capable of rescuing cells from ERL-induced cytotoxicity (Fig. 4A–C) and [3] NOX4 knockdown was capable of significantly suppressing the increase in DCFH oxidation induced by ERL (Fig. 5F). Moreover, since DHE oxidation was significantly decreased with ERL treatment (Fig. 4D), and SOD induced HNSCC cell killing alone and in the presence of ERL (Fig 4C), it is possible that EGFR induces O2.− production via NOX1, NOX2 and NOX5 which may activate downstream pro-survival pathways. On the other hand EGFR may suppress NOX4 expression and NOX4-induced H2O2 production probably due to the deleterious intracellular effects of H2O2. Based on our data, we speculate that inhibition of EGFR signaling with ERL downregulates O2.− -induced prosurvival signaling by suppressing NOX1, NOX2 and NOX5 expression and upregulates H2O2 -induced prodeath signaling by increasing NOX4 expression. However, definitive proof for this speculation will be the subject of future work.
Efforts to determine the intracellular location of H2O2 production induced by ERL were attempted by overexpression of CAT targeted to the mitochondria (AdMCAT) and cytosol (AdCAT). Since AdMCAT (and not AdCAT) was able to significantly rescue cells from ERL (Fig. 4B), these data suggest that H2O2 production originates in the mitochondria and perhaps NOX4 is located in the mitochondria. In support of this, studies by Block et al. (36) showed that NOX4 was present in crude and purified mitochondria, was localized with the mitochondrial marker Mitotracker, and Nox4 knockdown reduced NOX activity in pure mitochondria from mesangial cells and kidney cortex (36).
In summary, this report provides clear evidence in HNSCC models supporting the hypothesis that EGFR inhibition with ERL induces clonogenic cell killing via NOX4-mediated H2O2 production. These findings identify a novel mechanism of action for potentially increasing the biological activity observed with the combination of EGFR inhibitors and conventional antineoplastic agents that increase oxidative stress including cisplatin and ionizing radiation. This biochemical rationale also potentially represents a novel therapeutic strategy for reducing cancer cell resistance to therapy commonly seen with EGFR inhibitors in the clinic.
Acknowledgements
Supported by the Doris Duke Charitable Foundation (KPO, in vitro studies), the Department of Radiation Oncology and NIH K01CA134941, R01CA133114, T32CA078586, and P30CA086862. The authors thank the Central Microscopy Research Facilities for their help with confocal microscopy experiments.
References
- 1.Toyokuni S, Okamoto K, Yodoi J, Hiai H. Persistent oxidative stress in cancer. FEBS Lett. 1995;358(1):1–3. doi: 10.1016/0014-5793(94)01368-b. [DOI] [PubMed] [Google Scholar]
- 2.Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991;51(3):794–798. [PubMed] [Google Scholar]
- 3.Bize IB, Oberley LW, Morris HP. Superoxide dismutase and superoxide radical in Morris hepatomas. Cancer Res. 1980;40(10):3686–3693. [PubMed] [Google Scholar]
- 4.Oberley LW, Oberley TD, Buettner GR. Cell differentiation, aging and cancer: the possible roles of superoxide and superoxide dismutases. Med Hypotheses. 1980;6(3):249–268. doi: 10.1016/0306-9877(80)90123-1. [DOI] [PubMed] [Google Scholar]
- 5.Oberley LW, Oberley TD, Buettner GR. Cell division in normal and transformed cells: the possible role of superoxide and hydrogen peroxide. Med Hypotheses. 1981;7(1):21–42. doi: 10.1016/0306-9877(81)90018-9. [DOI] [PubMed] [Google Scholar]
- 6.Pani G, Galeotti T, Chiarugi P. Metastasis: cancer cell's escape from oxidative stress. Cancer Metastasis Rev. 2010;29(2):351–378. doi: 10.1007/s10555-010-9225-4. [DOI] [PubMed] [Google Scholar]
- 7.Weinberg F, Chandel NS. Reactive oxygen species-dependent signaling regulates cancer. Cell Mol Life Sci. 2009;66(23):3663–3673. doi: 10.1007/s00018-009-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, McCullough KD, Franke TF, Holbrook NJ. Epidermal growth factor receptor-dependent Akt activation by oxidative stress enhances cell survival. J Biol Chem. 2000;275(19):14624–14631. doi: 10.1074/jbc.275.19.14624. [DOI] [PubMed] [Google Scholar]
- 9.Fischer OM, Hart S, Gschwind A, Prenzel N, Ullrich A. Oxidative and osmotic stress signaling in tumor cells is mediated by ADAM proteases and heparin-binding epidermal growth factor. Mol Cell Biol. 2004;24(12):5172–5183. doi: 10.1128/MCB.24.12.5172-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prenzel N, Fischer OM, Streit S, Hart S, Ullrich A. The epidermal growth factor receptor family as a central element for cellular signal transduction and diversification. Endocr Relat Cancer. 2001;8(1):11–31. doi: 10.1677/erc.0.0080011. [DOI] [PubMed] [Google Scholar]
- 11.Rocha-Lima CM, Soares HP, Raez LE, Singal R. EGFR targeting of solid tumors. Cancer Control. 2007;14(3):295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 12.Ang KK, Berkey BA, Tu X, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62(24):7350–7356. [PubMed] [Google Scholar]
- 13.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor alpha and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53(15):3579–3584. [PubMed] [Google Scholar]
- 14.Zimmermann M, Zouhair A, Azria D, Ozsahin M. The epidermal growth factor receptor (EGFR) in head and neck cancer: its role and treatment implications. Radiat Oncol. 2006;1:11. doi: 10.1186/1748-717X-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta PR, Maity A. Cellular responses to EGFR inhibitors and their relevance to cancer therapy. Cancer Lett. 2007;254(2):165–177. doi: 10.1016/j.canlet.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang SM, Harari PM. Modulation of radiation response after epidermal growth factor receptor blockade in squamous cell carcinomas: inhibition of damage repair, cell cycle kinetics, and tumor angiogenesis. Clin Cancer Res. 2000;6(6):2166–2174. [PubMed] [Google Scholar]
- 17.Simons AL, Parsons AD, Foster KA, Orcutt KP, Fath MA, Spitz DR. Inhibition of glutathione and thioredoxin metabolism enhances sensitivity to perifosine in head and neck cancer cells. J Oncol. 2009;2009:519563. doi: 10.1155/2009/519563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peus D, Vasa RA, Meves A, Beyerle A, Pittelkow MR. UVB-induced epidermal growth factor receptor phosphorylation is critical for downstream signaling and keratinocyte survival. Photochem Photobiol. 2000;72(1):135–140. doi: 10.1562/0031-8655(2000)072<0135:uiegfr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 19.Spitz DR, Malcolm RR, Roberts RJ. Cytotoxicity and metabolism of 4-hydroxy-2-nonenal and 2-nonenal in H2O2-resistant cell lines: do aldehydic by-products of lipid peroxidation contribute to oxidative stress? Biochem J. 1990;267(2):453–459. doi: 10.1042/bj2670453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson ME. Handbook of Methods for Oxygen Radical Research. Florida: CRC Press Inc; 1985. pp. 317–323. [Google Scholar]
- 21.Griffth OW. Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem. 1980;106(1):207–212. doi: 10.1016/0003-2697(80)90139-6. [DOI] [PubMed] [Google Scholar]
- 22.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 23.Lam EW, Zwacka R, Seftor EA, et al. Effects of antioxidant enzyme overexpression on the invasive phenotype of hamster cheek pouch carcinoma cells. Free Radic Biol Med. 1999;27(5–6):572–579. doi: 10.1016/s0891-5849(99)00109-4. [DOI] [PubMed] [Google Scholar]
- 24.Bai J, Rodriguez AM, Melendez JA, Cederbaum AI. Overexpression of catalase in cytosolic or mitochondrial compartment protects HepG2 cells against oxidative injury. J Biol Chem. 1999;274(37):26217–26224. doi: 10.1074/jbc.274.37.26217. [DOI] [PubMed] [Google Scholar]
- 25.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 26.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30(11):1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 27.Colabufo NA, Contino M, Niso M, Berardi F, Leopoldo M, Perrone R. EGFR tyrosine kinase inhibitors and multidrug resistance: perspectives. Front Biosci. 2011;16:1811–1823. doi: 10.2741/3823. [DOI] [PubMed] [Google Scholar]
- 28.Sok JC, Coppelli FM, Thomas SM, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12(17):5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 29.Sutter AP, Höpfner M, Huether A, Maaser K, Scherübl H. Targeting the epidermal growth factor receptor by erlotinib (Tarceva) for the treatment of esophageal cancer. Int J Cancer. 2006;118(7):1814–1822. doi: 10.1002/ijc.21512. [DOI] [PubMed] [Google Scholar]
- 30.Ling YH, Li T, Yuan Z, Haigentz M, Jr, Weber TK, Perez-Soler R. Erlotinib, an effective epidermal growth factor receptor tyrosine kinase inhibitor, induces p27KIP1 up-regulation and nuclear translocation in association with cell growth inhibition and G1/S phase arrest in human non-small-cell lung cancer cell lines. Mol Pharmacol. 2007;72(2):248–258. doi: 10.1124/mol.107.034827. [DOI] [PubMed] [Google Scholar]
- 31.Huether A, Höpfner M, Sutter AP, Schuppan D, Scherübl H. Erlotinib induces cell cycle arrest and apoptosis in hepatocellular cancer cells and enhances chemosensitivity towards cytostatics. J Hepatol. 2005 Oct;43(4):661–669. doi: 10.1016/j.jhep.2005.02.040. [DOI] [PubMed] [Google Scholar]
- 32.Kamata T. Roles of Nox1 and other Nox isoforms in cancer development. Cancer Sci. 2009;100(8):1382–1388. doi: 10.1111/j.1349-7006.2009.01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serrander L, Cartier L, Bedard K, et al. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem J. 2007;15:406(1):105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dikalov SI, Dikalova AE, Bikineyeva AT, Schmidt HH, Harrison DG, Griendling KK. Distinct roles of Nox1 and Nox4 in basal and angiotensin II-stimulated superoxide and hydrogen peroxide production. Free Radic Biol Med. 2008;45(9):1340–1351. doi: 10.1016/j.freeradbiomed.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basuroy S, Bhattacharya S, Leffler CW, Parfenova H. Nox4 NADPH oxidase mediates oxidative stress and apoptosis caused by TNF-alpha in cerebral vascular endothelial cells. Am J Physiol Cell Physiol. 2009;296(3):C422–C432. doi: 10.1152/ajpcell.00381.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Block K, Gorin Y, Abboud HE. Subcellular localization of Nox4 and regulation in diabetes. Proc Natl Acad Sci U S A. 2009;106(34):14385–14390. doi: 10.1073/pnas.0906805106. [DOI] [PMC free article] [PubMed] [Google Scholar]