Abstract
Sirtuins are NAD-dependent deacetylases that regulate important biological processes. Mammals have seven sirtuins, Sirt1-7. Four of them (Sirt4-7) have no detectable or very weak deacetylase activity. Here we found that Sirt5 is an efficient protein lysine desuccinylase and demalonylase in vitro. The preference for succinyl and malonyl groups was explained by the presence of an arginine residue (Arg105) and tyrosine residue (Tyr102) in the acyl pocket of Sirt5. Several mammalian proteins were identified to have succinyl or malonyl lysine modifications by mass spectrometry. Deletion of Sirt5 in mice appeared to increases the level of succinylation on carbamoyl phosphate synthase 1, a known target of Sirt5. Thus protein lysine succinylation may represent a posttranslational modification that can be reversed by Sirt5 in vivo.
Silent Information Regulator 2 (Sir2) proteins, or sirtuins, are a family of evolutionally conserved enzymes with nicotinamide adenine dinucleotide (NAD)-dependent deacetylase activity (Fig. S1) (1-3). Since the initial discovery of the deacetylase activity (4, 5), many important biological functions of sirtuins have been revealed (2, 3). Based on sequence similarity, sirtuins can be grouped into different classes (6). Mammals have seven sirtuins, Sirt1-7. Among them, only Class I sirtuins (Sirt1-3) have robust deacetylase activity. Sirt4-7 have either no detectable or very weak deacetylase activity (7-11).
To check whether the lack of robust deacetylase activity for Sirt4-7 is due to strict requirement for the peptide sequence, the activities of six human sirtuins (all except Sirt4, which could not be expressed in soluble forms in E. coli) were monitored using 16 different acetyl peptides (Table S1). Under the experimental conditions used, Sirt1-3 and 5 showed deacetylase activity, but Sirt6 and Sirt7 did not. All 16 peptides could be deacetylated by Sirt1-3, while only eight could be deacetylated slowly by Sirt5 (Fig. S2). A histone H3K9 acetyl peptide was one of the best substrates for Sirt1-3 and Sirt5. With this peptide, the kcat and Km of different sirtuins were determined (Table S2). The catalytic efficiency (kcat/Km) of Sirt5 was about 500-fold lower than that of Sirt1.
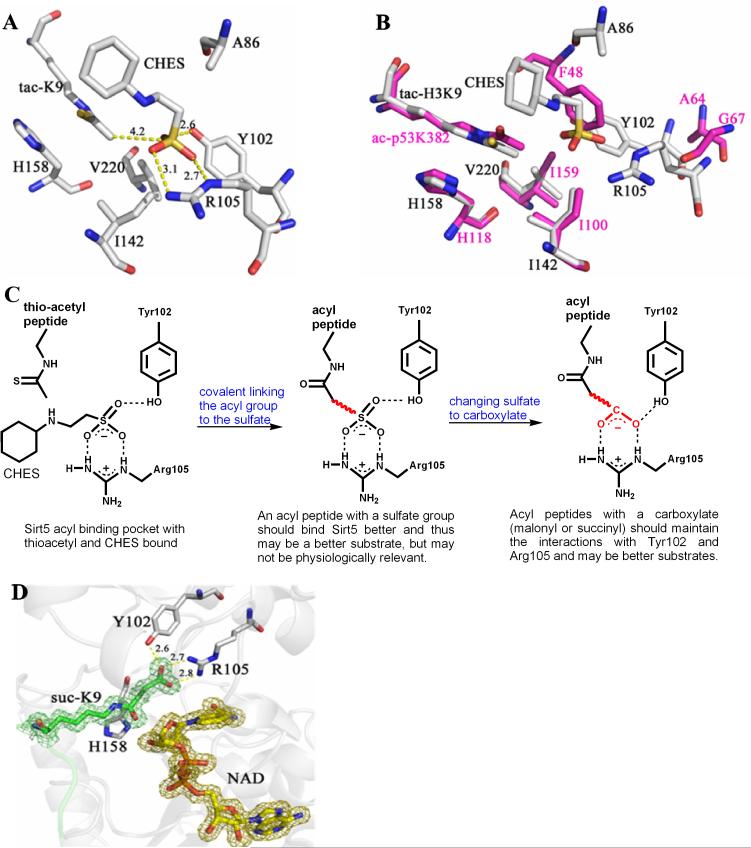
To understand why the deacetylase activity of Sirt5 is weak, a crystal structure of Sirt5 in complex with a thioacetyl peptide was obtained (the corresponding acetyl peptide could not be crystallized with Sirt5). A buffer molecule, CHES (N-cyclohexyl-2-aminoethanesulfonic acid), was also bound to Sirt5 (Fig. 1A and S3A). The interactions between the thioacetyl peptide and Sirt5 involved mostly backbone hydrogen bonding (Fig. S3B), similar to what was observed for Thermotoga maritime Sir2 (Sir2Tm), a sirtuin with robust deacetylase activity (12). Thus, the selectivity for peptide sequences is unlikely to be the major reason for the lack of robust deacetylase activity for Sirt5.
Fig. 1.
The structure of Sirt5 revealed an unusual acyl pocket. (A) The acyl pocket of Sirt5 was partially occupied by the sulfate from the CHES molecule via interactions with Arg105 and Tyr102. The sulfur was 4.2 Å away from the thioacetyl group. (B) Alignment of Sirt5-thioacetyl peptide structure (grey) and Sir2Tm-acetyl peptide structure (PDB 2h4f, magenta). (C) The rationale for predicting that malonyl/succinyl peptides could be better substrates for Sirt5. (D) Sirt5-succinyl peptide-NAD ternary structure showing that the succinyl group interacted with Tyr102 and Arg105.
When the Sirt5 structure was superimposed with the structure of Sir2Tm in complex with an acetyl peptide and NAD (PDB 2h4f) (13) (Fig. 1B), the positions of the thioacetyl lysine in Sirt5 and the acetyl lysine in Sir2Tm were almost identical in the superimposed structures. The acetyl group in the Sir2Tm structure was surrounded by three hydrophobic residues, Phe48, Ile100, and Ile159. In contrast, the corresponding pocket in Sirt5 was larger and bound by CHES. The sulfate group of CHES interacted with Tyr102 and Arg105 of Sirt5 and was ~4 Å away from the thioacetyl group (Fig. 1A). In the reported Sirt5 structure with HEPES bound (9), the sulfate from HEPES also interacted with Arg105 and Tyr102.
Based on the above structural analysis, we reasoned that if the acetyl group was replaced with an acyl group bearing a negatively-charged carboxylate, the acyl peptide should bind Sirt5 better than the acetyl peptide and thus may be a better substrate for Sirt5 (Fig. 1C). In cells, the most common acyl-CoA molecules with a carboxylate group are malonyl-CoA and succinyl-CoA (14). Malonyl-CoA, made from acetyl-CoA by acetyl-CoA carboxylase in the cytosol and the mitochondria, is a precursor for fatty acid biosynthesis (15, 16). Succinyl-CoA is an intermediate in the Krebs cycle in the mitochondria. Since acetyl-CoA is used to modify proteins in cells, it is possible that malonyl- and succinyl-CoA could also be used to modify proteins. Thus, H3K9 malonyl and succinyl peptides were synthesized and tested for hydrolysis by Sirt5.
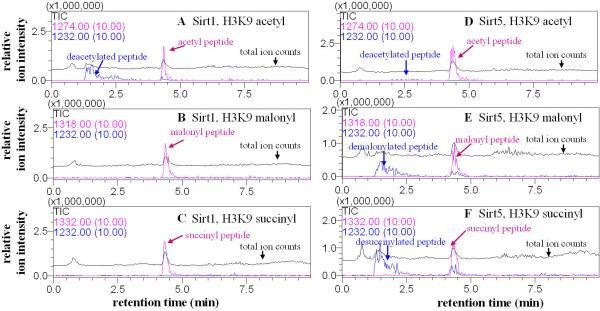
Liquid chromatography–mass spectrometry (LC-MS) was used to monitor the reactions. Sirt1, 2, and 3 catalyzed the hydrolysis of the acetyl peptide, but not the malonyl or succinyl peptide (Fig. 2A-C and S4, S5, S7, S9). In contrast, with Sirt5, little hydrolysis of the acetyl peptide was observed, but the malonyl and succinyl peptides were hydrolyzed significantly (Fig. 2D-F, Fig. S4-S7). The deacylated peptides have identical masses to the synthetic unmodified peptide (Fig. S4 and S9). Sirt6 and Sirt7 had no detectable activity on the acyl peptides under the conditions tested (Fig. S8). Thus, Sirt5 is a desuccinylase and demalonylase.
Fig. 2.
Sirt5 catalyzed the hydrolysis of malonyl and succinyl lysine. The enzymatic reactions were analyzed by LC-MS. Pink traces showed the ion intensities (10× magnified) for the masses of the acyl peptides (acetyl, m/z 1274.0; malonyl, m/z 1318.0; succinyl, m/z 1332.0) and blue traces showed the ion intensities (10× magnified) for the mass of the deacylated peptide (m/z 1232.0). Black traces showed the ion intensity for all masses from 100-2000 (total ion counts or TIC). With Sirt1, hydrolysis was observed for the acetyl peptide (A), but not malonyl (B) or succinyl (C) peptide. With Sirt5, hydrolysis of the acetyl peptide was barely detectable (D) while hydrolysis of the malonyl (E) and succinyl (F) peptides were obvious.
The deacetylation mechanism of sirtuins has been well studied (Fig. S1) (1, 17-20). If Sirt5 uses the same mechanism to catalyze demalonylation and desuccinylation, O-malonyl-ADPR (O-Ma-ADPR) or O-succinyl-ADPR (O-Su-ADPR) should be produced. These products were indeed detected by mass spectroscopy (Fig. S10). As controls, Sirt1 with malonyl and succinyl peptides did not generate these products (Fig. S10, A and B). The formation of O-Ma-ADPR and O-Su-ADPR was also detected using 32P-NAD (Fig. 3). Thus, the mechanism for Sirt5-catalyzed desuccinylation or demalonylation is similar to the deacetylation mechanism of Class I sirtuins.
Fig. 3.
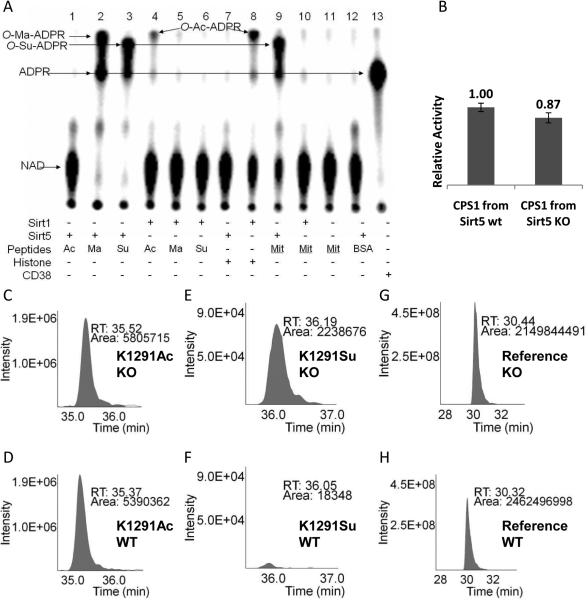
(A) Succinyl lysine was detected in bovine liver mitochondria. Sirt5-catalyzed hydrolysis of malonyl and succinyl peptides could be detected using 32P-NAD, which formed 32P-labeled O-Ma-ADPR (lane 2) and O-Su-ADPR (lane 3). No reaction occurred with acetyl peptide (lane 1). The formation of O-Ac-ADPR catalyzed by Sirt1 was detected (lanes 4 and 8). O-Su-ADPR was formed when bovine liver mitochondria peptides were incubated with Sirt5 (lane 9), but not with Sirt1 (lane 10). The control with BSA peptides and Sirt5 did not generate O-Su-ADPR (lane 12). CD38-catalzyed hydrolysis of NAD was used to generate the standard 32P-ADPR spot (lane 13). (B) The CPS1 activities were measured using the liver lysates from Sirt5 wt and KO mice. (n=3, p<0.05). Relative quantitation analysis was achieved by extracted ion chromatograms (XICs) for peak areas of CPS1 K1291 acetyl peptides from Sirt5 KO (C) and wt mice (D); of CPS1 K1291 succinyl peptides from Sirt5 KO (E) and wt mice (F), and of a reference peptide from Sirt5 KO (G) and wt mice (H).
The kcat and Km values for Sirt5-catalyzed deacetylation, demalonylation, and desuccinylation were determined (Table 1, Fig. S11) with three different peptide sequences (Table S4). With all three peptide sequences, the catalytic efficiencies for demalonylation and desuccinylation were much (29 to >1000 fold) higher than that for deacetylation (Table 1).
Table 1.
The kinetic parameters of Sirt5 on acetyl, malonyl, and succinyl peptides with different sequences.
| Peptide | kcat (s-1) | Km for peptide (μM) | kcat/Km (s-1M-1) | |
|---|---|---|---|---|
| H3 K9 (4-13 + WW*) | deacetylation | ND | ND (>750) | 7.8 |
| demalonylation | 0.037 ± 0.003 | 6.1 ± 2.8 | 6.1 × 103 | |
| desuccinylation | 0.025 ± 0.002 | 5.8 ± 2.7 | 4.3 × 103 | |
| GDH K503 (498-509 + WW*) | deacetylation | ND | ND (>750) | <2** |
| demalonylation | 0.014 ± 0.001 | 8.7 ± 1.3 | 1.6 × 103 | |
| desuccinylation | 0.028 ± 0.002 | 14 ± 4 | 2.0 × 103 | |
| ACS2 K628 (623-632 + WW*) | deacetylation | ND | ND (>750) | 18 |
| demalonylation | 0.079 ± 0.008 | 150 ± 40 | 5.2 × 102 | |
| desuccinylation | 0.268 ± 0.051 | 450 ± 150 | 6.0 × 102 |
Two tryptophan residues were added at the C-terminal of the peptide to facilitate the detection by UV-Vis absorption during the HPLC assay.
No activity. The value was estimated based on the detection limit.
ND: cannot be determined either because no activity was observed or because V versus [S] was linear (even the highest [S] of 750 μM used was much smaller than the Km).
A crystal structure of Sirt5 in complex with a succinyl peptide and NAD was obtained (Table S5). The structure (Fig. 1D) showed that the carboxylate from succinyl interacted with Tyr102 and Arg105, consistent with what was predicted based on the structure of Sirt5 with CHES bound (Fig. 1C). Changing Arg105 to Met or Tyr102 to Phe significantly increased the Km for desuccinylation (Table S3). Thus, Tyr102 and Arg105 were important for binding succinyl and malonyl groups.
We next determined whether lysine malonylation or succinylation exists in vivo. Lysine succinylation was reported to occur on E. coli homoserine trans-succinylase (21), but lysine malonylation has not been reported. We thought that detecting Sirt5-catalyzed formation of O-Ma-ADPR or O-Su-ADPR using 32P-NAD could be a sensitive assay to detect the presence of malonyl or succinyl lysine. H3K9 acetyl, malonyl, and succinyl peptides were incubated with Sirt5 or Sirt1 in the presence of 32P-NAD. The small molecule products were then separated by thin-layer chromatograph (TLC) and detected by autoradiography. With malonyl and succinyl peptides, Sirt5 consumed all the NAD molecules and new spots that corresponded to O-Ma-ADPR and O-Su-ADPR appeared (Fig. 3A). With the acetyl peptide, essentially no NAD was consumed by Sirt5 (Fig. 3A). Incubation of the acetyl peptide or calf thymus histones with Sirt1 produced O-Ac-ADPR (Fig. 3A). The O-Su-ADPR spot was separated from the O-Ac-ADPR and O-Ma-ADPR spots. Thus, Sirt5-catalyzed formation of O-Su-ADPR from the hydrolysis of succinyl peptides could be detected using 32P-NAD.
The 32P-NAD assay was then used to detect whether succinyl lysine was present in bovine liver mitochondrial proteins because Sirt5 is localized in mitochondria (7). When bovine liver mitochondrial peptides were treated with Sirt5 and 32P-NAD, the formation of O-Su-ADPR was detected (Fig. 3A), suggesting that bovine liver mitochondrial proteins contained succinyl lysine. Control reactions with Sirt1, without sirtuins, or with BSA peptides did not produce O-Su-ADPR or O-Ac-ADPR (Fig. 3A). The identity of O-Su-ADPR was further confirmed by LC-MS/MS (m/z 657, [M-2H]-, detected in negative mode, Fig. S12).
To identify succinylated proteins, succinyl peptides from bovine liver mitochondria were affinity purified using a FLAG-tagged Sirt5 (Sirt5-FLAG) and then identified by LC-MS/MS. Three succinylated proteins were identified: HMG-CoA synthase 2, thiosulfate sulfurtransferase, and aspartate aminotransferase. The sites of succinyl modification were identified by MS/MS (Fig. S13). Furthermore, LC-MS/MS identified succinyl lysine from three commercial mitochondrial enzymes purified from animal tissues: glutamate dehydrogenase (GDH), malate dehydrogenase, and citrate synthase (Table S6 and S7). Thus, lysine succinylation occurs on mammalian mitochondrial proteins.
LC-MS/MS of commercial mitochondrial enzymes identified three and two malonyl lysine residues on GDH (Table S6) and malate dehydrogenase (Table S7), respectively. Thus, protein lysine malonylation exists in mammalian cells.
Sirt5 is known to regulate the activity of carbamoyl phosphate synthase 1 (CPS1) (22). We therefore sought to test whether CPS1 was a desuccinylation target of Sirt5. Trypsin-digested CPS1 peptides from bovine liver mitochondria contained succinyl lysine (Fig. S14A). To confirm that Sirt5's desuccinylase functions in vivo, a Sirt5 knockout (KO) mouse strain was generated using standard technology. Consistent with earlier reports (22), CPS1 activity was 15% higher in Sirt5 wt than in Sirt5 KO mice (Fig. 3B). Using MS, we identified three lysine residues of CPS1 that are both acetylated and succinylated: Lys44, Lys287, and Lys1291 (Fig. S17). For Lys44 and Lys287, the levels of acetylation and succinylation did not change significantly in Sirt5 KO mice (Fig S16). For Lys1291, succinylation level increased dramatically in Sirt5 KO mice compared with the level in wt mice (Fig 3, E and F). In contrast, acetylation levels of Lys1291 did not change in Sirt5 KO compared with wt (Fig 3, C and D). Thus protein lysine succinylation may represent a posttranslational modification that can be reversed by Sirt5 in vivo.
Our data suggest that Sirt5 is an NAD-dependent demalonylase and desuccinylase. The demalonylase or desuccinylase activity is much higher than its deacetylase activity. The preference for negatively charged acyl groups can be explained by the presence of Tyr102 and Arg105 in the active site of Sirt5, which are conserved in most Class III sirtuins (Fig. S15) (6). Presumably all Class III sirtuins with the conserved Arg and Tyr should have NAD-dependent desuccinylase and/or demalonylase activity.
We showed that lysine malonylation and succinylation occur on several mammalian proteins. Protein lysine succinylation has been observed recently in bacteria (23). Protein lysine malonylation has not been reported previously.
Acetylation occurs on numerous metabolic enzymes and regulates their activities in mammals and bacteria (24, 25). Given that succinyl-CoA and malonyl-CoA are common metabolites like acetyl-CoA (14), and all the succinylated and malonylated proteins we found are metabolic enzymes, it is likely that protein lysine succinylation and malonylation function similarly to acetylation and regulate metabolism (24, 25).
Supplementary Material
Acknowledgements
This work is supported in part by Dreyfus Foundation (H.L.), NIH R01GM086703 (H.L.), Hong Kong GRF766510 (Q.H.), and NIH RR01646 (R.A.C and Q.H.), NIH PPG DK58920 (J.A.), the EU Ideas program (sirtuins; ERC-2008-AdG-23118, J.A.), and the Ecole Polytechnique Fédérale de Lausanne (J.A.). Atomic coordinates and structure factors were deposited in the Protein Data Bank (accession codes 3RIG and 3RIY).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Supporting Online Material: Materials and Methods; Table S1 to S7, Fig. S1 to S16; references; MS/MS spectra (Fig. S17).
References and Notes
- 1.Sauve AA, Wolberger C, Schramm VL, Boeke JD. Annu. Rev. Biochem. 2006;75:435. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 2.Imai S.-i., Guarente L. Trends Pharmacol. Sci. 2010;31:212. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haigis MC, Sinclair DA. Annu. Rev. Pathol. 2010;5:253. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai S.-i., Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 5.Tanner KG, Landry J, Sternglanz R, Denu JM. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14178. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frye RA. Biochem. Biophys. Res. Commun. 2000;273:793. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 7.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Mol. Biol. Cell. 2005;16:4623. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigis MC, et al. Cell. 2006;126:941. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 9.Schuetz A, et al. Structure. 2007;15:377. [Google Scholar]
- 10.Liszt G, Ford E, Kurtev M, Guarente L. J. Biol. Chem. 2005;280:21313. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 11.Michishita E, et al. Nature. 2008;452:492. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosgrove MS, et al. Biochemistry. 2006;45:7511. doi: 10.1021/bi0526332. [DOI] [PubMed] [Google Scholar]
- 13.Hoff KG, Avalos JL, Sens K, Wolberger C. Structure. 2006;14:1231. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Gao L, et al. J. Chromatogr. B. 2007;853:303. doi: 10.1016/j.jchromb.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Kim K-H. Ann. Rev. Nutr. 1997;17:77. doi: 10.1146/annurev.nutr.17.1.77. [DOI] [PubMed] [Google Scholar]
- 16.Saggerson D. Ann. Rev. Nutr. 2008;28:253. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 17.Sauve AA, et al. Biochemistry. 2001;40:15456. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 18.Smith BC, Denu JM. Biochemistry. 2006;45:272. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith BC, Denu JM. Biochemistry. 2007;46:14478. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- 20.Hawse WF, et al. Structure. 2008;16:1368. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen R, et al. FEBS Letters. 2004;577:386. doi: 10.1016/j.febslet.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. Cell. 2009;137:560. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, et al. Nat. Chem. Biol. 2011;7:58. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao S, et al. Science. 2010;327:1000. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q, et al. Science. 2010;327:1004. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.