Abstract
The spontaneously hypertensive rat (SHR) is a widely used rodent model of hypertension and metabolic syndrome. Previously we identified thousands of cis-regulated expression quantitative trait loci (eQTLs) across multiple tissues using a panel of rat recombinant inbred (RI) strains derived from Brown Norway and SHR progenitors. These cis-eQTLs represent potential susceptibility loci underlying physiological and pathophysiological traits manifested in SHR. We have prioritized 60 cis-eQTLs and confirmed differential expression between the parental strains by quantitative PCR in 43 (72%) of the eQTL transcripts. Quantitative trait transcript (QTT) analysis in the RI strains showed highly significant correlation between cis-eQTL transcript abundance and clinically relevant traits such as systolic blood pressure and blood glucose, with the physical location of a subset of the cis-eQTLs colocalizing with “physiological” QTLs (pQTLs) for these same traits. These colocalizing correlated cis-eQTLs (c3-eQTLs) are highly attractive as primary susceptibility loci for the colocalizing pQTLs. Furthermore, sequence analysis of the c3-eQTL genes identified single nucleotide polymorphisms (SNPs) that are predicted to affect transcription factor binding affinity, splicing and protein function. These SNPs, which potentially alter transcript abundance and stability, represent strong candidate factors underlying not just eQTL expression phenotypes, but also the correlated metabolic and physiological traits. In conclusion, by integration of genomic sequence, eQTL and QTT datasets we have identified several genes that are strong positional candidates for pathophysiological traits observed in the SHR strain. These findings provide a basis for the functional testing and ultimate elucidation of the molecular basis of these metabolic and cardiovascular phenotypes.
Keywords: expression quantitative trait locus, spontaneously hypertensive rat, quantitative trait transcript, sequence variation
the spontaneously hypertensive rat (SHR) strain is a widely used rodent model for the study of hypertension and metabolic syndrome, displaying a range of physiological and metabolic traits that can be related to human disorders including high blood pressure, dyslipidemia and insulin resistance (49, 52). Elucidation of the molecular basis of these traits has proven difficult as they are under the control of multiple genes and genetic loci. The standard approach to gene identification involves mapping by linkage analysis in experimental crosses, and this has led to the localization in the rat genome of hundreds of quantitative trait loci (QTLs) underlying trait variation (68). We refer to these loci as physiological quantitative trait loci (pQTLs). The confidence intervals for such QTLs are often large and contain tens to hundreds of candidate genes meaning that the identification of the underlying genes and genetic variants therefore remains a challenge.
Recombinant inbred (RI) strains represent a type of experimental cross in which traits can be mapped in a panel of inbred strains derived from two progenitor strains (9). Because each animal within an RI strain is genetically identical, repeated measurements can be made in multiple animals from the same strain. In addition, because the inbred lines can be propagated almost unchanged over many years, phenotypic and genetic data generated in the RI panel is cumulative and can be used to infer correlation and linkage between traits generated many years apart in a given RI panel.
The BXH/HXB RI strains were derived from the SHR/OlaIpcv and BN.Lx progenitor strains in the 1980's and have been used to map many SHR phenotypes to the rat genome (1, 3, 11, 46, 47). In addition to the mapping of pQTLs, the BXH/HXB panel have been used to map genetic determinants of gene expression. By measuring genome-wide gene expression using microarrays and treating each expression profile as a quantitative trait, thousands of genetic control points for gene expression, or expression QTLs (eQTLs), have been mapped to the genome in seven tissues of BXH/HXB RI animals (24, 26, 43, 44). A particular advantage of the eQTL study design is that it distinguishes cis- and trans-regulated control of gene expression. Because cis-regulated control of gene expression represents control of gene expression that is regulated by genomic sequence variation within or close to the gene whose expression is being measured, cis-regulated eQTL genes are attractive candidates for pQTLs that map to the same location (26).
Previously, we combined the use of genetic linkage and gene expression profiling to identify the Cd36 gene as a cause of defective insulin action and fatty acid metabolism in the SHR (2, 3). More recently, we have systematically combined genome-wide eQTL data with extensive physiological phenotyping data generated using the BXH/HXB panel in quantitative trait transcript (QTT) studies (42) which identify cis-eQTL transcripts whose expression correlates with a physiological trait, enabling us to identify Ogn as a susceptibility gene for cardiac hypertrophy (44) and renal Cd36 expression as a cause of hypertension (45). Integration of this analysis with other datasets such as pQTL data can successfully identify cis-regulated transcript expression, which correlates with physiological traits and whose gene colocalizes with pQTLs for these same traits. These genes can be considered strong candidates for these pQTLs. We designate such colocalizing, correlated, cis-eQTLs as “c3-eQTLs,” and these are the subject of the present study.
Here, we have used eQTL data from four tissues of BXH/HXB RI strain animals to carry out QTT analysis with 207 (patho)physiological and metabolic traits previously characterized in this RI panel. By combining the results of these studies with data from the recently generated SHR genome sequence (7) we have identified positional candidates for a range of well-characterized SHR phenotypes that provide a basis for prioritized functional testing and ultimate elucidation of the molecular basis of these traits.
MATERIALS AND METHODS
Prioritization of cis-eQTLs.
The generation of eQTL data from adrenal gland, kidney, retroperitoneal fat pad, and soleus muscle from 29 strains (four animals per strain) of the BXH/HXB RI panel have been described previously (24, 26, 43). Microarray expression data was deposited in Array Express (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-AFMX-7 and E-TABM-458. All data for eQTLs with a genome-wide significance (PGW) of P < 0.05, calculated as defined in Refs. 26, 44, were accessed via the eQTL Explorer database and software (39) (http://web.bioinformatics.ic.ac.uk/eqtlexplorer/). Expression data for the adrenal gland, retroperitoneal fat pad, and kidney datasets can also be accessed via the WebQTL database (69) (http://www.webqtl.org/). Transcripts were identified from Affymetrix probe-set IDs using Ensembl (http://www.ensembl.org/Rattus_norvegicus/Info/Index) and the Rat Genome Database (RGD) (http://rgd.mcw.edu/).
Cis-eQTLs were defined as previously, as eQTLs whose genetic control point maps to within 10 Mb of the associated gene (26), and were selected from four tissues that are relevant to the pathogenesis of the metabolic syndrome: adrenal, kidney, retroperitoneal fat, and soleus muscle. These cis-eQTLs were prioritized for further study using a filtering protocol that selected the most robust cis-eQTLs based on criteria relating to expression levels, differential transcript expression between parental strains (44), genome-wide P values and probe-set mapping to unique loci (See Fig. 1). From cis-QTLs that had not been excluded by the above criteria, we selected the 10 most significant cis-eQTLs by allelic P value for each tissue (40 in total). This “allelic P value” represents the PGW for expression differences between strains across the RI strain panel separated by genotype at the peak marker of linkage of the cis-eQTL. Furthermore, we selected 20 additional highly significant cis-eQTLs (allelic P value P < 0.001) with a transcript expression fold change >2.5 or <0.4 (2.5-fold downregulated in SHR) between the parental strains. Affymetrix microarray probe-set locations identified using Ensembl were checked for the presence of sequence variants using the SHR (7) and reference Brown Norway (BN) genome sequences (22). Sequence variants were sequenced with a minimum read depth of 3 and a minimum consensus quality value of 30 (7).
Fig. 1.
Schematic illustration of prioritization of cis-expression quantitative trait loci (eQTLs) for expression validation and quantitative trait transcript analysis from the BHX/HXB-derived multi-tissue eQTL dataset.
Animals and tissues.
BN.Lx/Cub and SHR/Ola rats were housed in an air conditioned animal facility with unrestricted access to standard laboratory chow and water at the Czech Academy of Sciences, Prague, Czech Republic. All procedures were approved by the Ethics Committee of the Institute of Physiology, Czech Academy of Sciences, in accordance with the Animal Protection Law of the Czech Republic (311/1997). All physiological trait measurements were carried out in male rats, unless females were specifically required (e.g., number of offspring etc.). Tissues were harvested from BN.Lx/Cub and SHR/Ola male rats at 6 wk of age as described previously (26).
RNA extraction.
RNA was extracted as described previously (29) except that 50 mg of crushed whole kidney tissue was homogenized, while adrenal gland, fat pad, and skeletal muscle tissues were homogenized whole. Additionally, total RNA was isolated from fat and skeletal muscle tissues using the RNeasy Lipid tissue and RNeasy fibrous tissue kits (Qiagen) respectively in accordance with the manufacturers' instructions. Individual tissues from four animals per strain were used without pooling.
Validation of microarray data by quantitative PCR.
Primers were designed using the Primer3 software package (Supplementary Table S1).1 Reverse transcription and subsequent qPCR were carried out as described previously (29), except reactions were run on an ABI 7900 Fast Real Time PCR system (Applied Biosystems). Appropriate housekeeping genes for normalization controls for each tissue were selected based on equal expression between parental strains in that tissue [β-actin, adrenal and left kidney; HPRT, retroperitoneal fat pad; and GAPDH, soleus muscle (skeletal muscle)] as assayed by quantitative PCR (qPCR). Expression data could not be obtained for seven transcripts due to a combination of low transcript expression and the presence of highly repetitive sequence making the design of suitable primers impossible. These transcripts were omitted from further analysis. Statistical significance was defined in the first instance as P < 0.05 using an uncorrected one-tailed Mann-Whitney test, before Bonferroni correction for multiple testing (53 transcripts tested).
Identification of correlating cis-eQTLs by QTT analysis.
This analysis used the microarray expression array values of cis-eQTLs from 29 RI strains and phenotype measurements for 207 physiological traits (Supplementary Table S2). Physiological and metabolic trait measurements have been reported previously (14, 17, 20, 28, 32, 44, 46, 48, 50, 51, 53, 72).
We assessed association between gene expression levels and phenotypic variation across the BXH/HXB RI strain panel by correlating transcript abundance with the values of physiological traits as described (42). Pearson correlation coefficients (r) and Westfall-Young corrected P values based on a minimum of 1,000 permutations were calculated using MatLab version 6.
Identification of c3-eQTLs.
Physiological QTLs (pQTL) that map to the same loci as cis-eQTLs correlating with physiological traits were identified using the RGD (68). This resource provides information regarding the physiological trait studied, strain combination used, associated linkage statistics, and the genomic coordinates of the pQTL region. For pQTL regions identified from RGD, the original data (Supplementary Table S3) were examined, and the 99% confidence interval [within the 2 logarithm of the odds (LOD) drop from the peak of linkage] was estimated. Cis-eQTLs were classed as colocalizing if the entire cis-eQTL gene falls within this interval. We limited our search to pQTLs defined using either the BXH/HXB RI strain panel or a BN or SHR-derived strain, with a minimum LOD score of 2.4.
Identification of sequence variants in the SHR strain.
Sequence variants arising between the SHR/Olalpcv and BN/SsNHsd/Mcwi rat strains were identified using the recently available SHR (7) and reference BN genome sequences (22). Classification of genetic variants by location relative to the relevant transcript was assigned using gene annotations in the Ensembl database (Rat RGSC 3.4, release 56, Sept. 2009). All sequence variants reported were sequenced with a minimum read depth of 3 and a minimum consensus quality value of 30 (7). We selected 22 sequence variants affecting the coding region and upstream regions for validation by standard capillary sequencing. Primers were designed using the Primer3 software package (Supplementary Table S8), while sequences were analyzed using the Sequencher DNA analysis software (Gene Codes).
Prediction of differential transcription factor binding.
We employed the sTRAP method (36) to identify putative transcription factor (TF) binding sites whose binding affinity may be altered by potential regulatory single nucleotide polymorphisms (SNPs). The binding affinity was computed for both alleles (including 100 bp flanking sequence) using a biophysical model (54) for each SNP and vertebrate TF with a position specific frequency matrix in the TRANSFAC database (38) release 12.1. To compare different TFs with each other we transformed the raw affinities to P values (37). Finally we quantified the change of affinity by the absolute log ratio of the P values of the two alleles. We used a threshold of 1.2 for the absolute log ratio, which was shown previously to yield a high specificity (36). Additionally we required that at least one allele has a high quality motif (min. P < 0.01).
Identification of evolutionarily conserved residues.
Species-specific protein sequences were retrieved from the GenBank sequence repository (http://www.ncbi.nlm.nih.gov/genbank/). Briefly, protein sequence derived from the BN strain for each of four c3-eQTL genes in which nonsynonymous SNPs were found [acyl coenzyme A oxidase 2, branched chain (Acox2); aldo-ketoreductase family 1, member C1 (Akr1c1); chymotrypsin-like elastase family, member 1 (Cela1); and peroxisomal biogenesis factor 11 beta (Pex11b)] were aligned to corresponding human and mouse protein sequences using the bl2seq specialized BLAST sequence alignment tool using default settings (http://blast.ncbi.nlm.nih.gov) (5). GenBank accession numbers for protein sequences used are: Acox2; NP_665713.1 (rat), AAH21339 (mouse) and NP_003491 (human), Akr1c1; BC088227.1 (rat), AC091817.6 (human), Cela1; NP_036684 (rat), NP_291090 (mouse) and NP_001962 (human), Pex11b; NP_001020855 (rat), AAC78661 (mouse) and CAG46844 (human). Amino acids (aa) that are retained between human, mouse, and rat were considered conserved.
RESULTS
Prioritization of cis-eQTLs.
Using the eQTL Explorer database (39), we identified 3217 cis-eQTL transcripts across four rat tissues [adrenal gland, kidney, retroperitoneal fat pad, and soleus (skeletal) muscle] (Table 1). Filtering based on criteria including transcript expression fold change and statistical significance in the parental and RI strains identified 480 transcripts representing the strongest cis-eQTLs (Supplementary Table S4). Of this group, we selected 60 cis-eQTLs for qPCR analysis based on statistical significance and parental fold change (Fig. 1 and Supplementary Table S4).
Table 1.
Cis-acting eQTLs identified in each of four rat tissues at a genome-wide P < 0.01
| Tissue |
|||||
|---|---|---|---|---|---|
| Cis-eQTLs | Adrenal | Fat | Kidney | Skeletal muscle | Total |
| cis-eQTLs, n | 537 | 491 | 600 | 1,589 | 3,217 |
| cis-eQTLs after filtering, n | 69 | 87 | 62 | 262 | 480 |
| Prioritized cis-eQTLs, n | 18 | 14 | 15 | 13 | 60 |
Microarray expression data for adrenal, fat, and kidney tissue were generated using the RAE230A Genechip (Affymetrix) which has probe-sets for 15,923 transcripts. Microarray expression data for skeletal muscle were generated using the RAE230_2 Genechip (Affymetrix), which has probe-sets for 31,099 transcripts. Protocols for filtering and prioritization were carried out as described in materials and methods.
eQTL, expression quantitative trait locus.
qPCR validation of differential expression of cis-eQTL transcripts.
Differences in cis-eQTL transcript expression observed in microarray analyses between parental strains were validated by qPCR analysis. Differential transcript expression between the parental strains (BN.Lx/Cub and SHR/Ola) was confirmed with raw P values <0.05 in 43 of the 60 cis-eQTL transcripts tested (72%) (Fig. 2), of which 36 were in the same direction as observed by microarray (Table 2). All of these transcripts remained statistically significant after correction for multiple testing. Validated transcripts exhibited expression dysregulation of between 1.3- (Trak2) and 11.1-fold (Acox2) between parental strains. Sequence analysis found that 13/60 transcripts contained an SNP that could potentially affect microarray probe binding; however, differential expression was subsequently confirmed by qPCR in eight of these cases (data not shown). No common factor (tissue, expression level, fold change) could be found linking cis-eQTLs whose differential expression could not be validated.
Fig. 2.
Validation by quantitative (q) PCR analysis of differential expression of 53 prioritized cis-eQTL transcripts in the Brown Norway (BN) and spontaneously hypertensive rat (SHR) parental strains for which qPCR assays could be developed. A: cis-eQTLs identified in kidney and skeletal muscle. B: cis-eQTLs identified in adrenal gland and retroperitoneal fat pad. *Raw P value <0.05, **P < 0.01 and ***P < 0.001. All transcript comparisons are shown with expression in the strain showing lower expression normalized to 1. Absolute expression data (microarray) is shown in Supplementary Table S4. Bonferroni corrected P values are shown in Table 2.
Table 2.
Microarray and qPCR expression fold change of prioritized cis-eQTLs with validated differential expression in parental strains
| Affymetrix Probe-set ID | Gene Symbol | Gene Name | Tissue | Microarray Fold Change | Microarray P Value | qPCR Fold Change | qPCR P Value | Corrected P Value |
|---|---|---|---|---|---|---|---|---|
| 1370148_at | Hp | haptoglobin | adrenal | 6.45 | 2.0E-02 | 7.89 | <1.0E-04 | <5.0E-03 |
| 1370065_at | Hpx | hemopexin | adrenal | 4.64 | 1.0E-03 | 6.72 | <1.0E-04 | <5.0E-03 |
| 1372923_at | Pex11b | peroxisomal biogenesis factor 11 beta | adrenal | 2.69 | 1.0E-02 | 1.68 | <1.0E-04 | <5.0E-03 |
| 1368304_at | Fmo3 | flavin containing monooxygenase 3 | adrenal | 2.47 | 9.0E-04 | 2.51 | <1.0E-04 | <5.0E-03 |
| 1388617_at | Bphl | biphenyl hydrolase like | adrenal | 2.23 | 6.0E-03 | 2.96 | <1.0E-04 | <5.0E-03 |
| 1376249_at | Fuco2 | fucosidase 2 | adrenal | 1.72 | 1.0E-02 | 4.42 | <1.0E-04 | <5.0E-03 |
| 1374921_at | Rtel1 | regulator of telomere elongation helicase 1 | adrenal | 1.57 | 7.0E-04 | 2.49 | <1.0E-04 | <5.0E-03 |
| 1373864_at | Map4k4 | mitogen activated protein kinase kinase kinase kinase 4 | adrenal | 0.60 | 7.0E-05 | 0.64 | <1.0E-04 | <5.0E-03 |
| 1398908_at | Stoml2 | stomatin like 2 | adrenal | 0.55 | 1.0E-02 | 0.66 | <1.0E-04 | <5.0E-03 |
| 1388366_at | Mrpl4 | mitochondrial ribosomal protein L4 | adrenal | 0.51 | 6.0E-03 | 0.37 | <1.0E-04 | <5.0E-03 |
| 1389572_at | Me3 | malic enzyme 3 | adrenal | 0.49 | 3.0E-04 | 0.46 | 1.0E-04 | <5.0E-03 |
| 1367986_at | Ptgfrn | prostaglandin F2 receptor negative regulator precursor | adrenal | 0.25 | 9.0E-06 | 0.26 | <1.0E-04 | <5.0E-03 |
| 1370881_at | Tst | thiosulfate sulfurtransferase | adrenal | 0.22 | 4.0E-02 | 0.27 | <1.0E-04 | <5.0E-03 |
| 1387808_at | Slc7a7 | solute carrier family 7 member 7 | fat | 3.30 | 5.0E-03 | 5.38 | <1.0E-04 | <5.0E-03 |
| 1388617_at | Bphl | biphenyl hydrolase like | fat | 1.62 | 2.0E-02 | 2.07 | <1.0E-04 | <5.0E-03 |
| 1388912_at | Rexo4 | RNA exonuclease 4 homolog | fat | 0.59 | 5.0E-04 | 0.42 | <1.0E-04 | <5.0E-03 |
| 1368891_at | Gnpat | glyceronephosphate O-acyltransferase | fat | 0.56 | 1.0E-02 | 0.77 | 1.3E-03 | 0.03 |
| 1388366_at | Mrpl4 | mitochondrial ribosomal protein L4 | fat | 0.49 | 3.0E-04 | 0.37 | <1.0E-04 | <5.0E-03 |
| 1369171_at | Mst1 | macrophage stimulating 1 | fat | 0.21 | 6.0E-04 | 0.15 | <1.0E-04 | <5.0E-03 |
| 1377452_at | Clec3b | C-type lectin domain family 3, member b | fat | 0.02 | 2.0E-03 | 0.29 | <1.0E-04 | <5.0E-03 |
| 1371137_at | Acox2 | acyl coenzyme A oxidase 2, branched chain | kidney | 9.80 | 4.0E-03 | 11.08 | <1.0E-04 | <5.0E-03 |
| 1383303_at | Acsm3 | acyl-CoA synthetase medium chain family member 3 | kidney | 9.00 | 3.0E-04 | 10.39 | <1.0E-04 | <5.0E-03 |
| 1370401_at | Ly6b | lymphocyte antigen 6 complex, locus B | kidney | 5.53 | 1.0E-03 | 3.86 | <1.0E-04 | <5.0E-03 |
| 1387314_at | Sult1b1 | sulfotransferase family cytosolic, 1B, member 1 | kidney | 4.60 | 9.0E-03 | 2.28 | <1.0E-04 | <5.0E-03 |
| 1388366_at | Mrpl4 | mitochondrial ribosomal protein L4 | kidney | 0.52 | 2.0E-03 | 0.46 | <1.0E-04 | <5.0E-03 |
| 1367917_at | Cyp2d2 | cytochrome P450, family 2, subfamily d, polypeptide 2 | kidney | 0.45 | 3.0E-04 | 0.44 | <1.0E-04 | <5.0E-03 |
| 1386985_at | Gstm | glutathione S-transferase, mu | kidney | 0.42 | 7.0E-06 | 0.24 | <1.0E-04 | <5.0E-03 |
| 1368292_at | Dnm1 | dynamin 1 | kidney | 0.37 | 7.0E-06 | 0.20 | <1.0E-04 | <5.0E-03 |
| 1387819_at | Cela1 | chymotrypsin-like elastase family, member 1 | kidney | 0.28 | 9.0E-05 | 0.18 | <1.0E-04 | <5.0E-03 |
| 1370269_at | Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 | kidney | 0.25 | 3.0E-05 | 0.17 | 2.5E-04 | 6.6E-03 |
| 1393902_at | Akr1c1 | aldo-ketoreductase family 1, member C1 | kidney | 0.24 | 3.0E-08 | 0.23 | <1.0E-04 | <5.0E-03 |
| 1370176_at | Trak2 | trafficking protein, kinesin binding 2 | muscle | 2.02 | 4.0E-03 | 1.28 | <1.0E-04 | <5.0E-03 |
| 1385201_at | Tmem182 | transmembrane protein 182 | muscle | 1.57 | 1.0E-02 | 1.85 | <1.0E-04 | <5.0E-03 |
| 1374467_at | Trap1 | TNF receptor associated protein 1 | muscle | 1.56 | 5.0E-03 | 2.35 | <1.0E-04 | <5.0E-03 |
| 1388508_at | Fam32a | family with sequence similarity 32, member a | muscle | 0.58 | 9.0E-03 | 0.65 | <1.0E-04 | <5.0E-03 |
| 1388366_at | Mrpl4 | mitochondrial ribosomal protein L4 | muscle | 0.52 | 2.0E-05 | 0.55 | <1.0E-04 | <5.0E-03 |
Microarray fold change is the mean spontaneously hypertensive rat (SHR) parental strain expression/mean Brown Norway (BN) parental strain expression by microarray. qPCR fold change is the mean SHR parental strain expression/mean BN parental strain expression by qPCR. Corrected P value is the Bonferroni corrected P value.
QTT analysis.
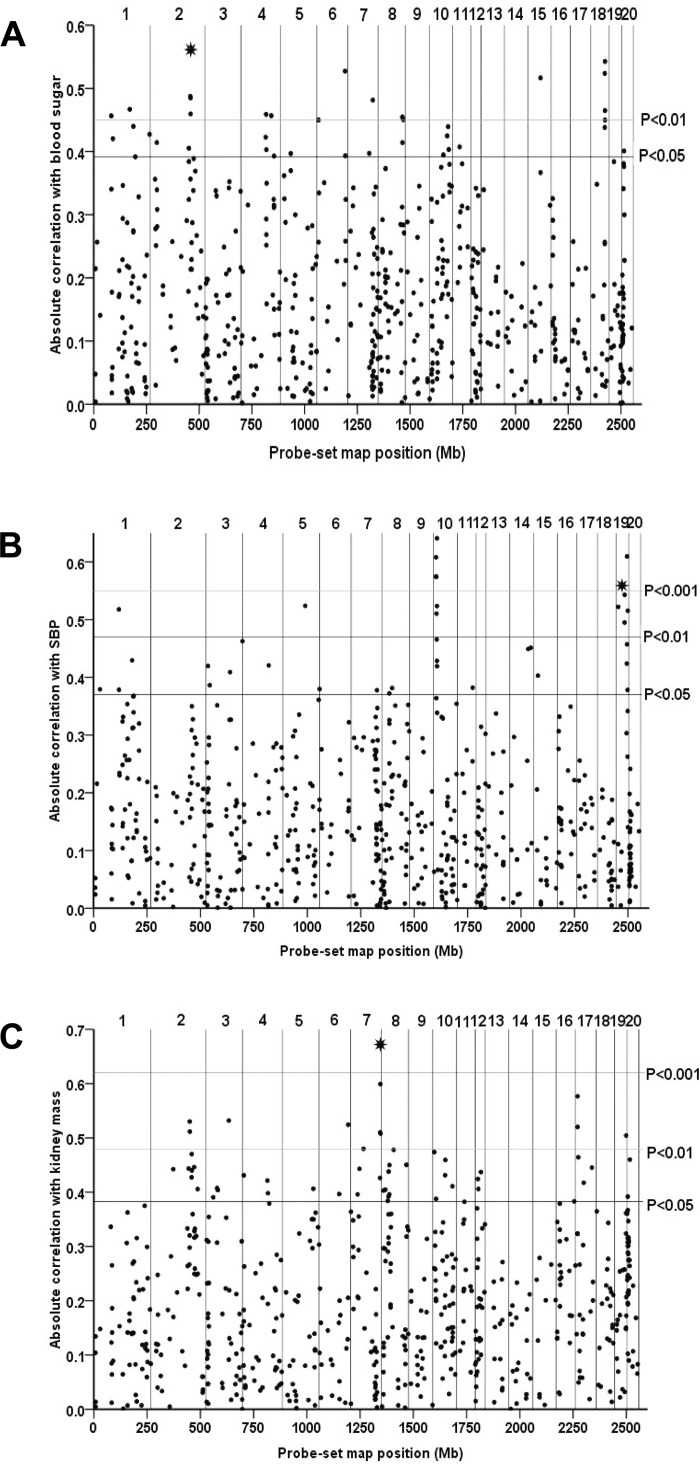
Of the 36 qPCR validated cis-eQTLs, we identified 22 transcripts that significantly correlated (P < 0.01, Westfall-Young P value corrected for multiple testing) with at least one of 207 physiological and metabolic traits measured across the BXH/HXB RI strain panel (Table 3 and Supplementary Table S5). Transcript abundance of these cis-eQTLs was found to correlate with a variety of traits including those relating to blood pressure, cardiac mass, stress response, and response to oxidative stress (Supplementary Table S5). Examples of three of the most significant correlations with associated scatter-plots are illustrated in Fig. 3 and Supplementary Fig. S1. Five cis-eQTLs correlated strongly with several functionally related traits, such as haptoglobin (Hp) and malic enzyme 3 (Me3), which correlate with traits relating to both lipid peroxidation in the kidney and blood pressure (Supplementary Table S5), and thiosulfate sulfurtransferase (Tst), which correlates with both isoproterenol-induced lipolysis and body weight. Transcript abundance of both lymphocyte antigen 6 complex, locus B (Ly6b) and peroxisomal biogenesis factor 11 beta (Pex11b) was found to correlate with multiple insulin and blood glucose traits, with Ly6b expression also correlating with lipolysis in adipocytes (Supplementary Table S5).
Table 3.
Quantitative trait transcript analysis showing the highest correlating trait in the tissue in which the expression trait was observed for each cis-eQTL (n = 22) whose expression correlates highly (P ≤ 0.01) with physiological, metabolic, or cardiovascular traits
| Gene | Chromosome | Tissue in Which Correlation is Found | Trait | Pearson Correlation Coefficient | P Value |
|---|---|---|---|---|---|
| Acox2 | 15 | kidney | night time telemetry DBP after captopril | 0.71 | <1.0E-04 |
| Acsm3 | 1 | kidney | weight of the fat pad relative to body weight | 0.53 | 6.0E-03 |
| Akr1c1 | 17 | kidney | LDL phospholipids in rats at the age of 11 wk in rats fed a high fat, high cholesterol diet for 4 wk | 0.65 | <1.0E-04 |
| Cela1 | 7 | kidney | basal glucose uptake by adipocytes | 0.50 | 1.0E-02 |
| Cyp1a1 | 8 | kidney | HDL3 cholesterol in rats at the age of 7 wk in rats when fed a normal diet | 0.60 | <1.0E-04 |
| Cyp2d2 | 7 | kidney | HDL3 cholesterol in rats at the age of 7 wk in rats when fed a normal diet | 0.57 | 1.0E-03 |
| Dnm1 | 3 | kidney | adrenal dopamine beta hydroxylase (pmol/h/mg protein) | 0.54 | 7.0E-04 |
| Fmo3 | 13 | fat | serum ACTH (corticotropin) levels after 20 min of immobilization stress | 0.54 | 1.0E-02 |
| Fuco2 | 1 | fat | liver cholesterol concentrations in rats fed a high fat, high cholesterol diet from the age of 7–11 wk | 0.65 | 2.0E-04 |
| Gnpat | 19 | skeletal muscle | daytime telemetry SBP after captopril | 0.68 | <1.0E-04 |
| Hp | 19 | adrenal | kidney cortex conjugated dienes (nM/mg protein) | 0.69 | <1.0E-04 |
| Hpx | 1 | adrenal | serum dopamine levels | 0.48 | 8.0E-03 |
| Ly6b | 7 | kidney | lipolysis glycerol levels (mmol/g tissue/2 h) | 0.57 | 9.0E-03 |
| Map4k4 | 9 | adrenal | serum corticosterone levels after 140 min of immobilization stress | 0.71 | 4.0E-04 |
| Me3 | 1 | adrenal | mean arterial pressure (mmHg) | 0.57 | 1.0E-03 |
| Pex11b | 2 | adrenal | blood sugar [120 min during oral glucose tolerance test (fructose diet) (mmol] | 0.56 | 2.0E-03 |
| Rexo4 | 3 | adrenal | hematocrit (proportion of blood volume occupied by red blood cells) at the age of 30 days | 0.83 | 8.0E-03 |
| Rtel1 | 3 | adrenal | LDL cholesterol in rats at the age of 7 wk in rats fed a normal diet | 0.57 | 2.0E-03 |
| Slc7a7 | 15 | fat | serum ACTH (corticotropin) levels after 20 min of immobilization stress | 0.54 | 1.0E-02 |
| Trak2 | 9 | skeletal muscle | insulin stimulated glucose uptake by adipocytes | 0.58 | 3.0E-03 |
| Trap1 | 10 | skeletal muscle | serum corticosterone levels after 80 min of immobilization stress | 0.67 | 1.0E-03 |
| Tst | 7 | adrenal | weight of the fat pad relative to body weight | 0.58 | 2.0E-03 |
A full list of trait names is given in Supplementary Table S2. Full details of all traits that were significantly correlated with a cis-eQTL are given in Supplementary Table S5.
Fig. 3.
Examples of quantitative trait transcript analysis of cis-eQTL genes and physiological and metabolic traits in the rat. Expression profiles of cis-eQTL genes (genome-wide P value ≤ 0.05) across the BXH/HXB recombinant inbred (RI) strains were correlated with values for blood sugar (oral glucose tolerance test, 120 min after glucose) (adrenal cis-eQTL dataset) (A), systolic blood pressure (adrenal cis-eQTL dataset) (SBP) (B), and kidney mass (fat cis-eQTL dataset) (C). For each cis-eQTL, the absolute Pearson correlation coefficient between expression values and phenotype was plotted against the location of the probe set (Mb). Vertical lines mark the physical position of the end of each chromosome. The number of each chromosome is shown above each plot. Horizontal lines indicate empirical significance levels P = 0.05, P = 0.01, and P = 0.001 of the correlations. Prioritized, qPCR validated cis-eQTL genes with transcripts showing strong Pearson correlation coefficients for the respective traits are shown by an asterisk [A: Pex11b (chromosome 2), B: Hp (chromosome 19), C: Cela1 (chromosome 7)].
Identification of c3-eQTLs.
Of the 22 correlating cis-eQTLs, we identified 12 that also colocalized within a pQTL region (99% confidence interval) (Supplementary Table S6). We found seven cis-eQTLs whose expression was found to correlate with blood pressure or related traits [aldo-ketoreductase family 1, member C1 (Akr1c1); chymotrypsin-like elastase family, member 1 (Cela1); cytochrome P450, family 1, subfamily a, polypeptide 1 (Cyp1a1); Hp; Ly6b; Me3; and Tst], which also colocalized with pQTLs for blood pressure or renal function. Furthermore, our analysis demonstrated that Ly6b and Tst as well as an additional five cis-eQTLs [acyl coenzyme A oxidase 2, branched chain (Acox2); cytochrome P450, family 2, subfamily d, polypeptide 2 (Cyp2d2); hemopexin (Hpx); Pex11b; and trafficking protein, kinesin binding 2 (Trak2)] correlated with fat metabolism or blood sugar-related traits, while colocalizing with pQTLs underlying differences in lipid levels, blood sugar levels, and/or body weight. Such c3-eQTL transcripts are strong candidate genetic determinants of these (patho)physiological traits.
Identification of sequence variation in c3-eQTL genes.
We used the SHRbase sequence database (7) to identify sequence variants with the potential to affect the expression and/or function of cis-eQTL transcripts. We detected sequence variation in the upstream and downstream noncoding regions in all 12 c3-eQTL genes (Table 4 and Supplementary Table S7, a and b). We successfully confirmed 90.9% (20/22) of the SNPs selected for validation by capillary sequencing (See Supplementary Table S9). Of the two SNPs that did not validate, one (Cela1 SNP2) was found not to vary between the parental strains, while the second (Trak2 SNP1) appeared to constitute part of a small deletion. On analysis of upstream region SNPs we identified 33 potential SNP/TF interactions in nine c3-eQTL genes, where predicted TF binding strength was changed significantly by an SNP between the SHR and BN strains (|log ratio| > 1.2) (Table 5). This included one c3-eQTL gene (Hp) that was found to carry a polymorphism affecting a putative binding site for a TF family known to play a role in its transcriptional regulation (CEBP) (40, 41). Three c3-eQTLs (Acox2, Trak2, and Pex11b) also show sequence variation affecting splice sites, potentially leading to aberrant splicing or altered transcript stability and protein function. Significantly, nonsynonymous coding region SNPs were identified in four of the 12 c3-eQTLs (Acox2, Akr1c1, Cela1, and Pex11b) (Table 4). Analysis of the corresponding protein sequences by the BLAST sequence alignment program (bl2seq) showed that several of these SNPs (Supplementary Table S7a) affect aa residues that are conserved between humans and rodents [ACOX2-aa 546, AKR1C1-aa 250, CELA1-aa 21, and PEX11b-aa 113 and 231]. Furthermore, small insertions and deletions (up to 7 bp) were found in 10 of the c3-eQTLs; however, none localized to either coding regions or to putative TF binding sites, and their functional significance remains unknown (Supplementary Table S7b).
Table 4.
Distribution of SHR sequence variants in c3-eQTL genes
| Flanking Region |
Transcript |
Intronic |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Upstreama | Downstreama | 5′-UTRb | Splice Sitec | Silentd | Nonsynonymouse | 3′-UTRf | Intron |
| Tst | 30 | 19 | 0 | 0 | 2 | 0 | 0 | 30 |
| Hp | 32 | 2 | 0 | 0 | 0 | 0 | 0 | 1 |
| Me3 | 3 | 30 | 0 | 0 | 1 | 0 | 0 | 142 |
| Acox2 | 23 | 9 | 0 | 1 | 5 | 2 | 0 | 84 |
| Ly6b | 21 | 9 | 0 | 0 | 0 | 0 | 1 | 19 |
| Cyp1a1 | 10 | 15 | 0 | 0 | 1 | 0 | 1 | 9 |
| Akr1c1 | 8 | 15 | 0 | 0 | 0 | 1 | 3 | 37 |
| Pex11b | 8 | 10 | 0 | 2 | 0 | 2 | 2 | 27 |
| Cela1 | 15 | 3 | 0 | 0 | 2 | 2 | 0 | 47 |
| Trak2 | 7 | 3 | 1 | 1 | 1 | 0 | 1 | 111 |
| Hpx | 4 | 5 | 1 | 0 | 0 | 0 | 1 | 17 |
| Cyp2d2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
Region located within 5 kb upstream of the transcription initiation site or 5 kb downstream of transcript end.
Number of sequence variants in the 5′-untranslated region (UTR).
Number of sequence variants occurring in splice sites, i.e., the region located 1-3 bp into exon or 1-8 bp into an intron.
Number of sequence variants located in coding region that do not affect amino acid sequence.
Number of sequence variants located in coding region that do affect amino acid sequence.
Number of sequence variants in the 3′-UTR. c3-eQTL, colocalizing, correlated cis-eQTL.
Table 5.
Putative transcription factor binding sites altered by 5′-UTR and regulatory region SNPs
| Affymetrix probe-set ID | c3-eQTL | SNPa | Distance from TSSb | Predicted Transcription Factor Site | P BNc | P SHRd | Ratio (BN/SHR)e | Log Ratiof |
|---|---|---|---|---|---|---|---|---|
| 1393902_at | Akr1c1 | T>A | −1716 | Hmgiγ | 7.90E-04 | 3.10E-02 | 0.026 | −1.59 |
| 1387819_at | Cela1 | T>C | −1747 | Xpf1 | 2.80E-03 | 4.90E-02 | 0.056 | −1.25 |
| 1387819_at | Cela1 | A>T | −2463 | Pax | 8.70E-03 | 1.80E-01 | 0.049 | −1.31 |
| 1370269_at | Cyp1a1 | A>G | −563 | Ahr | 2.50E-04 | 8.80E-06 | 28.4 | 1.45 |
| 1370269_at | Cyp1a1 | C>T | −1735 | Spz1 | 1.30E-03 | 8.50E-02 | 0.015 | −1.83 |
| 1370148_at | Hp | G>T | −1735 | Gata1 | 2.90E-01 | 2.10E-03 | 137.9 | 2.14 |
| 1370148_at | Hp | G>T | −2882 | Gata1 | 2.40E-01 | 1.80E-03 | 135 | 2.13 |
| 1370148_at | Hp | A>T | −3293 | Gata1 | 2.10E-01 | 2.40E-03 | 88.5 | 1.95 |
| 1370148_at | Hp | A>T | −3293 | Lmo2com | 2.40E-01 | 3.60E-03 | 68.1 | 1.83 |
| 1370148_at | Hp | A>T | −3293 | Gata2 | 3.30E-01 | 4.90E-03 | 67.2 | 1.83 |
| 1370148_at | Hp | G>A | −79 | Cebp | 8.70E-02 | 2.00E-03 | 43.1 | 1.63 |
| 1370148_at | Hp | G>T | −3789 | Pax | 3.30E-04 | 5.40E-03 | 0.061 | −1.21 |
| 1370148_at | Hp | C>T | −2290 | Taxcreb | 2.00E-03 | 5.10E-02 | 0.038 | −1.41 |
| 1370148_at | Hp | A>T | −3293 | Cdpcr3 hd | 5.00E-03 | 2.70E-01 | 0.018 | −1.73 |
| 1370148_at | Hp | A>T | −3293 | Pbx1 | 1.50E-03 | 1.50E-01 | 0.01 | −1.99 |
| 1370065_at | Hpx | A>G | −4986 | Oct1 | 7.20E-03 | 1.30E-01 | 0.058 | −1.24 |
| 1370401_at | Ly6b | A>G | −4396 | Atf4 | 3.60E-01 | 8.70E-03 | 41.3 | 1.62 |
| 1370401_at | Ly6b | T>C | −3505 | Sry | 1.40E-03 | 4.00E-05 | 36.3 | 1.56 |
| 1370401_at | Ly6b | T>C | −3505 | Creb | 6.70E-02 | 1.90E-03 | 35.8 | 1.55 |
| 1370401_at | Ly6b | A>G | −2224 | E4F1 | 2.20E-01 | 7.90E-03 | 27.3 | 1.44 |
| 1370401_at | Ly6b | A>G | −2224 | Crebp1 | 3.10E-02 | 1.20E-03 | 25.7 | 1.41 |
| 1370401_at | Ly6b | A>G | −2224 | Creb | 9.80E-02 | 4.20E-03 | 23 | 1.36 |
| 1370401_at | Ly6b | G>A | −3866 | Hnf4a | 5.50E-02 | 3.20E-03 | 17.4 | 1.24 |
| 1370401_at | Ly6b | A>G | −2224 | Barbie | 7.60E-03 | 2.40E-01 | 0.031 | −1.50 |
| 1370401_at | Ly6b | A>C | −1048 | Titf1 | 3.80E-03 | 2.30E-01 | 0.016 | −1.78 |
| 1389572_at | Me3 | T>A | −3512 | Nkx3A | 4.20E-02 | 2.20E-03 | 18.8 | 1.27 |
| 1372923_at | Pex11b | G>A | −554 | Rfx | 3.80E-01 | 4.00E-03 | 97 | 1.99 |
| 1370881_at | Tst | T>C | −3925 | E2F1 | 2.50E-01 | 8.70E-03 | 28.7 | 1.46 |
| 1370881_at | Tst | T>C | −580 | Cp2 | 1.50E-02 | 6.40E-04 | 23.9 | 1.38 |
| 1370881_at | Tst | C>A | −1907 | AP2rep | 5.80E-02 | 2.80E-03 | 20.7 | 1.32 |
| 1370881_at | Tst | T>C | −1192 | CACCC binding factor | 1.10E-01 | 5.90E-03 | 17.9 | 1.25 |
| 1370881_at | Tst | T>C | −1192 | Cdpcr3 | 8.4E-03 | 1.5E-01 | 0.055 | −1.26 |
| 1370881_at | Tst | T>A | −161 | AP2rep | 3.2E-04 | 8.2E-03 | 0.039 | −1.41 |
Single nucleotide polymorphism (SNP) observed in SHR compared with BN reference sequence.
Distance of SNP from transcription start site (TSS) in Ensembl (http://www.ensembl.org/Rattus-norvegicus/Info/Index).
Normalized measure of binding affinity of the transcription factor to the putative site in BN.
Normalized measure of binding affinity of the transcription factor to the putative site in SHR.
The ratio of binding affinity of the transcription factor between the strains (BN/SHR).
Log10 transformed ratio (BN/SHR).
DISCUSSION
Advancing our understanding of the genetic susceptibility to common health problems such as high blood pressure, obesity, and insulin resistance could lead to significant improvements in their detection and treatment. We have studied traits related to a variety of common disease phenotypes in the rat model. We combined the analysis of transcript expression, physiological phenotyping, and sequence datasets to identify candidate genes for pathophysiological traits in the SHR, and sequence variants with the potential to underlie differential transcript expression and altered gene function of these candidates. This could provide insight into the molecular basis underlying not just cis-regulation of transcript abundance, but that of downstream physiological or metabolic traits.
After filtering microarray expression data in four tissues (Fig. 1), we selected 60 of the most robust cis-eQTLs and validated differential expression in the SHR and BN parental strains for 36 of these (Fig. 2). No common factor could be identified between prioritized transcripts that did not validate. Of this subset, we identified 22 cis-eQTL transcripts whose expression across the RI strains correlated significantly with measurements of physiological traits relating to disorders manifested in the SHR model (Fig. 3, Table 3, and Supplementary Table S5). Three highly correlating transcripts, Cela1, Hp, and Pex11b, exhibited Pearson correlation coefficients (R) between expression and physiological trait measurement of between 0.56 and 0.67, strongly suggesting a correlation between transcript expression and the individual physiological trait measurements, identifying these correlating cis-eQTLs as attractive candidates for the correlated trait (Supplementary Fig. S1). Incidences where QTT analysis identifies several closely localizing correlating cis-eQTLs are often the consequence of a correlated strain distribution pattern. Previous studies have already demonstrated a significant relationship between the absolute correlation coefficient of expression of eQTL pairs residing on the same chromosome and the distance between them (34, 56). Likewise, pair-wise correlation analysis of our cis-eQTL dataset found that >80% of the correlation between expression of individual cis-eQTL transcripts is explained by the shared genotypes of cis-eQTL genes at tightly linked loci (23). We found 12 of the correlating cis-eQTLs to colocalize with pQTLs previously mapped using either the RI strain panel or a BN or SHR strain. These 12 c3-eQTLs represent strong candidate genes for the associated traits, showing high genome-wide statistical significance of linkage, high differential expression between strains, and strong correlations to the traits in question. Seven of these c3-eQTLs, Akr1c1; Cyp1a1; Ly6b; and Tst (correlations found in kidney); Cela1 (fat and kidney); and Hp and Me3 (adrenal) correlate with blood pressure-related traits. Those identified in kidney are of particular interest given the role played by the kidney in the regulation of blood pressure. In addition, transcript levels of Hp correlate with lipid peroxidation traits in the kidney. Although the Hp expression phenotype occurs in a different tissue (adrenal) to the correlating physiological trait (kidney), this finding could potentially be a reflection of a more global role for this secreted plasma protein in the prevention of oxidative stress and tissue lipid peroxidation (Supplementary Table S5).
Likewise, we have shown that Tst (in adrenal) and Trak2 (skeletal muscle) transcript levels correlate with traits associated with fat metabolism and colocalize with pQTLs for lipid level and body weight, while Hpx (adrenal) and Pex11b (fat) represent c3-eQTLs relating to blood sugar regulation and traits recognized as downstream consequences of abnormal blood sugar regulation (4, 10, 55, 64) (Supplementary Table S6). These c3-eQTLs are particularly striking given that they also occur in tissues that are physiologically relevant for the control of these traits, suggesting that they represent strong candidate genes underlying these phenotypes in the SHR. The correlations of certain transcripts with traits associated with fat metabolism and body weight (Acox2 and Cyp2d2, both in kidney) and blood pressure/kidney mass (Me3, adrenal) did not occur in tissues normally associated with the correlating trait. These associations could indicate additional as yet unknown mechanisms of control, possibly as a consequence of the transcript forming cis-eQTLs in a number of tissues (Supplementary Table S4), not all of which were prioritized for expression validation and QTT analysis. A possible example of this is Cela1 whose expression correlates with glucose uptake in adipocytes despite being prioritized as a cis-eQTL in kidney. Further analysis of the entire cis-eQTL dataset found that Cela1 is also a cis-eQTL in fat tissue, albeit below our prioritization threshold.
The availability of the SHR genome sequence has enabled the analysis of sequence variation between the SHR and BN strains on a genome-wide scale. The high sequence concordance between BN substrains (60) meant that we were able to successfully validate 90.9% of selected SNPs. We investigated sequence changes that disrupt TF binding sites (TFBSs) that could represent candidate quantitative trait nucleotides (QTNs) underlying c3-eQTL expression traits. We found several SNPs in the upstream regions of nine c3-eQTLs that are predicted to disrupt TFBSs (Table 5). Eight c3-eQTL genes have polymorphisms affecting the coding region, four of which have nonsynonymous changes potentially affecting protein activity and stability. Five of these nonsynonymous SNPs affecting four c3-eQTLs (Acox2, Akr1c1, Cela1, and Pex11b) lead to substitution of highly conserved aa residues (ACOX2, aa 546 Y>H; AKR1C1, aa 250 W>R; CELA1, aa 21 P>A; and PEX11b, aa 113 R>H and aa 231 P>R) (Table 4 and Supplementary Table S7a), suggesting likely effects on protein activity. Furthermore, none of the 12 c3-eQTLs localized to regions identified as being subject to copy number variation in the SHR (18), strengthening the view that sequence variation rather than copy number variation close to or within dysregulated transcripts is the major cause of differential expression observed in cis-eQTL genes.
Although the c3-eQTLs represent a diverse group of genes with a range of different functions, we noted that several c3-eQTLs [Akr1c1 (16), Cyp1a1 (19, 61), Hp (6, 15), Hpx (66), Me3 (59, 70), Pex11b (58), and Tst (31)] have previously been demonstrated to play a role in oxidative stress pathways, suggesting they may be important factors controlling the extent of tissue and DNA damage in response to oxidative stress. This link is of interest given that oxidative stress has been linked to hypertension, cardiovascular disease, and Type 2 diabetes, among other pathological states (25). Indeed, the SHR parental strain has been shown to exhibit reduced endogenous antioxidant capabilities compared with other rat strains, with studies showing increased DNA and protein oxidation in SHR compared with WKY (12, 30, 62, 71). This suggests that an increased knowledge of the molecular mechanisms underlying reduced antioxidant capacity is of functional relevance.
For example, the IL-6-responsive acute-phase gene Hp (41) has been shown along with another c3-eQTL Hpx to prevent hemoglobin-mediated oxidative damage by promoting clearance of hemoglobin released as a consequence of red blood cell turnover or hemolysis (6, 15) via scavenger receptors (8, 27, 57). The Hp locus is polymorphic in humans, with variants found to show evidence of association with increased risk of cardiovascular disease in diabetic patients (6). Hp is considered a susceptibility factor for atherosclerosis in diabetic patients and has also been shown to reduce hemoglobin-mediated hypertension in other species (13). Human Hp promoter polymorphisms disrupting CEBPβ sites have been shown to significantly reduce transcriptional activation in response to IL-6 in human cell lines (41, 63). Here we show that Hp transcript levels in adrenal correlate with kidney oxidation traits and blood pressure, while the gene itself colocalizes with pQTLs for blood pressure (Table 3, Supplementary Tables S5 and S6). Expression of both Hp and Hpx is increased in SHR, suggesting that dysregulation of this mechanism may serve to act as a protective factor attenuating the oxidative stress response in this strain. We identified several disrupted putative TFBSs upstream of the Hp gene, including an SNP affecting a putative CEBP family binding site in SHR (Table 5). This finding is of note due to an earlier report identifying the CEBP family protein, CEBPβ, as a key TF in the control of Hp transcript levels in humans (40, 41), suggesting that SNPs within the Hp upstream region represent excellent candidate QTNs that regulate Hp expression and potentially the development of downstream physiological traits, though verification of the effects of these SNPs will entail functional analysis.
Analysis of sequence variants in c3-eQTL upstream regions showed an SNP that disrupts a putative PAX binding motif in the Cela1 gene whose expression in fat and kidney correlates with kidney mass traits and colocalizes with QTLs for blood pressure-related traits (Table 5 and Supplementary Tables S5 and S6). Although the correlation of Cela1 expression with kidney mass was detected primarily in the fat cis-eQTL dataset, this transcript was also found to correlate with this trait in the kidney cis-eQTL dataset, albeit at a lower level of statistical significance (P = 0.01), suggesting a role for this gene in the control of kidney mass traits. Members of the PAX TF family (Pax2 and Pax5) have been shown to be involved in the regulation of kidney development (21, 67) and repair (33). This raises the possibility that Cela1 may be a downstream target by which PAX TFs mediate these processes, with disruption of its expression leading to aberrant kidney mass and development. Indeed, alterations in the renal elastin-elastase system, leading to increased expression of elastins, have been linked to Type I diabetic nephropathy in mice and humans (65). Furthermore, Cela1 also contains two missense coding region SNPs, affecting codons 21 and 160 (See Supplementary Table S7a). The P21A change occurs in a residue that is conserved between humans and rodents, close to the cleavage site of the immature Cela1 proprotein product (35), potentially affecting transcript stability or protein function. This provides an additional means by which sequence variation at the Cela1 locus may contribute to the development of the observed expression and physiological traits. These observations, while suggestive, require confirmation in future functional studies.
In this study we have combined the use of transcript expression data, physiological trait measurements and the recently assembled SHR genome sequence along with curated data resources such as TRANSFAC to prioritize novel candidate genes underlying physiological traits manifested in the SHR strain. By integrating these resources, we have identified genes that not only are highly correlated with relevant physiological traits across the BXH/HXB RI strains and located within previously mapped QTL regions in this RI strain panel but also contain sequence variants that represent a plausible genetic basis for the differential expression observed and could affect downstream physiological and metabolic traits. While further studies would be required to confirm the physiological significance of the transcripts and sequence variants identified, the increasing availability of knockout and transgenic animal models makes studies a plausible next step. Indeed, knockout mice are available for a number of c3-eQTLs identified; however, these have yet to be tested for a relevant phenotype. These data therefore provide a starting point for systematic analysis of functional variants that potentially underlie these traits in the SHR model.
GRANTS
This work was primarily supported by intramural funding from the MRC Clinical Sciences Centre, by the Imperial College BHF Centre of Research Excellence, by a Wellcome Trust studentship (069962/Z/02/Z) to I. C. Grieve, and by the Grant Agency of the Czech Republic (grant 301/08/0166) and the Ministry of Education of the Czech Republic (grant ME10019) (M. Pravenec).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Aitman TJ, Critser JK, Cuppen E, Dominiczak A, Fernandez-Suarez XM, Flint J, Gauguier D, Geurts AM, Gould M, Harris PC, Holmdahl R, Hubner N, Izsvak Z, Jacob HJ, Kuramoto T, Kwitek AE, Marrone A, Mashimo T, Moreno C, Mullins J, Mullins L, Olsson T, Pravenec M, Riley L, Saar K, Serikawa T, Shull JD, Szpirer C, Twigger SN, Voigt B, Worley K. Progress and prospects in rat genetics: a community view. Nat Genet 40: 516–522, 2008. [DOI] [PubMed] [Google Scholar]
- 2.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St, Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet 21: 76–83, 1999. [DOI] [PubMed] [Google Scholar]
- 3.Aitman TJ, Gotoda T, Evans AL, Imrie H, Heath KE, Trembling PM, Truman H, Wallace CA, Rahman A, Dore C, Flint J, Kren V, Zidek V, Kurtz TW, Pravenec M, Scott J. Quantitative trait loci for cellular defects in glucose and fatty acid metabolism in hypertensive rats. Nat Genet 16: 197–201, 1997. [DOI] [PubMed] [Google Scholar]
- 4.Alberti KG, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, Fruchart JC, James WP, Loria CM, Smith SC., Jr Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 120: 1640–1645, 2009. [DOI] [PubMed] [Google Scholar]
- 5.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asleh R, Marsh S, Shilkrut M, Binah O, Guetta J, Lejbkowicz F, Enav B, Shehadeh N, Kanter Y, Lache O, Cohen O, Levy NS, Levy AP. Genetically determined heterogeneity in hemoglobin scavenging and susceptibility to diabetic cardiovascular disease. Circ Res 92: 1193–1200, 2003. [DOI] [PubMed] [Google Scholar]
- 7.Atanur SS, Birol I, Guryev V, Hirst M, Hummel O, Morrissey C, Behmoaras J, Fernandez-Suarez XM, Johnson MD, McLaren WM, Patone G, Petretto E, Plessy C, Rockland KS, Rockland C, Saar K, Zhao Y, Carninci P, Flicek P, Kurtz T, Cuppen E, Pravenec M, Hubner N, Jones SJ, Birney E, Aitman TJ. The genome sequence of the spontaneously hypertensive rat: Analysis and functional significance. Genome Res 20: 791–803, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachli EB, Schaer DJ, Walter RB, Fehr J, Schoedon G. Functional expression of the CD163 scavenger receptor on acute myeloid leukemia cells of monocytic lineage. J Leukoc Biol 79: 312–318, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Bailey DW. Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11: 325–327, 1971. [DOI] [PubMed] [Google Scholar]
- 10.Bate KL, Jerums G. 3: Preventing complications of diabetes. Med J Aust 179: 498–503, 2003. [DOI] [PubMed] [Google Scholar]
- 11.Bielavska E, Kren V, Musilova A, Zidek V, Pravenec M. Genome scanning of the HXB/BXH sets of recombinant inbred strains of the rat for quantitative trait loci associated with conditioned taste aversion. Behav Genet 32: 51–56, 2002. [DOI] [PubMed] [Google Scholar]
- 12.Binda D, Nicod L, Viollon-Abadie C, Rodriguez S, Berthelot A, Coassolo P, Richert L. Strain difference (WKY, SPRD) in the hepatic antioxidant status in rat and effect of hypertension (SHR, DOCA). Ex vivo and in vitro data. Mol Cell Biochem 218: 139–146, 2001. [DOI] [PubMed] [Google Scholar]
- 13.Boretti FS, Buehler PW, D'Agnillo F, Kluge K, Glaus T, Butt OI, Jia Y, Goede J, Pereira CP, Maggiorini M, Schoedon G, Alayash AI, Schaer DJ. Sequestration of extracellular hemoglobin within a haptoglobin complex decreases its hypertensive and oxidative effects in dogs and guinea pigs. J Clin Invest 119: 2271–2280, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bottger A, van Lith HA, Kren V, Krenova D, Bila V, Vorlicek J, Zidek V, Musilova A, Zdobinska M, Wang JM, van Zutphen BF, Kurtz TW, Pravenec M. Quantitative trait loci influencing cholesterol and phospholipid phenotypes map to chromosomes that contain genes regulating blood pressure in the spontaneously hypertensive rat. J Clin Invest 98: 856–862, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buehler PW, Abraham B, Vallelian F, Linnemayr C, Pereira CP, Cipollo JF, Jia Y, Mikolajczyk M, Boretti FS, Schoedon G, Alayash AI, Schaer DJ. Haptoglobin preserves the CD163 hemoglobin scavenger pathway by shielding hemoglobin from peroxidative modification. Blood 113: 2578–2586, 2009. [DOI] [PubMed] [Google Scholar]
- 16.Burczynski ME, Lin HK, Penning TM. Isoform-specific induction of a human aldo-keto reductase by polycyclic aromatic hydrocarbons (PAHs), electrophiles, and oxidative stress: implications for the alternative pathway of PAH activation catalyzed by human dihydrodiol dehydrogenase. Cancer Res 59: 607–614, 1999. [PubMed] [Google Scholar]
- 17.Buresova M, Zidek V, Musilova A, Simakova M, Fucikova A, Bila V, Kren V, Kazdova L, Di Nicolantonio R, Pravenec M. Genetic relationship between placental and fetal weights and markers of the metabolic syndrome in rat recombinant inbred strains. Physiol Genomics 26: 226–231, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Charchar FJ, Kaiser M, Bingham AJ, Fotinatos N, Ahmady F, Tomaszewski M, Samani NJ. Whole genome survey of copy number variation in the spontaneously hypertensive rat: relationship to quantitative trait loci, gene expression, and blood pressure. Hypertension 55: 1231–1238, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa C, Catania S, De Pasquale R, Stancanelli R, Scribano GM, Melchini A. Exposure of human skin to benzo[a]pyrene: role of CYP1A1 and aryl hydrocarbon receptor in oxidative stress generation. Toxicology 271: 83–86, 2010. [DOI] [PubMed] [Google Scholar]
- 20.de Wolf ID, Fielmich-Bouman XM, van Oost BA, Beynen AC, Kren V, Pravenec M, van Zutphen LF, van Lith HA. Genetic and correlation analysis of hepatic copper content in the rat. Biochem Biophys Res Commun 289: 1247–1251, 2001. [DOI] [PubMed] [Google Scholar]
- 21.Dressler GR. The specification and maintenance of renal cell types by epigenetic factors. Organogenesis 5: 73–82, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Cooney AJ, D'Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F; Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521, 2004. [DOI] [PubMed] [Google Scholar]
- 23.Grieve IC, Dickens NJ, Pravenec M, Kren V, Hubner N, Cook SA, Aitman TJ, Petretto E, Mangion J. Genome-wide co-expression analysis in multiple tissues. PLoS One 3: e4033, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, Lu H, Li Y, Sarwar R, Langley SR, Bauerfeind A, Hummel O, Lee YA, Paskas S, Rintisch C, Saar K, Cooper J, Buchan R, Gray EE, Cyster JG, Cardiogenics Consortium, Erdmann J, Hengstenberg C, Maouche S, Ouwehand W, Rice CM, Samani NJ, Schunkert H, Goodall AH, Schulz H, Roider HG, Vingron M, Blankenberg S, Munzel T, Zeller T, Szymczak S, Ziegler A, Tiret L, Smyth DJ, Pravanec M, Aitman T, Cambien F, Clayton D, Todd JA, Hubner N, Cook SA. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature 467: 460–464, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopps E, Noto D, Caimi G, Averna MR. A novel component of the metabolic syndrome: the oxidative stress. Nutr Metab Cardiovasc Dis 20: 72–77, 2010. [DOI] [PubMed] [Google Scholar]
- 26.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Muller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet 37: 243–253, 2005. [DOI] [PubMed] [Google Scholar]
- 27.Hvidberg V, Maniecki MB, Jacobsen C, Hojrup P, Moller HJ, Moestrup SK. Identification of the receptor scavenging hemopexin-heme complexes. Blood 106: 2572–2579, 2005. [DOI] [PubMed] [Google Scholar]
- 28.Jirout ML, Friese RS, Mahapatra NR, Mahata M, Taupenot L, Mahata SK, Kren V, Zidek V, Fischer J, Maatz H, Ziegler MG, Pravenec M, Hubner N, Aitman TJ, Schork NJ, O'Connor DT. Genetic regulation of catecholamine synthesis, storage and secretion in the spontaneously hypertensive rat. Hum Mol Genet, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson MD, He L, Herman D, Wakimoto H, Wallace CA, Zidek V, Mlejnek P, Musilova A, Simakova M, Vorlicek J, Kren V, Viklicky O, Qi NR, Wang J, Seidman CE, Seidman J, Kurtz TW, Aitman TJ, Pravenec M. Dissection of chromosome 18 blood pressure and salt-sensitivity quantitative trait loci in the spontaneously hypertensive rat. Hypertension 54: 639–645, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitts DD, Yuan YV, Godin DV. Plasma and lipoprotein lipid composition and hepatic antioxidant status in spontaneously hypertensive (SHR) and normotensive (WKY) rats. Can J Physiol Pharmacol 76: 202–209, 1998. [PubMed] [Google Scholar]
- 31.Krueger K, Koch K, Juhling A, Tepel M, Scholze A. Low expression of thiosulfate sulfurtransferase (rhodanese) predicts mortality in hemodialysis patients. Clin Biochem 43: 95–101, 2010. [DOI] [PubMed] [Google Scholar]
- 32.Kunes J, Kren V, Klir P, Zicha J, Pravenec M. Genetic determination of heart and kidney weights studied using a set of recombinant inbred strains: the relationship to blood pressure. J Hypertens 8: 1091–1095, 1990. [DOI] [PubMed] [Google Scholar]
- 33.Lindoso RS, Verdoorn KS, Einicker-Lamas M. Renal recovery after injury: the role of Pax-2. Nephrol Dial Transplant 24: 2628–2633, 2009. [DOI] [PubMed] [Google Scholar]
- 34.Lum PY, Chen Y, Zhu J, Lamb J, Melmed S, Wang S, Drake TA, Lusis AJ, Schadt EE. Elucidating the murine brain transcriptional network in a segregating mouse population to identify core functional modules for obesity and diabetes. J Neurochem 97, Suppl 1: 50–62, 2006. [DOI] [PubMed] [Google Scholar]
- 35.MacDonald RJ, Swift GH, Quinto C, Swain W, Pictet RL, Nikovits W, Rutter WJ. Primary structure of two distinct rat pancreatic preproelastases determined by sequence analysis of the complete cloned messenger ribonucleic acid sequences. Biochemistry 21: 1453–1463, 1982. [DOI] [PubMed] [Google Scholar]
- 36.Manke T, Heinig M, Vingron M. Quantifying the effect of sequence variation on regulatory interactions. Hum Mutat 31: 477–483, 2010. [DOI] [PubMed] [Google Scholar]
- 37.Manke T, Roider HG, Vingron M. Statistical modeling of transcription factor binding affinities predicts regulatory interactions. PLoS Comput Biol 4: e1000039, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller M, Goel A, Thimma M, Dickens NJ, Aitman TJ, Mangion J. eQTL Explorer: integrated mining of combined genetic linkage and expression experiments. Bioinformatics 22: 509–511, 2006. [DOI] [PubMed] [Google Scholar]
- 40.Oliviero S, Cortese R. The human haptoglobin gene promoter: interleukin-6-responsive elements interact with a DNA-binding protein induced by interleukin-6. EMBO J 8: 1145–1151, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliviero S, Morrone G, Cortese R. The human haptoglobin gene: transcriptional regulation during development and acute phase induction. EMBO J 6: 1905–1912, 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passador-Gurgel G, Hsieh WP, Hunt P, Deighton N, Gibson G. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat Genet 39: 264–268, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, Fischer J, Maatz H, Kren V, Pravenec M, Hubner N, Aitman TJ. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet 2: e172, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petretto E, Sarwar R, Grieve I, Lu H, Kumaran MK, Muckett PJ, Mangion J, Schroen B, Benson M, Punjabi PP, Prasad SK, Pennell DJ, Kiesewetter C, Tasheva ES, Corpuz LM, Webb MD, Conrad GW, Kurtz TW, Kren V, Fischer J, Hubner N, Pinto YM, Pravenec M, Aitman TJ, Cook SA. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Genet 40: 546–552, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pravenec M, Churchill PC, Churchill MC, Viklicky O, Kazdova L, Aitman TJ, Petretto E, Hubner N, Wallace CA, Zimdahl H, Zidek V, Landa V, Dunbar J, Bidani A, Griffin K, Qi N, Maxova M, Kren V, Mlejnek P, Wang J, Kurtz TW. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat Genet 40: 952–954, 2008. [DOI] [PubMed] [Google Scholar]
- 46.Pravenec M, Gauguier D, Schott JJ, Buard J, Kren V, Bila V, Szpirer C, Szpirer J, Wang JM, Huang H. Mapping of quantitative trait loci for blood pressure and cardiac mass in the rat by genome scanning of recombinant inbred strains. J Clin Invest 96: 1973–1978, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pravenec M, Klir P, Kren V, Zicha J, Kunes J. An analysis of spontaneous hypertension in spontaneously hypertensive rats by means of new recombinant inbred strains. J Hypertens 7: 217–221, 1989. [PubMed] [Google Scholar]
- 48.Pravenec M, Kunes J, Zicha J, Kren V, Klir P. Platelet aggregation in spontaneous hypertension: genetic determination and correlation analysis. J Hypertens 10: 1453–1456, 1992. [DOI] [PubMed] [Google Scholar]
- 49.Pravenec M, Kurtz TW. Recent advances in genetics of the spontaneously hypertensive rat. Curr Hypertens Rep 12: 5–9, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pravenec M, Zidek V, Musilova A, Kren V, Bila V, Di Nicolantonio R. Chromosomal mapping of a major quantitative trait locus regulating compensatory renal growth in the rat. J Am Soc Nephrol 11: 1261–1265, 2000. [DOI] [PubMed] [Google Scholar]
- 51.Pravenec M, Zidek V, Musilova A, Simakova M, Kostka V, Mlejnek P, Kren V, Krenova D, Bila V, Mikova B, Jachymova M, Horky K, Kazdova L, St, Lezin E, Kurtz TW. Genetic analysis of metabolic defects in the spontaneously hypertensive rat. Mamm Genome 13: 253–258, 2002. [DOI] [PubMed] [Google Scholar]
- 52.Pravenec M, Zidek V, Simakova M, Kren V, Krenova D, Horky K, Jachymova M, Mikova B, Kazdova L, Aitman TJ, Churchill PC, Webb RC, Hingarh NH, Yang Y, Wang JM, Lezin EM, Kurtz TW. Genetics of Cd36 and the clustering of multiple cardiovascular risk factors in spontaneous hypertension. J Clin Invest 103: 1651–1657, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pravenec M, Zidek V, Zdobinska M, Kren V, Krenova D, Bottger A, van Zutphen LF, Wang J, St, Lezin E. Mapping genes controlling hematocrit in the spontaneously hypertensive rat. Mamm Genome 8: 387–389, 1997. [DOI] [PubMed] [Google Scholar]
- 54.Roider HG, Kanhere A, Manke T, Vingron M. Predicting transcription factor affinities to DNA from a biophysical model. Bioinformatics 23: 134–141, 2007. [DOI] [PubMed] [Google Scholar]
- 55.Santiago JV. Overview of the complications of diabetes. Clin Chem 32: B48–B53, 1986. [PubMed] [Google Scholar]
- 56.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, Lum PY, Leonardson A, Thieringer R, Metzger JM, Yang L, Castle J, Zhu H, Kash SF, Drake TA, Sachs A, Lusis AJ. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37: 710–7, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaer DJ, Schaer CA, Buehler PW, Boykins RA, Schoedon G, Alayash AI, Schaffner A. CD163 is the macrophage scavenger receptor for native and chemically modified hemoglobins in the absence of haptoglobin. Blood 107: 373–380, 2006. [DOI] [PubMed] [Google Scholar]
- 58.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta 1763: 1755–1766, 2006. [DOI] [PubMed] [Google Scholar]
- 59.Singh R, Lemire J, Mailloux RJ, Appanna VD. A novel strategy involved in [corrected] anti-oxidative defense: the conversion of NADH into NADPH by a metabolic network. PLoS One 3: e2682, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.STAR Consortium, Saar K, Beck A, Bihoreau MT, Birney E, Brocklebank D, Chen Y, Cuppen E, Demonchy S, Dopazo J, Flicek P, Foglio M, Fujiyama A, Gut IG, Gauguier D, Guigo R, Guryev V, Heinig M, Hummel O, Jahn N, Klages S, Kren V, Kube M, Kuhl H, Kuramoto T, Kuroki Y, Lechner D, Lee YA, Lopez-Bigas N, Lathrop GM, Mashimo T, Medina I, Mott R, Patone G, Perrier-Cornet JA, Platzer M, Pravenec M, Reinhardt R, Sakaki Y, Schilhabel M, Schulz H, Serikawa T, Shikhagaie M, Tatsumoto S, Taudien S, Toyoda A, Voigt B, Zelenika D, Zimdahl H, Hubner N. SNP and haplotype mapping for genetic analysis in the rat. Nat Genet 40: 560–566, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stohs SJ. Oxidative stress induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Free Radic Biol Med 9: 79–90, 1990. [DOI] [PubMed] [Google Scholar]
- 62.Tanito M, Nakamura H, Kwon YW, Teratani A, Masutani H, Shioji K, Kishimoto C, Ohira A, Horie R, Yodoi J. Enhanced oxidative stress and impaired thioredoxin expression in spontaneously hypertensive rats. Antioxid Redox Signal 6: 89–97, 2004. [DOI] [PubMed] [Google Scholar]
- 63.Teye K, Quaye IK, Koda Y, Soejima M, Tsuneoka M, Pang H, Ekem I, Amoah AG, Adjei A, Kimura H. A-61C and C-101G Hp gene promoter polymorphisms are, respectively, associated with ahaptoglobinaemia and hypohaptoglobinaemia in Ghana. Clin Genet 64: 439–443, 2003. [DOI] [PubMed] [Google Scholar]
- 64.Thomas RJ, Palumbo PJ, Melton LJ, 3rd, Roger VL, Ransom J, O'Brien PC, Leibson CL. Trends in the mortality burden associated with diabetes mellitus: a population-based study in Rochester, Minn, 1970–1994. Arch Intern Med 163: 445–451, 2003. [DOI] [PubMed] [Google Scholar]
- 65.Thongboonkerd V, Barati MT, McLeish KR, Benarafa C, Remold-O'Donnell E, Zheng S, Rovin BH, Pierce WM, Epstein PN, Klein JB. Alterations in the renal elastin-elastase system in type 1 diabetic nephropathy identified by proteomic analysis. J Am Soc Nephrol 15: 650–662, 2004. [DOI] [PubMed] [Google Scholar]
- 66.Tolosano E, Altruda F. Hemopexin: structure, function, regulation. DNA Cell Biol 21: 297–306, 2002. [DOI] [PubMed] [Google Scholar]
- 67.Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development 121: 4057–4065, 1995. [DOI] [PubMed] [Google Scholar]
- 68.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. The Rat Genome Database, update 2007–easing the path from disease to data and back again. Nucleic Acids Res 35: D658–D662, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics 1: 299–308, 2003. [DOI] [PubMed] [Google Scholar]
- 70.Winkler BS, Solomon F. High activities of NADP+-dependent isocitrate dehydrogenase and malic enzyme in rabbit lens epithelial cells. Invest Ophthalmol Vis Sci 29: 821–823, 1988. [PubMed] [Google Scholar]
- 71.Yuan YV, Kitts DD, Godin DV. Heart and red blood cell antioxidant status and plasma lipid levels in the spontaneously hypertensive and normotensive Wistar-Kyoto rat. Can J Physiol Pharmacol 74: 290–297, 1996. [PubMed] [Google Scholar]
- 72.Zidek V, Pintir J, Musilova A, Bila V, Kren V, Pravenec M. Mapping of quantitative trait loci for seminal vesicle mass and litter size to rat chromosome 8. J Reprod Fertil 116: 329–333, 1999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.