Abstract
Testosterone (T) has an anabolic effect on skeletal muscle and is believed to exert its local effects via the androgen receptor (AR). The AR harbors a polymorphic stretch of glutamine repeats demonstrated to inversely affect receptor transcriptional activity in prostate and kidney cells. The effects of AR glutamine repeat length on skeletal muscle are unknown. In this study we examined the effect of AR CAG repeat length on AR function in C2C12 cells. AR expression vectors harboring 14, 24, and 33 CAG repeats were used to assess AR transcriptional activity. C2C12 cell proliferation, differentiation, gene expression, myotube formation, and myonuclear fusion index were assessed. Transcriptional activity increased with increasing repeat length and in response to testosterone (AR14 = 3.91 ± 0.26, AR24 = 25.21 ± 1.72, AR33 = 36.08 ± 3.22 relative light units; P < 0.001). Ligand activation was increased for AR33 (2.10 ± 0.04) compared with AR14 (1.54 ± 0.09) and AR24 (1.57 ± 0.05, P < 0.001). AR mRNA expression was elevated in each stably transfected line. AR33 cell proliferation (20,512.3 ± 1,024.0) was decreased vs. AR14 (27,604.17 ± 1,425.3; P < 0.001) after 72 h. Decreased CK activity in AR14 cells (54.9 ± 2.9 units/μg protein) in comparison to AR33 (70.8 ± 8.1) (P < 0.05) was noted. The myonuclear fusion index was lower for AR14 (15.21 ± 3.24%) and AR33 (9.97 ± 3.14%) in comparison to WT (35.07 ± 5.60%, P < 0.001). AR14 and AR33 cells also displayed atypical myotube morphology. RT-PCR revealed genotype differences in myostatin and myogenin expression. We conclude that AR polyglutamine repeat length is directly associated with transcriptional activity and alters the growth and development of C2C12 cells. This polymorphism may contribute to the heritability of muscle mass in humans.
Keywords: skeletal muscle, testosterone, transcription, myoblast, polymorphism
androgens play an essential role in a number of physiological processes including muscle and bone development and the development and maintenance of secondary sexual characteristics. Testosterone (T) administration results in increased fat-free muscle mass and decreased fat mass in hypogonadal and aging men (2, 7), as well as increased upper and lower body strength in aging men (38) and increased bone density and bone mineral content in hypogonadal men (25) and women (31). Supraphysiologic T doses in healthy young men increase fat-free mass in a dose-dependent manner (5), and muscle mass is increased even further by the combination of T and resistance exercise (4). As a result T therapy is becoming a more commonly accepted treatment for aging and disease-related muscle and bone wasting conditions.
Though the mechanisms behind the effects of T on skeletal muscle are still poorly understood, it is believed that the majority of the anabolic effects are mediated via its interaction with the androgen receptor (AR) (6, 27, 35). The AR is a ligand-activated nuclear hormone receptor that upon ligand binding dissociates from binding proteins, moves into the nucleus, and acts as a transcription factor to regulate expression of androgen responsive genes (39). Structurally, the AR consists of three functional domains: an amino-terminal transactivation domain (NTD), a central DNA binding domain, and a carboxy-terminal ligand binding domain (LBD). The NTD harbors binding sites for a variety of additional cofactors (1, 8, 28), and mutational deletions demonstrate that the region is required for full receptor transcriptional activation (12), hence its designation as the transactivation domain.
The NTD also harbors a polyglutamine repeat polymorphism that has been demonstrated to affect AR transcriptional activity in a variety of cell types (3, 11, 32, 40). This repeat has also been associated with a number of androgen-related maladies including prostate cancer (20), prostate hypertrophy (19), and spinal bulbar muscular atrophy (14). Though well characterized in respect to prostate, the effect of AR polyglutamine length on skeletal muscle physiology is less clear. Results from genetic association studies examining AR polyglutamine repeat length in relation to skeletal muscle have been conflicting, with both longer repeat length (9, 42) and shorter repeat length (33) being correlated with greater fat-free mass. If we consider AR activity to exert an anabolic effect on muscle, then based on data from previous studies where AR activity is inversely correlated with repeat length in nonmuscle tissue one would hypothesize that AR repeat length would be inversely related to fat-free mass. Given these conflicting data further clarification on the role of the AR polyglutamine repeat polymorphism in skeletal muscle physiology is warranted.
The aim of the current study was to determine if AR polyglutamine repeat length affects AR transcriptional activity and skeletal muscle cell development in vitro. We hypothesized that AR transcriptional activity would decrease with increasing repeat length. Contrary to our hypothesis, our results clearly demonstrate that AR polyglutamine repeat length is directly related to transcriptional activity in skeletal muscle and alters the development of C2C12 cells.
EXPERIMENTAL PROCEDURES
DNA plasmids.
The human androgen receptor expression vector pCMVhAR (a kind gift from Dr. Elizabeth Wilson, Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC) was used to derive human (h) AR constructs encoding for 14, 24 (parent vector), or 33 AR CAG repeats. In healthy humans AR CAG repeat lengths range from 11 to 31 (17), so we chose to utilize a nested design. Genomic DNA from human subjects (42) carrying 14 and 33 CAG repeats, respectively (representing the smallest and largest repeat lengths available from our samples), was amplified via PCR using recombinant Taq DNA polymerase (Fermentas, Glen Burnie, MD) and the following primers: forward primer, 5′-TGCACCTACTTCAGTGGACACTGAAT-3′, reverse primer; 5′-GTATCTTCAGTGCTCTTGCCTGCG-3′. The reaction mix was denatured at 95°C for 5 min, followed by 40 cycles of 95°C denaturation for 30 s, 60°C annealing for 30 s, and 72°C extension for 75 s. The fragments and pCMVhAR vector were sequentially restriction enzyme-digested with BglII and BsmI (New England Biolabs, Ipswich, MA) at 37°C overnight and at 65°C for 2 h, respectively. Digested PCR products and vector backbone were resolved via agarose gel electrophoresis and purified using the PureLink quick gel extraction kit (Invitrogen, Carlsbad, CA). Genomic DNA inserts and cut vector were treated with shrimp alkaline phosphatase (Fermentas) before being ligated together using T4 DNA ligase (Invitrogen) at a 10:1 insert to vector ratio. Ligated products were transformed into MAX Efficiency DH5α competent Escherichia coli bacterial cells (Invitrogen) before being seeded on ampicillin-positive agar. Individual colonies were selected and expanded overnight at 37°C in Lysogeny broth media before plasmid DNA was isolated using QIAfilter Plasmid Midi Kit (Qiagen, Valencia, CA). The resulting pCMV-hAR14, pCMV-hAR33, and pCMV-hAR24 (original vector) plasmids were sequenced using an Applied Biosystems 3730 DNA sequencer to confirm the correct AR CAG repeat lengths and orientation. Additional plasmids, pRL-TK, pGL3-basic, and pCI-neo (Promega, Madison, WI) and p159pPr-luc (Addgene plasmid 8392; Addgene, Cambridge, MA) were obtained commercially. Use of human DNA for these experiments was approved by the University of Maryland Institutional Review Board.
Cell culture.
C2C12 mouse myoblasts (ATCC, Manassas, VA) were maintained in growth medium (GM) consisting of Dulbecco's modified Eagle's medium (DMEM) (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Mediatech) and 1% penicillin/streptomycin solution at 37°C and 5% CO2. C2C12 cell differentiation was initiated by incubating the cells in differentiation media (DM) consisting of DMEM supplemented with 2% horse serum (Mediatech) and 1% penicillin-streptomycin. Cell lines stably expressing hAR plasmids were maintained in GM supplemented with G418 (Invitrogen), an antibiotic that interferes with protein synthesis in both prokaryotic and eukaryotic cells (23), at 500 μg/ml (see section below for details on the creation of cell lines).
AR transcriptional activity.
The transcriptional activity of each androgen receptor construct was assessed using a commercial luciferase assay system. C2C12 cells were seeded into 24-well plates at a density of 4,000 cells per well and incubated in GM overnight. Cells were transiently transfected with 400 ng of pCMV-hAR14, pCMV-hAR24, or pCMV-hAR33, respectively, as well as a 400 ng of firefly luciferase reporter vector pPr-LUC driven by the highly androgen responsive probasin gene promoter (24) and 50 ng of pRL-TK Renilla luciferase normalization vector using Lipofectamine and Plus reagents (Invitrogen). Following transfection cells were treated with 100 nM testosterone (initial dose-response experiments demonstrated 100 nM T to induce the greatest degree of response without negative effects) or ethanol vehicle control in GM and incubated for 24 h. Cells were lysed in passive lysis buffer (Promega) for 20 min at room temperature with gentle rocking, spun at 3,000 g for 10 min, and 20 μl aliquots of supernatant were assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturer instructions. Luminescence was recorded over a 5 s period following a 2 s delay using a Modulus fluorometer (Turner Biosystems). Each experimental condition was measured in triplicate and on three separate occasions.
Stable line creation.
Approximately 10,000 C2C12 myoblasts were seeded into 35 mm plates and incubated in GM overnight before being cotransfected with 1.5 μg of pCMV-hAR14, pCMV-hAR24, or pCMV-hAR33, and 1.5 μg of the G418 antibiotic resistance vector pCl-neo. Transfections were carried out using Lipofectamine and Plus Reagent (Invitrogen) according to manufacturer instructions. G418 was applied to each well at a concentration of 1000 μg/ml in GM following an overnight incubation. Cells were maintained in GM+G418 for an additional 10–14 days for selection. Surviving colonies were expanded and RNA was collected and amplified via RT-PCR. PCR was performed using an AR primer set designed to amplify the CAG repeat region (Table 1) to confirm AR transgene expression.
Table 1.
Gene expression primers and conditions
| Gene | F Primer (5′-3′) | R Primer (5′-3′) | Anneal Temp. °C | Cycle # | Expected Size, bp |
|---|---|---|---|---|---|
| AR | accgaggagctttccagaat | cagctgagtcatcctcgtccg | 55 | 30 | 420 (21CAGs) |
| Myogenin | tccctgtccaccttcagggcttcg | taaggagtcagctaaattccctcg | 59 | 30 | 804 |
| Myostatin | taaccttcccaggaccagga | cactctccagagcagtaatt | 55 | 30 | 225 |
| MyoD | gtggcagaaagttaagacga | agtcgaaacacgggtcatca | 50 | 25 | 170 |
| ACTA1 | gcgcaagtactcagtgtgga | cacgattgtcgattgtggtc | 55 | 22 | 182 |
| GAPDH | gtgtccgtcgtggatctg | cctgcttcaccaccttcttg | 55 | 25 | 90 |
F, forward; R, reverse; AR, androgen receptor.
Cellular proliferation assay.
Stable lines and wild-type (WT) C2C12 cells were seeded into 24-well plates at 4,000 cells per well in GM and treated with either 100 nM T or ethanol vehicle (ethanol concentration did not exceed 0.1% in any experiment). Cell number was assayed at 24 h intervals using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay kit (Promega). Cells were incubated within the MTS solution for 3 h at 37°C before being assayed for absorbance at 490 nm using a Wallac Multiplate Reader (Perkin Elmer, Waltham, MA). The number of cells present per well was determined by plotting the absorbance value for each sample on a standard curve. The standard curves were generated for each individual sample plate by seeding differing number of cells between 2,000 and 30,000 per well on the same sample plate ∼6–8 h before each reading. Standard curve wells were measured simultaneously with the experimental samples, and each experimental value fell within the boundaries of the standard curve. Each experimental condition was measured in triplicate and on three individual experiments.
Cellular differentiation assay.
Stable lines and WT controls were seeded into 35 mm plates at a density of 25,000 cells per well and incubated in GM until ∼90% confluent, at which point the culture media was switched to DM. We added 100 nM T or ethanol vehicle, and the cells were incubated for up to 5 days. Cell lysates were obtained, and 10 μl of each sample was analyzed for creatine kinase (CK) activity using the EnzyChrom Creatine Kinase Assay Kit (BioAssay Systems, Hayward, CA) according to manufacturer instructions. The remaining lysate from each sample was assayed for total protein content via the BCA Protein Assay (Pierce, Rockford, IL). CK values were normalized to total protein. All experimental conditions were performed in duplicate and on three individual experiments.
Immunocytochemistry.
Cells were seeded on 35 mm plates in GM supplemented with 100 nM T or ethanol vehicle until they reached 90–95% confluence. Incubation medium was then switched to DM with 100 nM T or ethanol vehicle for the indicated period of time. Cells were washed 3× in PBS, fixed in ice-cold methanol for 10 min at room temperature, and again washed 3× in PBS. Cells were washed 3× with 3% notfat dry milk in Tris-buffered saline with Tween for 5 min at room temperature to block nonspecific interactions. Anti-sarcomeric myosin antibody MF-20 (Developmental Studies Hybridoma Bank, University of Iowa) was applied diluted 1:50 in PBS for 1 h at room temperature. After rinsing 3× in PBS, a fluoroscein-isothiocyanate-conjugated anti-mouse IgG secondary diluted 1:500 in PBS was applied for 1 h at room temperature in darkness. Cells were washed 3× in PBS, and 4,6-diamidino-2-phenylindole (DAPI) was applied at 500 ng/ml for 5 min at room temperature in darkness. The cells were again rinsed in PBS and visualized using a Nikon TI-U inverted fluorescent microscope (Nikon, Melville, NY) using the ×20 objective and appropriate filters.
Gene expression.
RNA was extracted from cells at the appropriate time points using TRIzol (Invitrogen) and standard techniques. RNA was treated with DNase I (Fermentas) for 30 min at 37°C to degrade any trace remaining DNA. cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Conditions for each individual gene are displayed in Table 1. GAPDH was selected as a normalization reference gene.
Statistical analysis.
Pair-wise comparisons of means were performed by two-tailed Student's t-test. Two-way ANOVA was used for multiple comparisons in dose-response experiments (e.g., luciferase activity) with drug and cell line as main effects. Three-way ANOVA with repeated measures was used for time course experiments with drug, cell line, and day as main effects, and Tukey's post hoc for comparison of means. All experiments were performed in triplicate and were repeated on three separate occasions, unless otherwise noted. All comparisons were carried out using the SPSS software package (SPSS).
RESULTS
AR transcriptional activity in C2C12 cells.
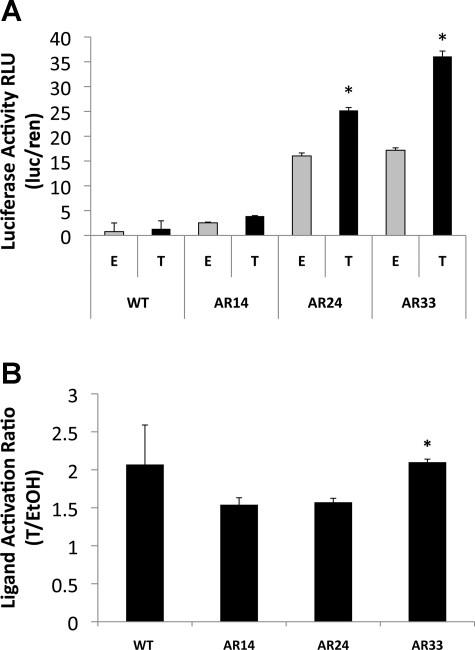
We observed a positive relationship between AR repeat length and transcriptional activity in T-treated C2C12 cells (Fig. 1A). AR33 and AR24 were significantly more active than AR14 even in the absence of T, indicating ligand-independent activation of AR at repeat lengths ≥ 24. When treated with T AR24 had transcriptional activity more than fivefold higher than AR14, and AR33 had 43% greater transcriptional activity than AR24 (P < 0.001). Across the full range of CAG repeat lengths, total transcriptional activity was increased more than ninefold (AR33 vs. AR14, P < 0.001). Mock transfections carried out with a promoter-less reporter vector displayed minimal luciferase activity (data not shown), indicating that AR-mediated activation of the probasin promoter was responsible for luciferase activity, rather than constitutive reporter vector activity. WT C2C12 cells displayed minimal luciferase activity. Interestingly, the ligand activation percentage (firefly/Renilla luciferase activity ratio in T-treated cells vs. the firefly/Renilla luciferase activity ratio in ethanol vehicle-treated cells) was considerably higher for AR33 compared with AR14 and AR24 (110% vs. 53.9% and 57.3%, respectively, P < 0.001, Fig. 1B), indicating that T activation was greatest with AR33.
Fig. 1.
Effects of repeat length on androgen receptor (AR) transcriptional activity in C2C12 myoblasts. A: C2C12 cells were transfected with a human AR expression vector harboring 14, 24, or 33 CAG repeats, respectively, along with pPr-Luc reporter vector and pRL-TK normalization vector. Wild-type (WT) and transfected cells were treated with 100 nM testosterone (T) or ethanol vehicle (E or EtOH) for 24 h. Data are expressed as the mean luciferase ratio, and bars represent the means and SE of triplicates from 3 separate experiments. RLU, relative light units. *Significant difference from WT, P < 0.001. B: ligand activation ratio was determined by dividing the average firefly/Renilla luciferase ratio of T-treated cells by the average firefly/Renilla luciferase ratio of E-treated cells. *Significant difference from AR14, AR24 lines; P < 0.001.
AR expression.
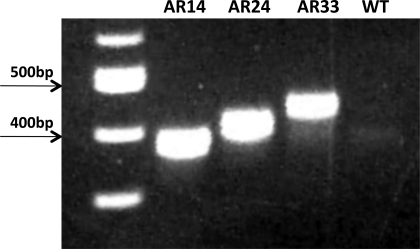
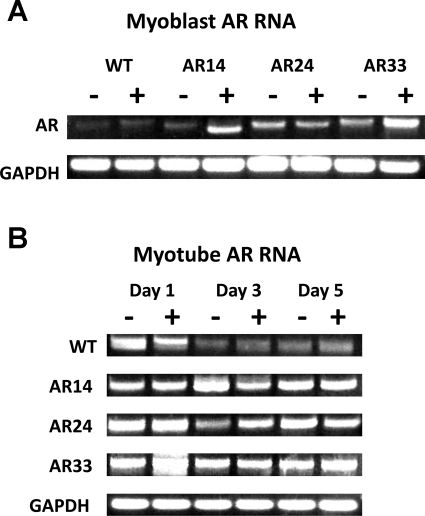
DNA was obtained from cells transfected with each respective AR vector and was sequenced to confirm the presence of the intended number of CAG repeats. RNA was isolated and RT-PCR performed to confirm increased AR mRNA expression (Fig. 2). RT-PCR revealed that AR mRNA expression in all of the stably transfected lines was readily detectable in myoblasts and throughout the differentiation process (Fig. 3), while AR mRNA expression was only apparent in WT cells 24 h after switching to DM.
Fig. 2.
RT-PCR from RNA extracted from stably transfected lines resolved on 1% agarose gels. Note the differences in migration distance for each construct (AR14, 399 bp; AR24, 429 bp; AR33, 456 bp). Ladder is shown on the far left lane.
Fig. 3.
A: AR RNA from myoblasts incubated in growth medium (GM) for 1 day. B: AR RNA from myotubes after 1, 3, and 5 days in differentiation medium (DM). GAPDH is included as a loading control.
Effect of AR repeat length on C2C12 proliferation and differentiation.
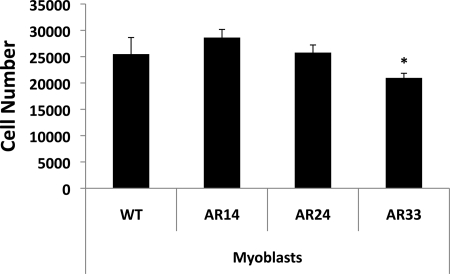
We created three cell lines, each stably expressing one of our AR vectors, to examine the effects of AR repeat length on cell growth and differentiation. It was recently demonstrated that T promotes both myoblast proliferation and differentiation (18), but the effect of altered AR activity is unknown. The impact of AR repeat length on C2C12 cell proliferation was assessed by incubating the different cell lines in GM for up to 3 days in the presence of T or ethanol vehicle. After 3 days in GM, there were fewer AR33 cells compared with AR14 and AR24 cells (20,512 ± 3,071 vs. 27,604 ± 4,275 vs. 25,375 ± 3,859, respectively; P < 0.001) (Fig. 4); however, T had no effect on proliferation at any point, indicated these results were induced by AR in a ligand-independent manner.
Fig. 4.
Effect of AR repeat length on C2C12 cell proliferation after 3 days in GM. Cell growth of the stably transfected and WT C2C12 lines was measured using a colorimetric proliferation assay. Cell number was calculated by measuring formazan absorbance at 490 nm, and the data were plotted on a standard curve derived from known cell densities. Error bars represent the mean and SE of triplicates from 3 separate experiments. *Significant difference from WT, P < 0.05.
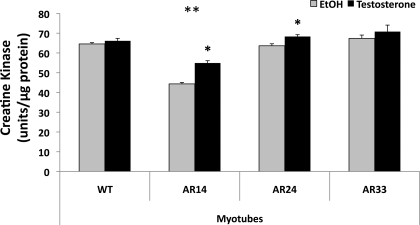
The impact of differing AR repeat length on C2C12 cell differentiation was assessed by measuring the activity of the enzyme CK, an enzyme typically expressed only upon the initiation of differentiation in skeletal muscle cells (10, 34). Cells were grown to ∼90% confluence in GM and then switched to DM in the presence of T or ethanol vehicle for 1–5 days before lysis and analysis of CK activity. By the 5th day of incubation CK activity of the AR14 line was decreased vs. all other lines: 54.9 ± 1.2 vs. 70.8 ± 3.3, 68.3 ± 0.9, and 66.1 ± 1.2 units/μg protein in AR33, AR24, and WT lines, respectively, P < 0.001 (Fig. 5). The AR33, AR24, and WT lines were not different from each other. CK activity was increased by T in AR14 and AR24 cells but not in AR33 cells, indicating a degree of ligand-independence of the AR in the latter (Fig. 5).
Fig. 5.
Influence of AR repeat length and T treatment on C2C12 cell differentiation. Creatine kinase activity was assessed from whole cell lysates of each stably transfected line and WT C2C12 cells incubated in differentiation medium for 5 days in the presence of 100 nM T (+) or ethanol vehicle (−). *Significant effect of T in that cell line, P < 0.001. **Significant difference from WT, P < 0.05.
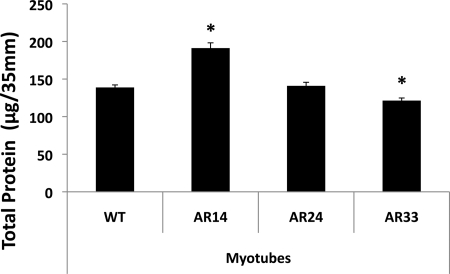
Notable differences in total protein content among the lines were also observed (Fig. 6). By the 5th day of differentiation significantly more protein was present in the AR14 line compared with the WT line (191.2 ± 18.5 vs. 138.8 ± 4.0, P < 0.001), while the AR33 line contained significantly less total protein than the WT line (121.3 ± 6.1 vs. 138.8 ± 4.0, P < 0.001). T treatment did not affect protein content in any of the lines.
Fig. 6.
Total protein content (per 35 mm plate) in differentiating myotubes after 5 days in differentiation medium. *Significant difference from WT, P < 0.001 and P < 0.05 in AR14 and AR33, respectively.
Myotube development and morphology.
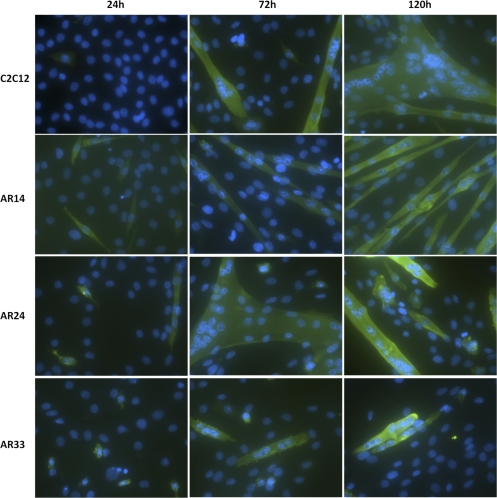
During the maintenance and culturing of our C2C12 lines we observed distinct phenotypic differences in the appearance and morphology of the cells, which we further analyzed via immunohistochemical staining. Cells were grown in GM until ∼90% confluence, when they were switched to DM with 100 nM T or ethanol vehicle for the indicated time period. The cells were stained with an anti-sarcomeric myosin antibody to identify differentiated cells and DAPI as a nuclear stain. Cells were designated as a myotube where the cell stained positive for myosin and contained a minimum of three myonuclei. After 1 day of incubation in DM very few myotubes were observed in any of the lines. However, the AR14 line displayed many myosin-positive, elongated, mononucleated cells (Fig. 7). After 3 days, WT and AR24 lines displayed typical myotube morphology: large, thick, myosin-positive, multinucleated cells. In contrast, by the 3rd day the AR14 line developed into very long, thin cells with few sparse nuclei, while the AR33 line developed into short, truncated myotubes with clustered nuclei.
Fig. 7.
Immunocytochemical staining revealing differences in myotube development among the 3 lines. Green, sarcomeric myosin protein; blue, 4,6-diamidino-2-phenylindole-stained nuclei. Criteria for classification as a myotube included a myosin-positive stain and a minimum of 3 myonuclei. The above images are representative of a minimum of 15 fields per condition. All images were captured using the ×20 objective.
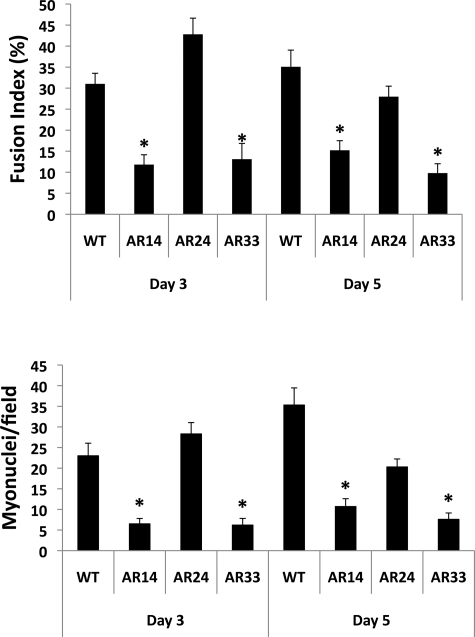
Fusion index (FI) is a relative estimate of the efficiency of myoblast fusion by determining the percentage of intra-myotube nuclei within a given field. The images captured suggest that fusion in both AR14 and AR33 lines is impaired, and FI analysis supports this finding (Fig. 8, top). There was virtually no indication of myotube formation after 1 day of incubation (data not shown). By the 3rd day the FI of the AR24 line was highest at 42.8 ± 3.8%, followed by WT at 31.0 ± 2.5%, with the FI of both the AR14 and AR33 lines far lower (11.8 ± 2.3% and 13.1 ± 3.8%, respectively; P < 0.001). By the 5th day the FI of the WT line was highest at 35.1 ± 4.0%, followed by AR24 at 28.0 ± 2.5, AR14 at 15.2 ± 2.3, and AR33 at 9.8 ± 2.2. After 3 days the average number of myonuclei per field was significantly reduced in both AR14 and AR33 cells: 6.5 ± 1.3 and 6.2 ± 1.6, respectively, vs. 23 ± 3.1 and 28.3 ± 2.7 in WT and AR24 cells, respectively (P < 0.001; Fig. 8, bottom); AR14 and AR33 lines were not different from each other. Similar results were observed after 5 days: 10.7 ± 1.9 and 7.6 ± 1.5 myonuclei/field for AR14 and AR33 cells respectively, vs. 35.3 ± 4.2 and 20.3 ± 1.9 in WT and AR24 cells, respectively (P < 0.001).
Fig. 8.
Myotube fusion index (top) and average myonuclei number per field (bottom) are reduced in AR14 and AR33 lines. Myonuclei were identified as nuclei within cells that 1) stained positive for myosin expression and 2) contained at least 3 nuclei. Data are expressed as the means and SE of 10 fields. *Significant difference from WT, P < 0.001.
Gene expression.
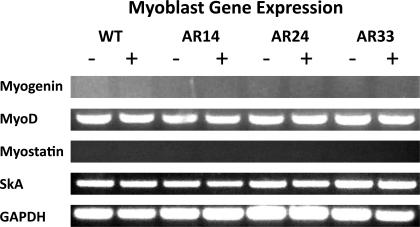
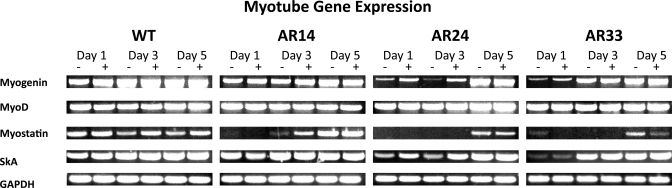
RT-PCR analysis was performed on RNA extracted from proliferating myoblasts and in differentiating myotubes 1, 3, and 5 days after switching to DM. Myogenin and MyoD were assessed as surrogate markers of the myogenic process (29, 36, 37). Skeletal α-actin (SkA) and myostatin were chosen since they are both muscle specific and sensitive to androgen exposure at the transcriptional level, and thus we utilized their expression as an endogenous indicator of AR activity (15, 21). Though gene expression was qualitatively similar among lines during the myoblast stage (Fig. 9), appreciable differences were observed in the expression of myogenin, myostatin, and SkA after the initiation of differentiation (Fig. 10). Myogenin expression was delayed in both AR24 and AR33 lines compared with WT, indicating a slower progression into a differentiated state. SkA expression was likewise delayed in the AR33 line. However, myostatin expression is delayed in all three lines overexpressing AR, and the delay is repeat-length dependent.
Fig. 9.
Myoblast mRNA gene expression. RT-PCR was performed on RNA extracted from cells harvested at ∼75% confluence and after a 24 h incubation with either ethanol (−) or 100 nM T (+). GAPDH is included as a reference gene. All images are representative of 3 separate experiments. SkA, skeletal α-actin.
Fig. 10.
Myotube mRNA gene expression. RT-PCR was performed on RNA extracted from each line 24, 72, and 120 h after switching to DM supplemented with either ethanol (−) or 100 nM T (+). GAPDH is included as a reference gene. All images are representative of 3 separate experiments.
DISCUSSION
This is the first study to examine the relationship of AR glutamine repeat length and transcriptional activity in skeletal muscle cells, and we find strong evidence of a relationship between AR repeat length and transcriptional activity in these skeletal muscle cells. The difference in total activation between AR14 and AR33 was also quite remarkable; the nearly ninefold difference being far greater than that demonstrated by other studies using a similar spread in AR repeat length (3, 40). In addition to its far greater transcriptional activity, the AR33 construct demonstrated a much greater ligand-dependent activation ratio compared with the AR14 and AR24 constructs, indicating that longer AR repeat lengths affect responsiveness to T treatment in C2C12 cells. However, both the AR24 and AR33 vectors demonstrated considerable transcriptional activity even in the absence of T, suggesting the presence of basal activity that we did not detect in the AR14 vector. Despite consistent evidence for higher transcriptional activity with longer repeat lengths, our data also show that extremes in AR activity, whether high or low, seem to have negative effects on C2C12 cell development.
The mechanism by which AR transcriptional activity is positively affected by repeat length in skeletal muscle, while the opposite appears to be true in prostate and kidney cells is unclear. It is possible that alterations of the repeat length induce conformational changes in the AR NTD that enhance or suppress transcriptional cofactor and AR-associated protein interactions. For example, a number of AR-associated proteins such as SRC1, ARNIP, ARA24, p300/CBP, and TFIIF, among others, are known to interact with the NTD. Many of these are known to have histone acetyltransferase activity (22), and altering the binding affinity for these proteins may lead to significant changes in AR transcriptional activity in the tissues in which they are expressed. However, as there are a large number of proteins known to interact with the AR and the exact binding sites are not elucidated, determining which cofactor(s) binding sites might be altered by AR polyglutamine repeat expansion was beyond the scope of the present study. In addition to effects on AR transcriptional activity, disruption of cofactor binding via polyglutamine repeat length variation may also affect AR-mediated expression of myogenic genes. For example, AR is known to induce expression of a skeletal muscle contractile protein indirectly by its association with serum response factor (SRF) (41), and disruption of AR-SRF interactions could affect AR-mediated gene expression of other gene targets. There is also the potential that AR nuclear localization is increased in skeletal muscle with expanded repeat lengths. NTD-LBD facilitates AR activation by allowing interaction of activation function 1 (AF1) and activation function 2 (AF2) domains located in the NTD and LBD, respectively, stabilizing the ligand-binding pocket and exposing the nuclear localization signal (NLS) found in the hinge region (43). Expansion of the glutamine repeat may result in increased exposure of the NLS and, facilitated by cofactor interaction, increased nuclear translocation and transcriptional activation in skeletal muscle cells but not in nonmuscle tissues. Clearly more work is warranted to identify the mechanisms driving these apparently tissue-specific differences in AR activity.
We assessed myogenin and MyoD as markers of proliferation and differentiation to determine if the different AR vectors alter the myogenic process in the C2C12 cells. Despite a significantly reduced rate of proliferation in the AR33 line, there were no substantial differences in MyoD mRNA expression in the myoblast stage, though we did detect differences in the temporal response of myogenin mRNA expression in the different cell lines. Specifically, the AR14 cells upregulated myogenin mRNA expression much quicker than the other lines, which conversely was associated with lower CK activity. However, it appears that the lower CK activity was in part driven by the higher protein content of those particular cells. There was no appreciable difference in myogenin levels after 5 days in DM across the cell lines, yet there were clear differences in the morphological appearances in the myotubes from each cell line. Thus, it appears that the difference in the appearances of myotubes derived from these cell lines are not mediated by alterations in expression of critical myogenic transcription factors.
In addition, we attempted to determine if the overexpression of the different AR vectors affected endogenous gene expression in the C2C12 cells. SkA and myostatin mRNAs were measured in all the cell lines since previous investigations have found these myogenic genes to be sensitive to AR (16, 21). Interestingly, the data demonstrate that overexpression of the AR reduced myostatin mRNA levels compared with WT cells at all the time points. Previous publications have shown that myostatin is negatively regulated by androgen treatment (30), and our data suggest that this is in part mediated by the AR and not due to secondary effects on the myogenic process. It appears that myostatin may be affected by the different repeats in the AR in that myostatin mRNA expression was repressed to a greater extent in AR24 and AR33 lines in line with the greater AR activity we detected in these same cell lines. In contrast, we found minimal differences in SkA mRNA across the cell lines, suggesting minimal effects of AR overexpression on SkA and no effect of the different repeat lengths on SkA expression. This may be in part due to the difference in binding activation of AR to both promoters. Specifically, AR can directly bind to the myostatin promoter, while the AR indirectly binds to the SkA promoter by way of its interaction with SRF (41). Thus, it is possible that the different repeat lengths do not effect transcription under conditions where AR is not directly binding to the promoter.
Our data prompt the question of whether or not the AR polyglutamine repeat polymorphism can affect skeletal muscle mass and serum androgen levels in humans, as several gene association studies indicate (9, 26, 33, 42). Our data indicate that this polymorphism does indeed affect the development of skeletal muscle cells in vitro. We reported the presence of a significantly lower number of AR33 cells after 3 days of culturing in GM, roughly 25 and 20% fewer cells compared with AR14 and AR24 lines, respectively. This indicates that the greatly increased transcriptional activity of the AR33 construct has a negative effect on myoblast proliferation, a finding that seems to be at odds with a correlation of increased AR repeat length and lean body mass (42). Activated satellite cells go through a phase of proliferation before fusing to one another or to existing myofibers, and decreasing the ability of myogenic cells to proliferate would limit the growth of existing fibers and the formation of new fibers. In contrast, the AR14 line demonstrated significantly lower CK activity after 5 days of differentiation compared with all other lines. As contractile protein content is limited in myoblasts, a reduced differentiation capacity would likely result in lower skeletal muscle protein content and force production in vivo. In fact, it was recently demonstrated that loss of functional AR results in a loss of myofilaments and Z-line disruptions in rodent hindlimb muscle (13). These data indicate that shorter AR repeat lengths, and subsequently lower AR transcriptional activity, may result in altered myofiber morphology as well as decreased muscle strength. This hypothesis was further supported by an examination of each line over 5 days of incubation in DM, where remarkable differences in the morphology of the AR14 and AR33 lines were apparent. AR14 cells developed into highly elongated, thin myofibers with very sparse myonuclei. In contrast, the AR33 cells developed into rather short, cylindrical myotubes with myonuclei clustered together but still far fewer in number than WT or AR24 cells. In addition, fusion index and myonuclear number were heavily suppressed in both the AR14 and AR33 lines, further indicating deficiencies in myotube development and fusion. Collectively these data support a hypothesis of decreased skeletal muscle mass in humans carrying exceptionally long or short AR CAG repeat lengths.
We set out to determine the effect of CAG repeat length on AR transcriptional activity in skeletal muscle cells and to elucidate in vitro mechanisms by which CAG repeat length may affect skeletal muscle mass in vivo. Walsh et al. (42) reported a positive correlation of lean body mass and repeat length in healthy men, but the effect was physiologically small, and a repeat length of 22 was used as the cut-off point between short and long repeat lengths, respectively. Though their sample size was reasonable, it is possible a larger sample size and grouping of “short” and “long” groups near the extremes of the normal range could provide a different outcome. In contrast, Nielsen et al. (33) reported ∼4% higher lean thigh mass per 10 CAG repeat reduction. Nielsen et al. also treated CAG repeat length as a continuous variable and subject age was limited to 20–29 yr, whereas Walsh et al. (42) studied subjects ranging from 19 to 90 yr of age and CAG repeat lengths were analyzed as either <22 or ≥ to 22 repeats. The data of Campbell et al. (9), which found higher lean mass with longer repeat length, were obtained from a small sample size of a very specific group of African men, which may or may not accurately represent a broader population. Linking these gene association findings directly to the present functional analysis of AR repeat length in vitro is challenging given the genetic complexity of skeletal muscle phenotypes and inconsistencies across studies; however, the present study provides support for the AR repeat polymorphism as a contributing gene to skeletal muscle traits, though very likely modified by a number of other gene variants.
In summary, our data demonstrate that extremes in AR activity, whether high or low, seem to have negative effects on C2C12 cell development, but that higher repeat lengths in the normal range demonstrate a positive relationship with AR transcriptional activity. Our study provides an important first step toward a more comprehensive in vivo analysis of the importance of altered AR signaling in relation to CAG repeat length in skeletal muscle.
GRANTS
Funding was provided by National Institute on Aging Grants AG-022791 and AG-021500.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Alen P, Claessens F, Verhoeven G, Rombauts W, Peeters B. The androgen receptor amino-terminal domain plays a key role in p160 coactivator-stimulated gene transcription. Mol Cell Biol 19: 6085–6097, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allan CA, Strauss BJ, Burger HG, Forbes EA, McLachlan RI. Testosterone therapy prevents gain in visceral adipose tissue and loss of skeletal muscle in nonobese aging men. J Clin Endocrinol Metab 93: 139–146, 2008 [DOI] [PubMed] [Google Scholar]
- 3. Beilin J, Ball EM, Favaloro JM, Zajac JD. Effect of the androgen receptor CAG repeat polymorphism on transcriptional activity: specificity in prostate and non-prostate cell lines. J Mol Endocrinol 25: 85–96, 2000 [DOI] [PubMed] [Google Scholar]
- 4. Bhasin S, Storer TW, Berman N, Callegari C, Clevenger B, Phillips J, Bunnell TJ, Tricker R, Shirazi A, Casaburi R. The effects of supraphysiologic doses of testosterone on muscle size and strength in normal men. N Engl J Med 335: 1–7, 1996 [DOI] [PubMed] [Google Scholar]
- 5. Bhasin S, Woodhouse L, Casaburi R, Singh AB, Bhasin D, Berman N, Chen X, Yarasheski KE, Magliano L, Dzekov C, Dzekov J, Bross R, Phillips J, Sinha-Hikim I, Shen R, Storer TW. Testosterone dose-response relationships in healthy young men. Am J Physiol Endocrinol Metab 281: E1172–E1181, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Bhasin S, Woodhouse L, Storer TW. Proof of the effect of testosterone on skeletal muscle. J Endocrinol 170: 27–38, 2001 [DOI] [PubMed] [Google Scholar]
- 7. Brodsky IG, Balagopal P, Nair KS. Effects of testosterone replacement on muscle mass and muscle protein synthesis in hypogonadal men–a clinical research center study. J Clin Endocrinol Metab 81: 3469–3475, 1996 [DOI] [PubMed] [Google Scholar]
- 8. Burd CJ, Petre CE, Moghadam H, Wilson EM, Knudsen KE. Cyclin D1 binding to the androgen receptor (AR) NH2-terminal domain inhibits activation function 2 association and reveals dual roles for AR corepression. Mol Endocrinol 19: 607–620, 2005 [DOI] [PubMed] [Google Scholar]
- 9. Campbell BC, Gray PB, Eisenberg DT, Ellison P, Sorenson MD. Androgen receptor CAG repeats and body composition among Ariaal men. Int J Androl 32: 140–148, 2009 [DOI] [PubMed] [Google Scholar]
- 10. Caravatti M, Perriard JC, Eppenberger HM. Developmental regulation of creatine kinase isoenzymes in myogenic cell cultures from chicken. Biosynthesis of creatine kinase subunits M and B. J Biol Chem 254: 1388–1394, 1979 [PubMed] [Google Scholar]
- 11. Chamberlain NL, Driver ED, Miesfeld RL. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucl Acids Res 22: 3181–3186, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chamberlain NL, Whitacre DC, Miesfeld RL. Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J Biol Chem 271: 26772–26778, 1996 [DOI] [PubMed] [Google Scholar]
- 13. Chambon C, Duteil D, Vignaud A, Ferry A, Messaddeq N, Malivindi R, Kato S, Chambon P, Metzger D. Myocytic androgen receptor controls the strength but not the mass of limb muscles. Proc Natl Acad Sci USA 107: 14327–14332, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dejager S, Bry-Gauillard H, Bruckert E, Eymard B, Salachas F, LeGuern E, Tardieu S, Chadarevian R, Giral P, Turpin G. A comprehensive endocrine description of Kennedy's disease revealing androgen insensitivity linked to CAG repeat length. J Clin Endocrinol Metab 87: 3893–3901, 2002 [DOI] [PubMed] [Google Scholar]
- 15. Diel P, Baadners D, Schlupmann K, Velders M, Schwarz JP. C2C12 myoblastoma cell differentiation and proliferation is stimulated by androgens and associated with a modulation of myostatin and Pax7 expression. J Mol Endocrinol 40: 231–241, 2008 [DOI] [PubMed] [Google Scholar]
- 16. Diel P, Friedel A, Geyer H, Kamber M, Laudenbach-Leschowsky U, Schanzer W, Thevis M, Vollmer G, Zierau O. Characterisation of the pharmacological profile of desoxymethyltestosterone (Madol), a steroid misused for doping. Toxicol Lett 169: 64–71, 2007 [DOI] [PubMed] [Google Scholar]
- 17. Edwards A, Hammond HA, Jin L, Caskey CT, Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics 12: 241–253, 1992 [DOI] [PubMed] [Google Scholar]
- 18. Fu R, Liu J, Fan J, Li R, Li D, Yin J, Cui S. Novel evidence that testosterone promotes cell proliferation and differentiation via G protein-coupled receptors in the rat L6 skeletal muscle myoblast cell line. J Cell Physiol [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19. Giovannucci E, Platz EA, Stampfer MJ, Chan A, Krithivas K, Kawachi I, Willett WC, Kantoff PW. The CAG repeat within the androgen receptor gene and benign prostatic hyperplasia. Urology 53: 121–125, 1999 [DOI] [PubMed] [Google Scholar]
- 20. Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, Hennekens CH, Kantoff PW. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci USA 94: 3320–3323, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hong MH, Sun H, Jin CH, Chapman M, Hu J, Chang W, Burnett K, Rosen J, Negro-Vilar A, Miner JN. Cell-specific activation of the human skeletal alpha-actin by androgens. Endocrinology 149: 1103–1112, 2008 [DOI] [PubMed] [Google Scholar]
- 22. Janne OA, Moilanen AM, Poukka H, Rouleau N, Karvonen U, Kotaja N, Hakli M, Palvimo JJ. Androgen-receptor-interacting nuclear proteins. Biochem Soc Trans 28: 401–405, 2000 [PubMed] [Google Scholar]
- 23. Jimenez A, Davies J. Expression of a transposable antibiotic resistance element in Saccharomyces. Nature 287: 869–871, 1980 [DOI] [PubMed] [Google Scholar]
- 24. Kasper S, Rennie PS, Bruchovsky N, Lin L, Cheng H, Snoek R, Dahlman-Wright K, Gustafsson JA, Shiu RP, Sheppard PC, Matusik RJ. Selective activation of the probasin androgen-responsive region by steroid hormones. J Mol Endocrinol 22: 313–325, 1999 [DOI] [PubMed] [Google Scholar]
- 25. Katznelson L, Finkelstein JS, Schoenfeld DA, Rosenthal DI, Anderson EJ, Klibanski A. Increase in bone density and lean body mass during testosterone administration in men with acquired hypogonadism. J Clin Endocrinol Metab 81: 4358–4365, 1996 [DOI] [PubMed] [Google Scholar]
- 26. Krithivas K, Yurgalevitch SM, Mohr BA, Wilcox CJ, Batter SJ, Brown M, Longcope C, McKinlay JB, Kantoff PW. Evidence that the CAG repeat in the androgen receptor gene is associated with the age-related decline in serum androgen levels in men. J Endocrinol 162: 137–142, 1999 [DOI] [PubMed] [Google Scholar]
- 27. MacLean HE, Chiu WS, Notini AJ, Axell AM, Davey RA, McManus JF, Ma C, Plant DR, Lynch GS, Zajac JD. Impaired skeletal muscle development and function in male, but not female, genomic androgen receptor knockout mice. FASEB J 22: 2676–2689, 2008 [DOI] [PubMed] [Google Scholar]
- 28. Markus SM, Taneja SS, Logan SK, Li W, Ha S, Hittelman AB, Rogatsky I, Garabedian MJ. Identification and characterization of ART-27, a novel coactivator for the androgen receptor N terminus. Mol Biol Cell 13: 670–682, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Megeney LA, Rudnicki MA. Determination versus differentiation and the MyoD family of transcription factors. Biochem Cell Biol 73: 723–732, 1995 [DOI] [PubMed] [Google Scholar]
- 30. Mendler L, Baka Z, Kovacs-Simon A, Dux L. Androgens negatively regulate myostatin expression in an androgen-dependent skeletal muscle. Biochem Biophys Res Commun 361: 237–242, 2007 [DOI] [PubMed] [Google Scholar]
- 31. Miller KK, Biller BM, Beauregard C, Lipman JG, Jones J, Schoenfeld D, Sherman JC, Swearingen B, Loeffler J, Klibanski A. Effects of testosterone replacement in androgen-deficient women with hypopituitarism: a randomized, double-blind, placebo-controlled study. J Clin Endocrinol Metab 91: 1683–1690, 2006 [DOI] [PubMed] [Google Scholar]
- 32. Nenonen H, Bjork C, Skjaerpe PA, Giwercman A, Rylander L, Svartberg J, Giwercman YL. CAG repeat number is not inversely associated with androgen receptor activity in vitro. Mol Hum Reprod 16: 153–157, 2010 [DOI] [PubMed] [Google Scholar]
- 33. Nielsen TL, Hagen C, Wraae K, Bathum L, Larsen R, Brixen K, Andersen M. The impact of the CAG repeat polymorphism of the androgen receptor gene on muscle and adipose tissues in 20–29-year-old Danish men: Odense Androgen Study. Eur J Endocrinol 162: 795–804, 2010 [DOI] [PubMed] [Google Scholar]
- 34. Perriard JC, Caravatti M, Perriard ER, Eppenberger HM. Quantitation of creatine kinase isoenzyme transition in differentiating chicken embryonic breast muscle and myogenic cell cultures by immunoadsorption. Arch Biochem Biophys 191: 90–100, 1978 [DOI] [PubMed] [Google Scholar]
- 35. Rogozkin V. Metabolic effects of anabolic steroid on skeletal muscle. Med Sci Sports 11: 160–163, 1979 [PubMed] [Google Scholar]
- 36. Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary MyoD-/- myogenic cells derived from adult skeletal muscle. J Cell Biol 144: 631–643, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet 57: 16–25, 2000 [DOI] [PubMed] [Google Scholar]
- 38. Sattler FR, Castaneda-Sceppa C, Binder EF, Schroeder ET, Wang Y, Bhasin S, Kawakubo M, Stewart Y, Yarasheski KE, Ulloor J, Colletti P, Roubenoff R, Azen SP. Testosterone and growth hormone improve body composition and muscle performance in older men. J Clin Endocrinol Metab 94: 1991–2001, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shank LC, Paschal BM. Nuclear transport of steroid hormone receptors. Crit Rev Eukaryot Gene Expr 15: 49–73, 2005 [DOI] [PubMed] [Google Scholar]
- 40. Tut TG, Ghadessy FJ, Trifiro MA, Pinsky L, Yong EL. Long polyglutamine tracts in the androgen receptor are associated with reduced trans-activation, impaired sperm production, and male infertility. J Clin Endocrinol Metab 82: 3777–3782, 1997 [DOI] [PubMed] [Google Scholar]
- 41. Vlahopoulos S, Zimmer WE, Jenster G, Belaguli NS, Balk SP, Brinkmann AO, Lanz RB, Zoumpourlis VC, Schwartz RJ. Recruitment of the androgen receptor via serum response factor facilitates expression of a myogenic gene. J Biol Chem 280: 7786–7792, 2005 [DOI] [PubMed] [Google Scholar]
- 42. Walsh S, Zmuda JM, Cauley JA, Shea PR, Metter EJ, Hurley BF, Ferrell RE, Roth SM. Androgen receptor CAG repeat polymorphism is associated with fat-free mass in men. J Appl Physiol 98: 132–137, 2005 [DOI] [PubMed] [Google Scholar]
- 43. Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem 269: 13115–13123, 1994 [PubMed] [Google Scholar]