Abstract
Intrauterine growth retardation (IUGR) predisposes humans toward hippocampal morbidities, such as impaired learning and memory. Hippocampal dual specificity phosphatase 5 (DUSP5) may be involved in these morbidities because DUSP5 regulates extracellular signal-regulated kinase phosphorylation (Erk). In the rat, IUGR causes postnatal changes in hippocampal gene expression and epigenetic characteristics. However, the impact of IUGR upon hippocampal DUSP5 expression and epigenetic characteristics is not known. We therefore hypothesized that IUGR affects hippocampal 1) DUSP5 expression, DNA CpG methylation, and histone code, and 2) erk1/2 phosphorylation in a well-characterized rat model of IUGR. We found that IUGR significantly decreased DUSP5 expression in the day of life (DOL) 0 and 21 male rat, while decreasing only DUSP5 protein levels in the DOL21 female rat. Fluorescent in situ hybridization and immunohistochemistry analyses localized the changes in DUSP5 mRNA and protein, many of which occurred in the dentate gyrus. IUGR also caused sex-specific differences in DNA CpG methylation and histone code in two sites of the hippocampal DUSP5 gene, a 5′-flanking specificity protein-1 (SP1) site and exon 2. Finally, when IUGR decreased DUSP5 protein levels, Erk phosphorylation increased. We conclude that IUGR affects hippocampal DUSP5 expression and epigenetic characteristics in a sex-specific manner.
Keywords: hippocampus, DUSP5 expression, epigenetic determinants, uteroplacental insufficiency, learning and memory, extracellular signal-related kinases 1 and 2
uteroplacental insufficiency causes intrauterine growth restriction (IUGR) and a predisposition toward neurodevelopmental impairment. In humans, this predisposition often manifests itself through decreased academic and standardized test performance (14, 21, 30, 31, 40). These performance measures involve learning and memory, functions of the hippocampus. It is therefore relevant that IUGR also reduces preterm infant's hippocampal volume in association with functional behavioral differences (32). Therefore, determining the effect of IUGR on hippocampal molecular pathways that influence learning and memory is important for treatment and prognostic approaches.
In rats, uteroplacental insufficiency and subsequent IUGR affects hippocampal apoptosis, gene expression, and chromatin structure. The latter observation occurs both on a genome-wide and gene-specific level (22, 24). Moreover, these effects occur in a sex-specific manner. Missing from these studies is a focus upon pathways that directly impact learning and memory. Mitogen-activated protein kinase (MAPK) signaling through extracellular signal-related kinases 1 and 2 (Erk1/2) moderates hippocampal learning and memory (46, 49). A key regulator of MAP kinase signaling is dual specificity phosphatase 5 (DUSP5).
DUSP5 regulates MAPK signaling by specifically dephosphorylating Erk1/2 (17, 34). A characteristic of hippocampal DUSP5 that makes it an attractive target for study is that this gene's chromatin structure may respond to the IUGR insult by altering its epigenetic characteristics. This is suggested by previous studies in which IUGR altered hepatic DUSP5 expression, exon 2 histone 3 lysine 14 acetylation (H3K14ac) and DNA CpG methylation (12). Importantly, the differences between control (Con) and IUGR DNA CpG methylation persist into adulthood in a sex-specific manner. Specifically, IUGR decreased hepatic DUSP5 exon 2 DNA CpG methylation in adult male rats, while increasing hepatic DUSP5 exon 2 DNA CpG methylation in adult female rats (12).
Alterations in DNA CpG methylation and histone covalent modifications such as acetylation are common epigenetic mechanisms that regulate gene expression. These mechanisms can result in persistent differences in mRNA transcription by altering accessibility of DNA to transcription factor complexes. Because the question exists on whether it is reasonable to presume that differences in gene expression and epigenetic changes are generalizable from one tissue to another, we hypothesized that IUGR affects 1) hippocampal DUSP5 gene expression, DNA CpG methylation, and histone code and 2) hippocampal erk1/2 phosphorylation. We further hypothesized that IUGR affects these measures in a sex-specific fashion.
To test our hypotheses, we used the well-characterized model of uteroplacental insufficiency though bilateral uterine artery ligation in the pregnant Sprague-Dawley rat (3, 19, 24). The rat pup in this model suffers fetal hypoinsulinemia, hypoglycemia, acidosis, and hypoxia, similar to the human fetus in a gestation complicated by pregnancy induced hypertension (8–10). Litter size is not significantly affected in this model (29). Studies were performed at day of life 0 (DOL0) and day of life 21 (DOL21). DOL21 was selected to 1) allow direct comparison with the previous hepatic study, 2) to assess whether any changes induced at DOL0 were persistent, and 3) to minimize the confounding variable of the hormonal changes associated maturation and senescence.
We measured and localized DUSP5 gene expression through real-time RT-PCR, Western blotting, fluorescent in situ hybridization (FISH), and immunohistochemistry (IHC). DUSP5 hippocampal epigenetic characteristics (DNA CpG methylation and histone code) were assessed at the two sites assessed in the previous hepatic study: 1) a 5′-flanking specificity protein-1 (SP1) site in the promoter (SP1Pr) and 2) a region in exon 2 (Ex2) previously demonstrated in liver to be vulnerable to changes DNA CpG methylation in response to IUGR (12). SP1Pr site was considered a relevant target because Sp1 is a ubiquitously expressed transcription factor that regulates the expression of genes involved in almost all cellular processes, and methylation of the Sp1 site in the proximal promoter is related to gene silencing (4, 26, 39). Finally, because no direct measures of in vivo hippocampal DUSP5 function presently exist, and DUSP5 specifically dephosphorylates Erk1/2, we determined whether hippocampal erk1/2 phosphorylation using Western blotting.
MATERIALS AND METHODS
Animals.
All procedures were approved by the University of Utah Institutional Animal Care and Use Committee and are in accordance with the American Physiological Society's guiding principles (1). Surgical methods have been described previously (20, 24). In brief, on day 19.5 of gestation (term gestation in the rat is 21.5 days), pregnant rats were anesthetized with intraperitoneal xylazine (8 mg/kg) and ketamine (40 mg/kg), and both uterine arteries were ligated (IUGR) (n = 12 litters). Control animals underwent identical anesthesia (Con) (n = 12 litters). Day 0 pups (DOL0) were delivered by caesarian section at term (n = 6 litters Con and IUGR, respectively). Brains were quickly removed and either dissected to isolate the hippocampus and flash-frozen for hippocampi collection or processed for histological evaluation (23). DOL0 pups were genotyped using PCR for the spermatogenic gene (Sby) from the Y chromosome to ensure an equal distribution of each sex for each methodology (forward primer: 5′-ACTGTTCAAGCAGTCAGCCG; reverse primer: 5′-CTCCATGAACTTGGGGTC) (13, 33).
To obtain day 21 pups (DOL21), dams were allowed to deliver spontaneously, and litters were culled to six as previously described (20, 24). DOL21 rats were separated from their dams for 4 h without access to food, anesthetized, and killed (n = 6 litters con and IUGR, respectively). Brains were quickly removed, and hippocampi were dissected and flash-frozen in liquid nitrogen and stored in −80°C, unless tissue was to be used for IHC. For the latter, animals were perfused and the brain fixed with 10% formalin. Brains were then removed and paraffin embedded.
RNA isolation and real-time RT-PCR.
Total RNA was extracted from DOL0 and DOL21 hippocampi using RNeasy Mini Kit (Qiagen, Valencia, CA) and quantified using the NanoDrop spectrometer ND-1000 (NanoDrop Technologies, Wilmington, DE). RNA was treated with DNase (Ambion, Austin, TX). Gel electrophoresis was used to confirm the integrity of the samples.
Hippocampal mRNA levels of DUSP5 were measured using real-time RT-PCR at DOL0 and DOL21 as previously described (33). The probe and primers were designed using Primer Express (Applied Biosystems, Foster, CA) with the reporter dye FAM and the quencher dye TAMRA (Table 1). In brief, cDNA was synthesized from 2 μg of DNase-treated total RNA. cDNA, probe, and primers were added to Taqman universal PCR master mix (Applied Biosystems), and samples were run on an ABI Prism 7900. Real-time RT-PCR quantification was then performed using the Taqman glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. Relative quantification of PCR products was based on value differences between the target and GAPDH control, using the comparative CT method (Taqman Gold RT-PCR manual; PE Biosystems, Foster City, CA). Cycle parameters were 50°C, 2 min, 95°C, 10 min, and then 40 cycles at 95°C, 15 s, 60°C, 60 s. For each set of reactions, samples were run in triplicate.
Table 1.
PCR primers
| Primers and Probes | Accession # | |
|---|---|---|
| Real-time RT-PCR | ||
| DUSP5 exon 2 | For 5′ CGTCGCCTACAGACCAGCTT | |
| Rev 5′ TCCAAGGTAGAGGAAGGGAAGG | ||
| Probe 6FAM-TTTCAACTGGGCCACCCTGGTCA | ||
| GAPDH | For 5′ CAAGATGGTGAAGGTCGGTGT | M17701 |
| Rev 5′ CAAGAGAAGGCAGCCCTGGT | ||
| Probe 6FAM-GCGTCCGATACGGCCAAATCCG | ||
| In situ hybridization | ||
| DUSP5 | For 5′GCCCATTTCACAAGAGAAGC | |
| Rev 5′ATTCAGCAGGGCTGTGATGT | ||
| Bisulfite sequencing | ||
| DUSP5 SP1Pr | For 5′ GAAATAGTTGGTAAAGGGTTTGGAGG | NM_133578.1 |
| Rev 5′ CTATCTACCCCCACCATTCTCTC | ||
| DUSP5 exon 2 | For 5′ TTTTTATTTTTGAAGGTGGGTAYGAG | |
| Rev 5′ CACATACCTAATCATAAACTAATCTATAAAC | ||
| ChIP | ||
| DUSP5 SPPr | For 5′ AGCTAGCAGAGCCCTCAATAAATACT | |
| Rev 5′ TGCCCGCCCCCATT | ||
| Probe 6FAM_AGAGCAGACGAATCC | ||
| DUSP5 exon 2 | For 5′ ACAAGAGAAGCTCGAAGGTGAGA | |
| Rev 5′ AGGCGACGCTGAGAATGG | ||
| Probe 6FAM_AGGCCTCCTCAGCC | ||
Protein isolation and Western blotting.
Hippocampi were homogenized in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris pH 7.4, 1 mM EDTA, 0.25% Na-deoxycholate, 1% Igepal CA-630, 1 mM PMSF) with protease inhibitor cocktail (Roche). After centrifugation at 10,000 rpm for 15 min(4°C), the supernatants were transferred into new tubes and stored at −80°C until use. Protein concentrations were determined by the BCA method (Pierce, Rockford, IL). Proteins were separated by 10% SDS-PAGE ready gels (Bio-Rad, Hercules, CA) and transferred to PVDF membranes in standard transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol). After blocking the membranes with 5% milk in Tris buffered saline (TBS) for 1 h, we exposed bound proteins to the following antibodies overnight (4°C): DUSP5 (rabbit polyclonal; Animal Pharm Service, Healdsburg, CA), Erk1/Erk2, phospho-Erk1/Erk2 and GAPDH (rabbit polyclonal, Cell Signaling). GAPDH was used as an internal control. After extensive washing in TBS with 0.1% Tween 20 (TBST), a 1/2,000 dilution of goat anti-rabbit horseradish peroxidase (HRP) secondary antibody (Cell Signaling Technology) was applied, and the membrane was incubated for 1 h at room temperature. After extensive washing in TBST, blots were detected with Western Lightning ECL (PerkinElmer Life Sciences) quantification on a Kodak Image Station 2000R (Eastman Kodak/SIS, Rochester, NY).
FISH and IHC analysis for DUSP5 localization.
Coronal sections from DOL0 and DOL21 brains were deparaffinized and rehydrated in a graded series of ethanol and DEPC H2O for FISH or dH2O with a final wash in PBS for IHC.
For FISH, sections were refixed in 10% neutral buffered formalin for 60 min. After being washed in diethylpyrocarbonate (DEPC) H2O for 10 min, sections were treated with 4% H2O2 and 0.4% Tween 20 in DEPC H2O for 20 min, followed by a DEPC H2O wash for 10 min. Equilibrated tissue was then placed in proteinase K buffer (PK buffer: 50 mM Tris·HCl, 50 mM NaCl) for 20 min and digested tissue in PK solution (5 μg/ml proteinase K and 2 mM CaCl2 in PK buffer) for 60 min at room temperature (RT) with agitation. Sections were then washed in DEPC water for 20 min and air-dried thoroughly. Once dry, sections were treated with a hybridization solution and denatured digoxigenin (DIG)-labeled probe (made from PCR DIG probe synthesis kit, Roche, cat. #1636090) and incubated at 45°C overnight in a moisture chamber. The primers used for probe generation are listed in Table 1. The next day, sections were briefly rinsed with 4× SSC, washed in 4× SSC with 4% formamide with gentle agitation for 30 min (RT), and then washed in 4× SSC with 4% formamide again for 30 min at 50°C. After a brief rinse in 10 mM PBS for 5 min at RT, sections were incubated in a blocking solution (TSA Fluoresce system kit; PerkinElmer, Boston, MA) for 30 min at RT. The slides were then placed in an anti-DIG/HRP-conjugated solution (1:250 dilution) for 1 h at RT and then rinsed with PBS. Signals were amplified with tyramide rhodamine-conjugated solution (1:200 dilution) for 10 min. Finally, sections were rinsed with PBS, mounted, and coverslipped with DAPI mounting medium.
For IHC, sections were incubated in a 3% H2O2 solution for 30 min at room temperature (20–22°C) to quench endogenous peroxidase activity. Following a brief PBS wash, slides were subjected to an antigen retrieval procedure (Biogenex Laboratories, San Ramon, CA). For antigen retrieval, slides were put in a slide tray with 10 mM citrate solution (pH 6.0) and heated at high power in a microwave oven for 165 s followed by low power heating for 8 min. Slides were then cooled to room temperature, rinsed with tap water, washed in PBS for 10 min, and incubated in a blocking buffer (TSA Biotin System kit, PerkinElmer) at room temperature for 1 h. Following blocking, slides were exposed to DUSP5 rabbit polyclonal antibodies (1:200 dilution in blocking buffer) overnight at 4°C in a humidified chamber. The next day, sections were washed three times in PBS containing 0.2% Tween 20 (PBST) and exposed to biotinylated goat anti-rabbit immunoglobulins for 1 h at RT. After a 1 h incubation in Vectastain ABC mixture, slides were washed in PBST for 15 min and stained with 3,3-diaminobenzidine (Sigma, St. Louis, MO). Finally, slides were counterstained with hematoxylin, dehydrated, and coverslipped with Cytoseal 60 (Stephens Scientific, Kalamazoo, MI).
DNA isolation and sodium bisulfite sequencing.
Genomic DNA was extracted from DOL0 and DOL21 hippocampi. In brief, DOL0 and DOL21 hippocampi were digested overnight at 56°C in 670 μl of a proteinase K solution (10 mM Tris, pH 7.6, 25 mM EDTA, 75 mM NaCl, 1% SDS, 180 μg/ml proteinase K). Digests were then centrifuged for 15 min at 13,000 rpm. Supernatant was transferred into a fresh tube, 200 μl of saturated NaCl solution (∼6 M, RT) was added, and each tube was vortexed for 10–15 s. An equal volume of chloroform (∼870 μl) was added to each tube and then all samples were briefly vortexed. Samples were kept on ice for 10 min and then spun for 5 min at 13,000 rpm at 4°C. We transferred 500 μl supernatant to a fresh tube and added 1 ml of cold, absolute ethanol. The tube was gently inverted to mix and then spun for 2 min at 13,000 rpm. The supernatant was removed. The pellet was washed with cold 70% ethanol and rinsed with 95% ethanol. The resultant DNA was air dried and resuspended in 100 μl Tris-EDTA buffer (pH 7.4). Following resuspension, 1 μl of RNase (20 mg/ml) was added to each tube and all samples were incubated at 37°C for 30 min. DNA was quantified using the NanoDrop spectrometer ND-1000 (NanoDrop Technologies). To determine site specific CpG methylation, genomic DNA was subjected to sodium bisulfite modification according to the manufacture's protocol (CpGenome DNA modification kit; Chemicon International, Temecula, CA). DNA regions containing CpG sites that are within the SP1Pr site and exon 2 of DUSP5 were amplified. The PCR primers are listed in Table 1. Primers designed for exon 2 were spanned intron 1 to exon 2 and intron 2 to exon 2. PCR conditions were 95°C for 10 min, followed by 94°C for 30s, 56°C for 1 min, 72°C for 1 min, 35 cycles. The PCR products were cloned into pCR2.1 using the TOPO TA Cloning Kit (Invitrogen, San Diego, CA). Multiple colonies derived from the cloning procedure were sequenced according to the manufacturer's instructions for double-stranded plasmid DNA using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems).
Chromatin immunoprecipitation (ChIP) assay and real-time PCR.
Hippocampi were dissected from control and IUGR rat brains at DOL0 and DOL21. To perform the ChIP assay, 10 hippocampi from DOL0 rats needed to be pooled into each sex/treatment group(n = 4). ChIP with antibodies against histone H3 lysine 9 acetylation (H3K9ac), H3 lysine 14 acetylation (H3K14ac), H3 lysine 4 dimethylation (H3K4me2), H3 lysine 4 trimethylation (H3K4me3), H3K9me3, and H3K36me3 performed as described before (Cell Signaling Technologies, Beverly, MA) (12). DNA was quantified by measuring A260/A280. Real-time PCR was used to quantitate the amount of DNA from the DUSP5 SP1Pr, DUSP5 Ex2, and an intergenic sequence 263.8 kb upstream of IGF-1 gene. The PCR primers are listed in Table 1. The intergenic sequence which is not transcribed was used as an internal control. The amount of SP1Pr and Ex2 DNA in each sample was quantified relative to the amount of intergenic region (13). Two control experiments were performed simultaneously with our histone modification-specific ChIP experiments. First, we performed a “mock” ChIP that included input but did not utilize antibody. Second, we performed a ChIP that utilized a nonspecific antibody, which in this case was a secondary anti-rabbit negative control.
Statistics.
All data presented are expressed as the mean percentage of control ± SE. ANOVA determined statistical significance for data sets involving DUSP5 expression and epigenetic characteristics, including changes at specific histone or CpG sites (P < 0.05). We did this to allow for sex comparison. The student's t-test determined statistical significance for Erk phosphorylation. Different aged animals were split into separate analysis. To determine if differences existed between multiple groups, a post hoc comparison was conducted using Fisher's least protected square difference test.
RESULTS
Hippocampal DUSP5 expression.
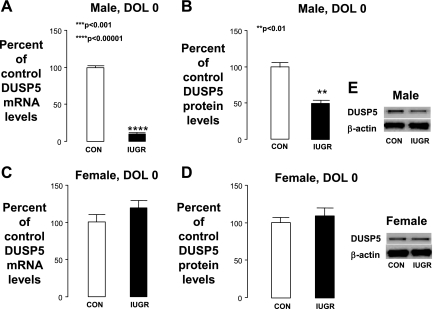
In DOL0 male pups, IUGR significantly decreased hippocampal DUSP5 mRNA and protein levels (Fig. 1, A and B). IUGR did not affect hippocampal DUSP5 mRNA and protein levels in the DOL0 IUGR female pups (Fig. 1, C and D).
Fig. 1.
Day of life (DOL) 0 hippocampal dual specificity phosphatase 5 (DUSP5) mRNA and protein levels. A–D: graphs representing DUSP5 mRNA and protein expressed as % of control ± SE for male pups (A, B) and female pups (C, D). Intrauterine growth restriction (IUGR) values are presented as black bars. E: representative Western blots for DUSP5 and β-actin (internal control). Protein from a control (Con) hippocampus is in the left lanes, and protein from an IUGR hippocampus is in the right. ****P < 0.0001, ***P < 0.001, **P < 0.01.
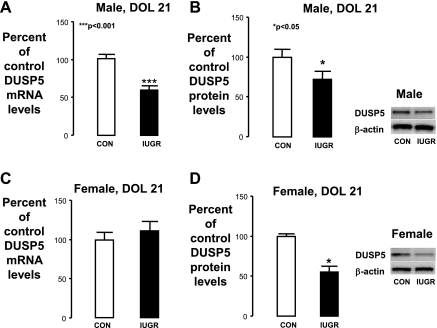
At DOL21, IUGR similarly decreased hippocampal DUSP5 mRNA and protein levels in male rats (Fig. 2, A and B). IUGR also decreased female hippocampal DUSP5 protein levels at DOL21 without affecting the associated mRNA levels (Fig. 2, C and D).
Fig. 2.
DOL21 hippocampal DUSP5 mRNA and protein levels. A–D: graphs representing DUSP5 mRNA and protein expressed as % of control ± SE for male pups (A, B) and female pups (C, D). IUGR values are presented as black bars. Right: representative Western blots for DUSP5 and β-actin (internal control). Protein from a Con hippocampus is in the left lanes, and protein from an IUGR hippocampus is in the right. ***P < 0.001, *P < 0.05.
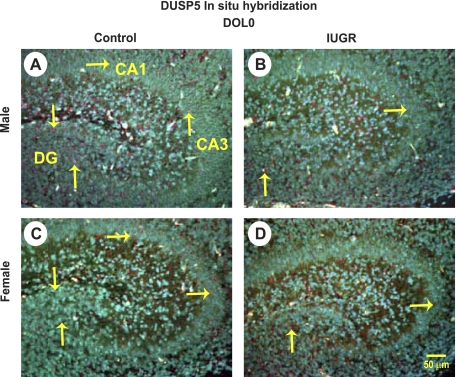
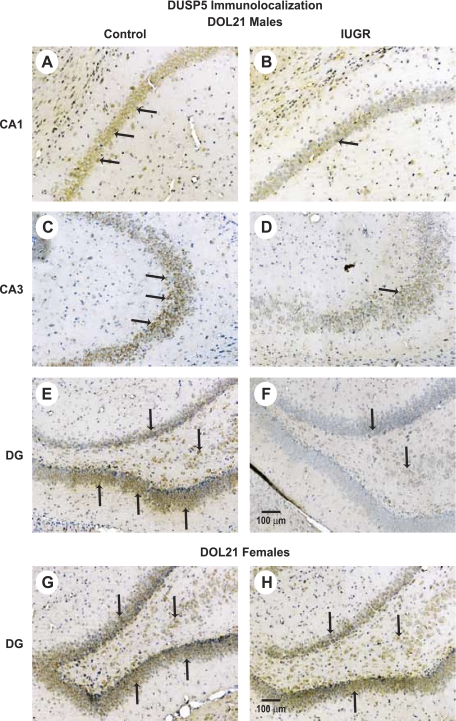
FISH and IHC were used to localize DUSP5 expression mRNA and protein, respectively. In general, decreased staining for DUSP5 expression was evident in the IUGR brains relative to sex-matched control brains. At DOL0, less expression of DUSP5 mRNA appeared to be localized in the dentate gyrus (DG) region in IUGR males (Fig. 3, A and B), while there was no difference observed in IUGR females (Fig. 3, C and D). At DOL21, IUGR appeared to decrease DUSP5 protein expression throughout all regions of the hippocampus in males (Fig. 4A) and in the DG region in IUGR females (Fig. 4B). There was no difference observed in the regions of cornu ammonis (CA)1 and CA3 between control and IUGR females (data no shown).
Fig. 3.
Representative fluorescent in situ hybridization of DUSP5 (red staining) from hippocampal cornu ammonis (CA)1, CA3, and dentate gyrus (DG) region of DOL0 control male pups (A), DOL0 IUGR pups (B), DOL0 control female pups (C), and DOL0 IUGR female pups (D). Scale bar = 50 μm (n = 6 litters). Arrows point to representative DUSP5-positive cells.
Fig. 4.
Representative immunohistochemistry of DUSP5 (brown staining) from the hippocampal DG region of DOL21 control male pups (A), DOL 21 IUGR pups (B), from hippocampal DG region of DOL21 control female pups (C), and DOL21 IUGR female pups (D). Scale bar = 100 μm (n = 6 litters). Arrows point to representative DUSP5-positive cells.
Hippocampal DUSP5 DNA CpG methylation.
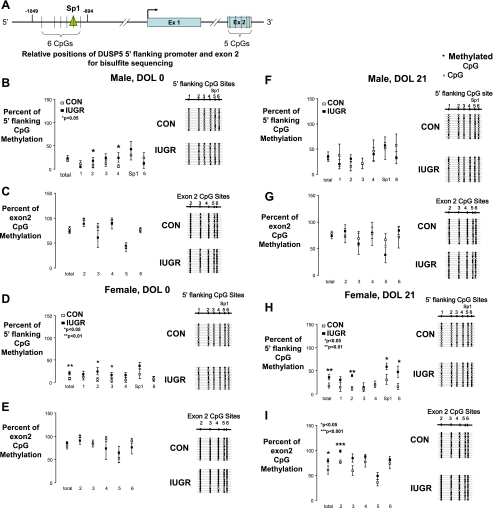
In DOL0 male rat hippocampus, IUGR significantly increased DUSP5 SP1Pr DNA CpG methylation at CpG “2” and “4” without affecting Ex2 DNA CpG methylation (Fig. 5, B and C). In DOL0 female rat hippocampus, IUGR significantly increased DUSP5 SP1Pr CpG methylation at CpG “2” and “3,” as well as total CpG methylation across the SP1Pr-targeted region. Similar to the males, IUGR did not affect Ex2 hippocampal female CpG methylation at DOL0 (Fig. 5, D and E).
Fig. 5.
Hippocampal CpG methylation of DUSP5 in 5′-flanking SP1Pr region and exon 2 (Ex2). A: schematic diagram of the DUSP5 gene 5′-region of Ex2. Numbers indicate the position relative to the transcription start site. The position of the 6 CpG sites on the 5′-flanking promoter and 5 CpG sites on Ex2 detected by bisulfite sequencing is indicated by a short vertical bar. B: percentage of methylation ± SE of 6 CpGs on the 5′-flanking SP1Pr region and 5 CpGs on Ex2 of the DUSP5 gene in rat brain from male at DOL0 (B, C), female at DOL0 (D, E), male at DOL21 (F, G) and female DOL21 (H, I). IUGR values are presented as black diamonds, and Con values are presented as squares. To the right of each graph is the methylation pattern of 14 representative control and IUGR on 5′-flanking SP1Pr region and Ex2 clones. Methylated CpGs are filled and nonmethylated CpGs are open. **P < 0.01, *P < 0,05.
In DOL21 male rat hippocampus, CpG methylation was not affected by IUGR within either the SP1Pr or the Ex2 targets (Fig. 5, F and G). In stark contrast to the male, IUGR significantly increased SP1Pr CpG methylation at site “2,” “5,” and “6,” as well as total CpG methylation across the SP1Pr-targeted region, in DOL21 female hippocampus. Furthermore, IUGR also increased Ex2 CpG methylation at site “2,” as well as total CpG methylation across the Ex2-targeted region, in the DOL21 female hippocampus (Fig. 5, H and I).
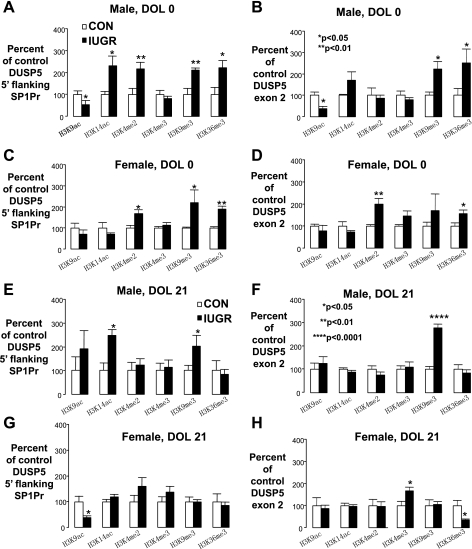
Hippocampal DUSP5 histone code.
In DOL0 male hippocampus, IUGR significantly increased SP1Pr H3K14ac, H3K4 me2, H3K9 me3, and H3K36 me3 occupancy. IUGR also significantly decreased SP1Pr H3K9ac (Fig. 6A). For Ex2, IUGR in the DOL0 male hippocampus significantly increased H3K9me3 and H3K36me3 occupancy, while decreasing H3K9ac occupancy (Fig. 6B). These results are interestingly similar to SP1Pr region. In DOL0 female hippocampus, IUGR significantly increased SP1Pr H3K4me2, H3K9me3, and H3K36me3 occupancy (Fig. 6C). IUGR also increased Ex2 H3K4me2 and H3K36me3 occupancy in the DOL0 female hippocampus (Fig. 6D).
Fig. 6.
Histone code on DUSP5 5′-flanking SP1Pr region and Ex2. Bar graphs representing histone markers expressed as % of control ± SE for DOL0 (male A, B; female C, D) and DOL21 (male E, F; female G, H) on 5′-flanking SP1Pr region (A, C, E, G) and Ex2 (B, D, F, H), IUGR values are presented as black bars. *P < 0.05, **P < 0.01, ****P < 0.0001
In DOL21 male hippocampus, IUGR significantly increased SP1Pr H3K14ac and trimethylation of H3K9me3 occupancy (Fig. 6E). In contrast to the SP1Pr region, IUGR significantly increased only Ex2 H3K9me3 occupancy, without affecting other marks (Fig. 6F). In DOL21 female hippocampus and in contrast to the male findings, IUGR significantly decreased SP1Pr H3K9ac occupancy only (Fig. 6G). For Ex2, IUGR increased significantly increased Ex2 H3K4me3 occupancy (Fig. 6H).
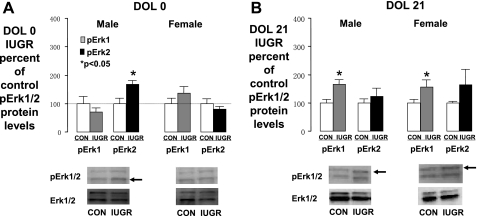
Hippocampal pErk 1 and pErk 2 protein levels.
Because DUSP5 specifically dephosphorylates Erk1/2, we determined whether changes in DUSP5 expression were associated with changes in hippocampal erk1/2 phosphorylation.
In DOL0 males, IUGR significantly increased hippocampal pErk2 levels, while the levels of pErk1 were unchanged (Fig. 7A). pErk 1 and pErk 2 were not altered in DOL0 IUGR female rat pups, consistent with the lack of impact of IUGR upon DOL0 female DUSP5 expression (Fig. 7A).
Fig. 7.
Phosphorylated extracellular signal-related kinases 1 and 2 (pErk1/Erk2) protein levels. Graphs representing pErk1/Erk2 levels in Con and IUGR hippocampus of male and female at DOL0 (A), male and female at DOL21 (B). Bottom: representative Western blots for pErk1/Erk2 and total Erk1/Erk2 (internal control). Protein from a Con hippocampus is in the left lanes, and protein from an IUGR hippocampus is in the right. pErk1 values are presented as gray bars, while pErk2 values are presented as black bars. *P < 0.05.
In DOL21 males and females, IUGR significantly increased hippocampal pErk1 protein levels, without affecting pErk2 levels (Fig. 7B). These findings are consistent with the observed decrease in DUSP5 protein in both sexes at DOL21.
DISCUSSION
The important findings of this study can be grouped into two categories. First, we demonstrate that IUGR significantly affects DUSP5 gene expression in the male hippocampus at both DOL0 and DOL21. The importance of these findings is that DUSP5 plays a role in regulating Erk signaling, which regulates hippocampal apoptosis and memory formation (6, 46). Second, we demonstrate that the impact of IUGR on DUSP5 DNA CpG methylation depends upon the tissue. Specifically, the impact of IUGR upon DUSP5 methylation patterns found in the hippocampus from this study differ from those previously found in the liver (12).
The present study focusing on hippocampal DUSP5 demonstrates that to presume that IUGR-induced epigenetic changes in one tissue type generalize to another is not reasonable. We initially focused upon the hepatic DUSP5 expression and epigenetic characteristics in the previous study because of the known impact of IUGR upon postnatal hepatic metabolism. As our field moves forward in translating epigenetic changes from animal studies using tissues such as liver and hippocampus to more accessible human samples such as cord and peripheral blood mononuclear cells, we need to determine the relevance of findings on a tissue-by-tissue basis. Furthermore, even within the category of blood mononuclear cells, significant epigenetic differences exist between lymphocytes and monocytes (36). Consequently, future studies will need to focus upon the impact of IUGR upon DUSP5 epigenetic characteristics in specific blood mononuclear cell type and the associated relevance to outcomes. Without this level of specificity, we run the risk of our results and conclusions being diluted or confounded by the multiple epigenomes found in the multiple mononuclear cell types that exist in blood. The contrast in our two studies between liver and hippocampus emphasizes this need.
In the liver, IUGR decreases DNA CpG methylation of DUSP5 Ex2 in both male and female rats though DOL21, and through DOL120 in male rats only (12). The decrease in hepatic Ex2 DNA CpG methylation occurred in conjunction with decreased hepatic mRNA levels of DUSP5 (12). Though dogma states that DNA CpG hypomethylation “causes” increased expression, DNA CpG hypomethylation often associates with decreased expression, particularly when the hypomethylation does not occur in a promoter region. This association between decreased expression and hypomethylation is elegantly introduced in “The DNA CpG methylation paradox” by P. A. Jones (18).
In the hippocampus, IUGR increases hippocampal DNA CpG methylation in the SP1Pr region at DOL0 in both sexes, as well as in the SP1Pr and Ex2 regions of the female hippocampus at DOL21. Though IUGR decreased both DUSP5 mRNA and protein levels in the DOL0 male hippocampus, a direct and tight relationship between DNA CpG methylation and DUSP5 expression does not appear to exist. Though it may provide some insight into mechanism, we speculate that the DNA CpG hypermethylation in this context functions only as a biomarker of the environment stress of IUGR.
However, a relationship between DUSP5 expression and increased trimethylation of lysine 9 (H3K9me3) in both the SP1Pr and Ex2 regions exists for the male hippocampal DUSP5 gene. When IUGR increased H3K9me3 in both the SP1Pr and Ex2 regions, mRNA and subsequent protein levels of hippocampal DUSP5 decreased. The significance of this finding is threefold. First, methylation of H3K9 is one of the most abundant and stable histone modifications (35). Methylation of H3K9 usually associates with gene silencing (27).
Second, similar to what was observed in this study, others find that methylation of H3K9 occurs not only in the promoters of suppressed genes, but in downstream regions (15, 47, 48). This phenomenon may be a consequence of a drop in RNA Pol II recruitment and/or a Krüppel-associated box domain zinc finger protein (KRAB-ZFP) and heterochromatin protein-1β repression (HP1β) (15). We speculate that the decreased expression of male hippocampal DUSP5 observed in the IUGR animals is most likely due to the decreased RNA Pol II recruitment as opposed KRAB-ZPF and HP1β repression (15). This speculation is based upon the fact that IUGR decreases DUSP5 expression but does not abolish it. The latter phenomenon is more likely with KRAB-ZFP/HP1β repression-driven formation of heterochromatin (15).
The third reason is that our finding of increased H3K9me3 occupancy at both SP1Pr and Ex2 regions potentially drives future studies. H3K9 methylation occurs through a series of methyltransferases. These methyltransferases include G9a/KMT1C, GLP/KMT1D, SETDB1/KMT1E, and Suv39h1/KMT1A, and these enzymes coexist in the same mega complex (11, 27). Future studies beyond the scope of this one will ask the question of what are characteristics of this megacomplex that provide fidelity to the hippocampal DUSP5 response to IUGR.
Understanding the factors that moderate DUSP5 expression in response to IUGR is important because of DUSP5's role in hippocampal cellular homeostasis. The rat DUSP5 cDNA was identified via a screen of candidate plasticity-related genes from hippocampus neurons stimulated with the glutamate analog kainate (16). Furthermore, DUSP5 is inducible by both heat shock proteins and growth factors (34). DUSP5 belongs to dual specificity MAPK phosphatases subfamily and preferentially dephosphorylates Erk1/Erk2. Erk1 and 2 are components of the MAPK pathway (5, 17, 25, 28, 34, 42). The reason we are intrigued by hippocampal DUSP5 is that Erk signaling is a common pathway for both apoptosis and memory establishment (Erk1/Erk2). In this study, IUGR increased phosphorylated Erk levels in males at DOL0 and 21, and females at DOL21.
Erk1/Erk2 perform important roles in regulating cell homeostasis, including the processes of cell survival and cell death (2). Sun et al. (45) showed that neuroprotection of brain-derived neurotrophic factor against hypoxic injury in vitro requires activation of Erk. However, a recent study by Subramaniam et al. (44) suggests that Erk1/Erk2 also has a key proapoptotic role in neuronal apoptosis induced by potassium (K+) withdrawal. The regulation of Erk phosphorylation and subsequent signaling are complex. However, it is evident that Erk signaling integrates central nervous system (CNS) functions requiring neuronal plasticity, such as long-term potentiation and depression, and memory (7). Therefore, we speculate that in the male IUGR hippocampus, altered Erk signaling homeostasis through decreased DUSP5 expression may contribute to an IUGR impact on hippocampal apoptosis and function.
Indeed, IUGR does increase hippocampal apoptosis in the IUGR rat. Furthermore human studies suggest that the hippocampus is among the regions of the brain vulnerable to uteroplacental insufficiency-driven IUGR. In the human, IUGR decreases hippocampal gray matter and decreases term infant performance on all six domains of the Assessment of Preterm Infants Behavior (32).
Caution is of course necessary when attempting to apply data from animal models to human pathophysiology. The brain of the newborn rat is immature relative to the term human, and the insult imposed on the fetal rat in this model of uteroplacental insufficiency is specific. In contrast, the timing and impact of uteroplacental insufficiency experienced by humans span a continuum and are impacted by environmental and genetic variables.
One arena that is similar between humans and rats is that of sex differences. In humans, sex influences moderate the IUGR impact upon neurodevelopment (38). In this rat model of uteroplacental insufficiency, sex moderates the IUGR impact upon hippocampal gene expression and chromatin structure (22, 24, 41). Unfortunately, little is definitively known about potential sex-dependent mechanisms that are active in the IUGR process. However, recent studies may to provide some early insight.
O'Grady et al. (37) found that IUGR decreases expression of Cyp19a variant 1.f in DOL0 rat male hippocampus. The Cyp19a gene encodes the protein aromatase, which catalyzes the biosynthesis of estrogens from androgens in brain. In non-CNS tissues, estrogen administration increases gene-specific H3K9me3 occupancy (43). We speculate that decreased Cyp19a variant 1.f expression may contribute to the increased H3K9me3 occupancy observed in the DOL IUGR male hippocampus.
IUGR also changed several other components of the hippocampal DUSP5 histone code in a sex-specific manner. Unfortunately, a direct relationship between either the sex or IUGR impact upon these DUSP5 histone codes (e.g., H3K9ac, K14ac, K4me3, K36me3) relative to DUSP5 hippocampal mRNA levels is unclear. It is likely these changes in histone code play a subtle or indirect role in the regulation of DUSP5 expression; however, considering the complexity of epigenetic regulation, we are reluctant to speculate without an obvious correlative observation. This reluctance is further supported by the present state of the field, which does not allow us to test the impact of each histone code modification for a specific nucleosome(s) and a specific gene.
In summary, IUGR affects hippocampal DUSP5 expression and epigenetic characteristics in a sex-specific manner. Furthermore, when IUGR decreased DUSP5 protein levels, Erk phosphorylation increased. Our finding that increased H3K9me3 occupancy consistently associated with decreased DUSP5 mRNA levels is particularly stimulating, in that it will drive future studies that focus upon “why” the epigenetic changes occurs versus “that” they occur.
GRANTS
This work was supported by National Institute of Diabetes and Digestive Kidney Diseases Grant R01DK-080558-04 (R. H. Lane).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Guiding principles for research involving animals, and human beings. Am J Physiol Regul Integr Comp Physiol 283: R281–R283, 2002 [DOI] [PubMed] [Google Scholar]
- 2. Agell N, Bachs O, Rocamora N, Villalonga P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca2+, and calmodulin. Cell Signal 14: 649–654, 2002 [DOI] [PubMed] [Google Scholar]
- 3. Baserga M, Kaur R, Hale MA, Bares A, Yu X, Callaway CW, McKnight RA, Lane RH. Fetal growth restriction alters transcription factor binding and epigenetic mechanisms of renal 11beta-hydroxysteroid dehydrogenase type 2 in a sex-specific manner. Am J Physiol Regul Integr Comp Physiol 299: R334–R342, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509, 2004 [DOI] [PubMed] [Google Scholar]
- 5. Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature 410: 37–40, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Cheung EC, Slack RS. Emerging role for ERK as a key regulator of neuronal apoptosis. Sci STKE 2004: PE45, 2004 [DOI] [PubMed] [Google Scholar]
- 7. Colucci-D'Amato L, Perrone-Capano C, di Porzio U. Chronic activation of ERK and neurodegenerative diseases. Bioessays 25: 1085–1095, 2003 [DOI] [PubMed] [Google Scholar]
- 8. Economides DL, Nicolaides KH. Blood glucose and oxygen tension levels in small-for-gestational-age fetuses. Am J Obstet Gynecol 160: 385–389, 1989 [DOI] [PubMed] [Google Scholar]
- 9. Economides DL, Nicolaides KH, Campbell S. Metabolic and endocrine findings in appropriate and small for gestational age fetuses. J Perinat Med 19: 97–105, 1991 [DOI] [PubMed] [Google Scholar]
- 10. Economides DL, Nicolaides KH, Gahl WA, Bernardini I, Bottoms S, Evans M. Cordocentesis in the diagnosis of intrauterine starvation. Am J Obstet Gynecol 161: 1004–1008, 1989 [DOI] [PubMed] [Google Scholar]
- 11. Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37: 46–56, 2010 [DOI] [PubMed] [Google Scholar]
- 12. Fu Q, McKnight RA, Yu X, Callaway CW, Lane RH. Growth retardation alters the epigenetic characteristics of hepatic dual specificity phosphatase 5. FASEB J 20: 2127–2129, 2006 [DOI] [PubMed] [Google Scholar]
- 13. Fu Q, Yu X, Callaway CW, Lane RH, McKnight RA. Epigenetics: intrauterine growth retardation (IUGR) modifies the histone code along the rat hepatic IGF-1 gene. FASEB J 23: 2438–2449, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geva R, Eshel R, Leitner Y, Fattal-Valevski A, Harel S. Memory functions of children born with asymmetric intrauterine growth restriction. Brain Res 1117: 186–194, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet 6: e1000869, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hevroni D, Rattner A, Bundman M, Lederfein D, Gabarah A, Mangelus M, Silverman MA, Kedar H, Naor C, Kornuc M, Hanoch T, Seger R, Theill LE, Nedivi E, Richter-Levin G, Citri Y. Hippocampal plasticity involves extensive gene induction and multiple cellular mechanisms. J Mol Neurosci 10: 75–98, 1998 [DOI] [PubMed] [Google Scholar]
- 17. Ishibashi T, Bottaro DP, Michieli P, Kelley CA, Aaronson SA. A novel dual specificity phosphatase induced by serum stimulation and heat shock. J Biol Chem 269: 29897–29902, 1994 [PubMed] [Google Scholar]
- 18. Jones PA. The DNA methylation paradox. Trends Genet 15: 34–37, 1999 [DOI] [PubMed] [Google Scholar]
- 19. Joss-Moore LA, Wang Y, Baack ML, Yao J, Norris AW, Yu X, Callaway CW, McKnight RA, Albertine KH, Lane RH. IUGR decreases PPARgamma and SETD8 Expression in neonatal rat lung and these effects are ameliorated by maternal DHA supplementation. Early Hum Dev 86: 785–791, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joss-Moore LA, Wang Y, Yu X, Campbell MS, Callaway CW, McKnight RA, Wint A, Dahl MJ, Dull RO, Albertine KH, Lane RH. IUGR decreases elastin mRNA expression in the developing rat lung and alters elastin content and lung compliance in the mature rat lung. Physiol Genomics 43: 499–505, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kan E, Roberts G, Anderson PJ, Doyle LW. The association of growth impairment with neurodevelopmental outcome at eight years of age in very preterm children. Early Hum Dev 84: 409–416, 2008 [DOI] [PubMed] [Google Scholar]
- 22. Ke X, Lei Q, James SJ, Kelleher SL, Melnyk S, Jernigan S, Yu X, Wang L, Callaway CW, Gill G, Chan GM, Albertine KH, McKnight RA, Lane RH. Uteroplacental insufficiency affects epigenetic determinants of chromatin structure in brains of neonatal and juvenile IUGR rats. Physiol Genomics 25: 16–28, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Ke X, McKnight RA, Wang ZM, Yu X, Wang L, Callaway CW, Albertine KH, Lane RH. Nonresponsiveness of cerebral p53-MDM2 functional circuit in newborn rat pups rendered IUGR via uteroplacental insufficiency. Am J Physiol Regul Integr Comp Physiol 288: R1038–R1045, 2005 [DOI] [PubMed] [Google Scholar]
- 24. Ke X, Schober ME, McKnight RA, O'Grady S, Caprau D, Yu X, Callaway CW, Lane RH. Intrauterine growth retardation affects expression and epigenetic characteristics of the rat hippocampal glucocorticoid receptor gene. Physiol Genomics 42: 177–189, 2010 [DOI] [PubMed] [Google Scholar]
- 25. Kelly A, Laroche S, Davis S. Activation of mitogen-activated protein kinase/extracellular signal-regulated kinase in hippocampal circuitry is required for consolidation and reconsolidation of recognition memory. J Neurosci 23: 5354–5360, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kitazawa SKR, Maeda S. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J Biol Chem 274: 28787–28793, 1999 [DOI] [PubMed] [Google Scholar]
- 27. Kouzarides T. Chromatin modifications and their function. Cell 128: 693–705, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Kwak SP, Dixon JE. Multiple dual specificity protein tyrosine phosphatases are expressed and regulated differentially in liver cell lines. J Biol Chem 270: 1156–1160, 1995 [DOI] [PubMed] [Google Scholar]
- 29. Lane RH, Chandorkar AK, Flozak AS, Simmons RA. Intrauterine growth retardation alters mitochondrial gene expression and function in fetal and juvenile rat skeletal muscle. Pediatr Res 43: 563–570, 1998 [DOI] [PubMed] [Google Scholar]
- 30. Leitner Y, Fattal-Valevski A, Geva R, Bassan H, Posner E, Kutai M, Many A, Jaffa AJ, Harel S. Six-year follow-up of children with intrauterine growth retardation: long-term, prospective study. J Child Neurol 15: 781–786, 2000 [DOI] [PubMed] [Google Scholar]
- 31. Leitner Y, Fattal-Valevski A, Geva R, Eshel R, Toledano-Alhadef H, Rotstein M, Bassan H, Radianu B, Bitchonsky O, Jaffa AJ, Harel S. Neurodevelopmental outcome of children with intrauterine growth retardation: a longitudinal, 10-year prospective study. J Child Neurol 22: 580–587, 2007 [DOI] [PubMed] [Google Scholar]
- 32. Lodygensky GA, Seghier ML, Warfield SK, Tolsa CB, Sizonenko S, Lazeyras F, Huppi PS. Intrauterine growth restriction affects the preterm infant's hippocampus. Pediatr Res 63: 438–443, 2008 [DOI] [PubMed] [Google Scholar]
- 33. MacLennan NK, James SJ, Melnyk S, Piroozi A, Jernigan S, Hsu JL, Janke SM, Pham TD, Lane RH. Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics 18: 43–50, 2004 [DOI] [PubMed] [Google Scholar]
- 34. Mandl M, Slack DN, Keyse SM. Specific inactivation and nuclear anchoring of extracellular signal-regulated kinase 2 by the inducible dual-specificity protein phosphatase DUSP5. Mol Cell Biol 25: 1830–1845, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev 6: 838–849, 2005 [DOI] [PubMed] [Google Scholar]
- 36. Miao F, Wu X, Zhang L, Riggs AD, Natarajan R. Histone methylation patterns are cell-type specific in human monocytes and lymphocytes and well maintained at core genes. J Immunol 180: 2264–2269, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Grady SP, Caprau D, Ke XR, Contreras Y, Haley S, Ermini F, Penn A, Moyer-Mileur L, McKnight R, Lane R. Intrauterine growth restriction alters hippocampal expression and chromatin structure of Cyp19a1 variants. Syst Biol Reprod Med 56: 292–302, 2010 [DOI] [PubMed] [Google Scholar]
- 38. Paz I, Gale R, Laor A, Danon YL, Stevenson DK, Seidman DS. The cognitive outcome of full-term small for gestational age infants at late adolescence. Obstet Gynecol 85: 452–456, 1995 [DOI] [PubMed] [Google Scholar]
- 39. Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucl Acids Res 27: 2991–3000, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaap AH, Wolf H, Bruinse HW, Smolders-de Haas H, van Ertbruggen I, Treffers PE. School performance and behaviour in extremely preterm growth-retarded infants. Eur J Obstet Gynecol Reprod Biol 86: 43–49, 1999 [DOI] [PubMed] [Google Scholar]
- 41. Schober ME, McKnight RA, Yu X, Callaway CW, Ke X, Lane RH. Intrauterine growth restriction due to uteroplacental insufficiency decreased white matter and altered NMDAR subunit composition in juvenile rat hippocampi. Am J Physiol Regul Integr Comp Physiol 296: R681–R692, 2009 [DOI] [PubMed] [Google Scholar]
- 42. Senn R, Keren O, Hefetz A, Sarne Y. Long-term cognitive deficits induced by a single, extremely low dose of tetrahydrocannabinol (THC): behavioral, pharmacological and biochemical studies in mice. Pharmacol Biochem Behav 88: 230–237, 2008 [DOI] [PubMed] [Google Scholar]
- 43. Starlard-Davenport A, Tryndyak VP, James SR, Karpf AR, Latendresse JR, Beland FA, Pogribny IP. Mechanisms of epigenetic silencing of the Rassf1a gene during estrogen-induced breast carcinogenesis in ACI rats. Carcinogenesis 31: 376–381, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Subramaniam S, Zirrgiebel U, von Bohlen Und Halbach O, Strelau J, Laliberté C, Kaplan DR, Unsicker K. ERK activation promotes neuronal degeneration predominantly through plasma membrane damage and independently of caspase-3. J Cell Biol 165: 357–369, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun X, Zhou H, Luo X, Li S, Yu D, Hua J, Mu D, Mao M. Neuroprotection of brain-derived neurotrophic factor against hypoxic injury in vitro requires activation of extracellular signal-regulated kinase and phosphatidylinositol 3-kinase. Int J Dev Neurosci 26: 363–370, 2008 [DOI] [PubMed] [Google Scholar]
- 46. Sweatt JD. Mitogen-activated protein kinases in synaptic plasticity and memory. Curr Opi Neurobiol 14: 311–317, 2004 [DOI] [PubMed] [Google Scholar]
- 47. Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 19: 381–391, 2005 [DOI] [PubMed] [Google Scholar]
- 48. Wiencke JK, Zheng S, Morrison Z, Yeh RF. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene 27: 2412–2421, 2008 [DOI] [PubMed] [Google Scholar]
- 49. Yan XB, Hou HL, Wu LM, Liu J, Zhou JN. Lithium regulates hippocampal neurogenesis by ERK pathway and facilitates recovery of spatial learning and memory in rats after transient global cerebral ischemia. Neuropharmacology 53: 487–495, 2007 [DOI] [PubMed] [Google Scholar]