Abstract
A new line of neuroscience research suggests that epigenetics may be the site of nature and nurture integration by providing the environment with a mechanism to directly influence the read-out of our genome. Epigenetic mechanisms in the brain are a series of post-translational chromatin and DNA modifications driven by external input. Given the critical hub that epigenetics appears to be, neuroscientists have come to suspect its fundamental influence on how our minds change in response to our unique environment and, in turn, how these changes can then impact our future interactions with the environment. The field of learning and memory is becoming particularly interested in understanding the cognitive influence of epigenetics. With the majority of us working with an eye toward therapeutics, the question naturally arises: “Has neuroepigenetics gotten us closer to treating memory disorders and if so, where do we go from here?” This review will begin with a brief exploration of recent advances in our understanding of how epigenetic mechanisms contribute to learning and memory processes that are susceptible to failure. Next the implications for disorders of cognition, such as Alzheimer’s Disease, will be discussed. Finally, we will use parallels from the field of cancer to speculate on where we should consider heading from here in the pursuit of therapeutics.
Mechanistically, epigenetics regulate transcription through post-translational modification of the N-terminus of core histone proteins and cytosine residues of DNA. These modifications influence transcription factor accessibility to gene promoters by controlling the organization of chromatin’s structure. Core histones are highly basic alkaline proteins that align and order DNA (~147 bp) into structural units termed nucleosomes. The nucleosome is comprised of protein octamers containing a pair of each core histone (H2A, H2B, H3, H4). These histones contain N-terminal tails that provide sites for a range of covalent chemical modifications that regulate the overall chromatin structure. These histone modifications include acetylation, phosphorylation, methylation, ubiquitination and sumoylation, though most research to date has focused on the first three. The addition and removal of these functional groups results in changes to chromatin structure that can either facilitate or repress gene expression. Histone acetylation, a transcriptional activator, is catalyzed by histone acetyltransferases (HATs), whereas histone deacetylases (HDACs) are responsible for removal of the acetyl group. Similarly, histone phosphorylation of serine, threonine and sometimes arginine residues, most often leads to transcriptional activation. As with histone acetylation, methylation can also occur on lysine residues. The transcriptional effects of histone methylation depend on which histone and lysine residues are involved, as well as the degree of methylation (i.e. mono-, di- or trimethylation).
The methylation of genomic DNA at CpG dinucleotides is predominantly associated with transcriptional silencing. DNA methylation is catalyzed by DNA methyltransferases (DNMTs) and usually involves the recruitment methyl-DNA binding proteins, such as MeCP2. Although DNA methylation has long been perceived to be a static modification, with changes only occurring in disease states (e.g. cancer), recent evidence suggests the process can be highly dynamic (Bhattacharya et al., 1999; Miller & Sweatt, 2007; Kangaspeska et al., 2008; Metivier et al., 2008; Kim et al., 2009; Ma et al., 2009).
The role of epigenetic mechanisms in memory
Most work in the field of epigenetics has focused on its involvement in the development and differentiation of both normal and cancerous cells. However, in recent years, an increasing number of neuroscientists have begun to acknowledge the important role of epigenetic mechanisms in neuronal function, cognition and behavior; as these molecular regulators have found their place at the nexus of environment-genome interplay. The young field of neuroepigenetics is discovering unanticipated, but critical, roles in post-mitotic neurons.
Accordingly, there has been an explosion in the number of studies investigating the involvement of epigenetic mechanisms in learning and memory. The findings indicate that various stages of memory, from acquisition to extinction, undergo epigenetic regulation and that epigenetic dysregulation may contribute to and modulate memory disorders.
Memory formation and consolidation require a long-lasting increase in synaptic strength that is supported by transcription. In the last few decades, considerable effort has gone into identifying the numerous transcription factors and genes that participate in this neuronal plasticity. Only recently has attention shifted towards the chromatin modifications that control access of these transcription factors to the appropriate gene promoters.
Histone modifications and memory
Some of the earliest evidence to suggest a relationship between chromatin modifications and memory formation came from a 2001 study in which novel taste learning in rats resulted in heightened acetylation of histone H2A and H4 in the insular cortex in an ERK/MAPK-dependent manner (Swank & Sweatt). This general notion of epigenetic involvement in memory was further extended to show that hippocampus-dependent fear memory is associated with histone acetylation (specifically H3) in an ERK/MAPK-dependent manner (Levenson et al., 2004). A series of studies at this same time utilized transgenic and knockout mouse-models targeting well-known transcription coactivator proteins with HAT activity, CREB binding protein (CBP) and P300, to demonstrate their necessity for hippocampal synaptic plasticity and long-term memory for novel objects and contextual fear (Alarcón et al., 2004; Korzus et al., 2004; Woods et al., 2005 and 2006; Oliveira et al., 2007;). The importance of histone acetylation to memory was further highlighted by studies in which HDAC inhibitors (HDACi’s) elevated histone acetylation and ameliorated impairments of neuronal plasticity and memory (Levenson et al., 2004; Alarcón et al., 2004; Korzus et al., 2004; Vecsey et al., 2007; Stefanko et al., 2009). HDACi’s have even been shown to support the formation of long-term memories following a training protocol that is too weak to support memory in the absence of HDACi treatment (Stefanko et al., 2009). In addition, CREB-CBP interaction has been indicated as a likely prerequisite for this HDACi-driven enhancement of synaptic plasticity and memory (Vecsey et al., 2007). Consistent with these findings, selective overexpression of HDAC2 decreases dendritic spine density, synapse number, synapse plasticity and contextual fear memory in mice (Guan et al., 2009). Conversely, HDAC2-deficient mice display elevated synapse number and fear memory. While the exact mechanism that HDACi’s use to improve synaptic plasticity and memory is still unclear, recent data indicate that they might act to induce formation of new synapses and dendritic sprouting (Fischer et al., 2007).
Other histone modifications implicated in memory include histone phosphorylation and methylation. A significant up-regulation of histone H3 phosphorylation occurs at the Ser10 residue in hippocampal Area CA1 during the formation of contextual fear memory (Chwang et al., 2006). This phosphorylation increase was found to be ERK/MAPK-dependent, as the effect was suppressed by administration of either NMDA receptor antagonists or the ERK/MAPK inhibitor SL327. In a similar vein, phosphorylation at the CREB gene promoter was elevated during novel object recognition, while it was reduced at the NF-κB promoter (Koshibu et al., 2009). Furthermore, the formation of contextual fear memory is also associated with increased H3K4 trimethylation (a transcriptionally active marker) at the zif268 and bdnf promoters, while the memory of mice lacking the H3K4-specific histone methyltransferase Mll was impaired (Gupta et al., 2010).
DNA methylation and memory
Another major mode of epigenetic regulation is the methylation of genomic DNA. While DNA methylation was initially thought to be a relatively static epigenetic marker in post-mitotic cells, this has become increasingly challenged by findings of recent studies that suggest DNA methylation can be a dynamic and reversible post-translational modification (Miller & Sweatt, 2007; Kangaspeska et al., 2008; Metivier et al., 2008; Kim et al., 2009; Ma et al., 2009).
The first indication of a role for DNA methylation in memory came from the work of Levenson et al. (2006), which demonstrated that DNMTs are critical for synaptic plasticity. Further, chemical activation of hippocampal slices results in altered methylation of the bdnf and reelin genes. This notion quickly gained in vivo support, demonstrating several points (Miller and Sweatt, 2007; Miller et al., 2008). First, hippocampal DNMT expression is up-regulated in rats during consolidation of contextual fear memory. Second, intra-hippocampal administration of DNMT inhibitors (DNMTi’s) blocks this memory consolidation. However, the DNMTi memory deficits can be overcome by HDACi pre-treatment (Miller and Sweatt, 2007; Miller et al., 2008). And third, rapid changes in DNA methylation at the time of learning provides bi-directional transcriptional regulation of memory promoting (reelin) and suppressing (PP1) genes. Importantly, the methylation changes associated with learning were prevented with DNMTi (Miller and Sweatt, 2007). Similar changes in DNA methylation have been noted for bdnf during contextual fear learning (Lubin et al., 2008). Additionally, conditional knockout mice lacking both DNMT1 and DNMT3a forebrain expression display deficits in long-term plasticity in the hippocampus, as well as hippocampal memory impairments (Feng et al., 2010). Interestingly, the hippocampal changes observed after learning in the Miller and Sweatt study (2007) were transient, lasting less than a day after training. This led to the examination of DNA methylation changes in the prefrontal cortex as the initially hippocampus-dependent fear memory underwent cortical integration during system consolidation. Hippocampal learning triggered gene-specific hypermethylation in the cortex that persisted for weeks. In addition, inhibiting this persistent DNA methylation in the anterior cingulate cortex thirty days after learning disrupted the memory (Miller et al., 2010). Taken together, these data indicate that DNA methylation can be both dynamic (to support synaptic consolidation) and stable (to support system consolidation).
Epigenetic mechanisms and memory disorders
Based on the accumulating evidence implicating epigenetic modifications in normal learning and memory processes, it stands to reason that some memory disorders may have epigenetic origins. Here we will focus on one neurologic disorder marked by memory failure, Alzheimer’s disease (AD).
AD is a common form of dementia, marked by a rapid decline in cognitive function and memory failure. It is characterized by accumulation of β-amyloid plaques and tau protein-related neurofibrillary tangles in the cortex and some subcortical regions (Wenk, 2003). The β-amyloid plaques are formed by deposition of neurotoxic β-amyloid peptides, which themselves are produced from the endoproteolysis of the amyloid precursor protein (APP) by β- and γ-secretases. Interestingly, this catalytic reaction also leads to the generation of an APP intracellular domain (AICD), which interacts with the nuclear adaptor protein Fe65 and the HAT TIP60. Together they work as a transcriptional regulator (Cao & Südhof, 2001). These results suggest that dysregulation of histone acetylation might be involved in some pathological features of AD. In further suppport of this, mutations of the gene responsible for coding of the catalytic subunit of the γ-secretase, presenilin 1 (PS1), maintains CBP activity in vitro; thus indicating potential hyperacetylation in cases of AD (Marambaud et al., 2003). Conversely, an increasing number of studies are highlighting hypoacetylation as a potential risk factor for AD.
For example, conditional knockout mice of PS1 and 2 show impairments in hippocampus-dependent synaptic plasticity and learning, as well as reduced expression of CBP and CREB-CBP contingent target genes (e.g. c-fos and bdnf) (Saura et al., 2004). In light of these findings from cell culture and animal models, histone acetylation likely plays a modulatory role in the development AD. However, the differences across studies suggest that the relationship between histone acetylation and AD may vary across brain regions, cell types and gene targets. Human evidence for the role of histone modifications in AD is sparse. However, preclinical animal work has gained some support from postmortem studies. Elevated levels of histone phophorylation at H2A serine 139 have been observed post-mortem in the hippocampus and cortex of patients diagnosed with AD (Myung et al., 2008). Moreover, there is evidence that changes in histone-DNA interplay during lipid perioxidation may contribute to the DNA damage induced by oxidative stress that is frequently noted in AD (Drake et al., 2004).
A link between aberrant DNA methylation and AD etiology has been observed in a wide range of studies; usually in terms hypomethylation. One of the earliest human studies to report epigenetic dysregulation in AD revealed hypomethylation of APP’s promoter in the parietal cortex of AD patients (West et al., 1995). Intriguingly, parietal cortex hypomethylation of this same promoter has been reported in individuals over the age of 70 (Tohgi, et al., 1999). More recently, a significant reduction in global DNA methylation was reported in layer II neurons of the entorhinal cortex with AD (Mastroeni et al., 2010). Finally, pharmacologically induced hypermethylation of the PS1 promoter region in vitro reduced PS1 expression and β-amyloid production (Scarpa et al., 2003). This highlights the use of a methyl-donor rich diet (such as folic acid and vitamins B6 and 12) as a promising therapeutic avenue.
Several studies have examined the therpaeutic effects of HDACi’s in animal models of aging and neurodegeneration. Age-dependent dysregulation of hippocampal H4K12 acetylation in mice is reported to contribute to memory decline by suppressing key learning and memory genes. Importantly, the authors demonstrate that administration of the HDACi SAHA (suberoylanilide hydroxamic acid) normalized H4K12 acetylation, reinstated gene expression and improved memory function in aged mice (Peleg et al., 2010). In a related study, both environmental enrichment and HDACi elevated histone acetylation and restored synaptic plasticity and learning in a neurodegenerative mouse model (Fischer et al., 2007). HDACi has also been demonstrated to improve memory performance in different mouse models of AD (Ricobaraza et al., 2009; Kilgore et al., 2010). The HDACi sodium 4-phenylbutyrate reduced tau phosphorylation and ameliorated spatial learning and memory deficits in the Tg2576 mouse model of AD (Ricobaraza et al., 2009). Similarly, the administration of three distinct HDACi’s rescued and maintained memory in the APPswe/PSI model of AD. This study further demonstrated unexpected class I HDAC selectivity of the inhibitors used in the study (Kilgore et al., 2010). This latter finding is particularly important. As we will discuss in more detail below, isoform-selectivity is crucial for the development of epigenetic therapeutics in order to reduce unwanted “off-target” effects. However, it is important to bear in mind that some HDACs can exert additional effects on neuronal function through interactions with non-histone proteins. For instance, while the HAT activity of P300 has been demonstrated to be crucial for normal memory function (Oliveira et al., 2007), this enzyme has also been associated with heightened acetylation of Tau proteins. This, in turn, prevents the degradation of phosphorylated Tau commonly associated with tauopathy (Min et al., 2010). Additionally, the NAD-dependent HDAC SIRT1 (Sir2, homolog 1) is thought to confer neuronal protection through the synergism of several different non-histone substrates, including deacetylation of Tau (Min et al., 2010), attenuation of β-amyloid production and inhibition of pro-apoptotic protein (e.g. P53 and FOXO proteins) functions (for more in depth review of this topic, see Wang et al., 2010; Outeiro et al., 2008; and Anekonda, 2006).
Developing epigenetic treatments for memory disorders
It is clear that the rapidly growing field of neuroepigenetics holds tremendous and far-reaching promise, particularly for the identification and treatment of memory-related disorders. In this final section, we will be more speculative as we discuss what the future of targeting epigenetic mechanisms for cognitive therapeutics may look like.
Epigenetic modifications as biomarkers?
The brain is a remarkable structure, complete with multiple “fail-safes” designed to protect tasks that are crucial to survival. This includes our ability to learn and remember. The downside to such excellent engineering is that the early stages of cognitive failure are difficult to detect with the disappointingly rudimentary means currently available to clinicians. These include patient interviews and cognitive assessment tests that require such substantive failures as the inability to complete the numbers on a clock face. Thus, individuals that can still remember the name of our country’s president and a list of three words are regularly sent home with the reassurance that they are fine, despite the sinking suspicion by the patients themselves that something is not right. And, because one of the brain’s best abilities is compensation, patients that do present with cognitive abnormalities are akin to patients with Stage III breast cancer. Treatment at this point is an uphill battle, at best. For this reason, there is a critical need for biomarkers of susceptibility to cognitive failure and early markers of the failure itself. There is, of course, a physical barrier to testing for any type of biomarker of neurologic disorders that must be overcome. Nevertheless, determining a memory disorder’s unique epigenetic signature may identify some of these biomarkers. Such an approach is proving to be useful in the cancer field. For example, acetylation of histone H3 and trimethylation of H3K9 enables discrimination between prostate cancer and non-malignant prostate tissue (Ellinger et al., 2010), while overexpression of the enhancer of zeste homolog-2 (EZH2), a histone demethylating component of the polycomb repressive complex-2 (PRC2), is a prognostic marker of heightened tumor cell proliferation in various types of cancer (Bachmann et al., 2006).
Epigenetic therapeutics as cognitive enhancers?
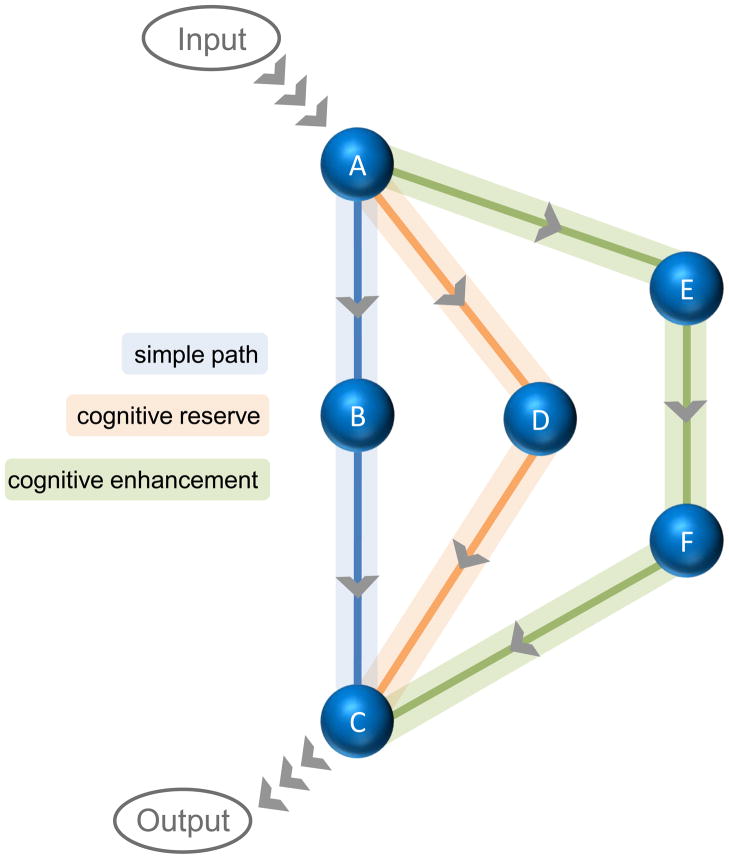
We have just finished stressing the potential importance of identifying epigenetic changes associated with cognitive failure. However, a theme that is integral to our own work is that the solution to memory disorders need not be the cause (Kilgore et al., 2010). For instance, a great deal of effort has been dedicated to understanding the etiology of AD, yet one of the primary complaints associated with AD is memory failure. While plaque and tangle pathology may be key players in producing memory deficits, epigenetic treatments have the potential to circumvent the damage by providing access to alternative pathways for memory traces (Fischer et al., 2007; Kilgore et al., 2010). This taps into the notion of cognitive reserve, which was first considered more than two decades ago, after a post-mortem study of patients diagnosed with AD revealed an unexpected finding. The degree of neuropathology did not always correlate with clinical symptoms of the disease (Katzman et al., 1988). One interpretation provided by the authors is that some patients have a greater ability to access alternate pathways for memory storage as cellular damage and loss occurs (Figure 1). Indeed, a 2008 study found a correlation between level of education (used by the authors as a proxy for cognitive reserve) and degree of dementia symptoms (Roe et al., 2008). And a study published just a few months later confirmed and extended this finding. The authors reported that, while level of education is correlated with dementia risk, it does not slow progression of memory loss once it begins (Wilson et al., 2009).
Figure 1. Multiple paths to memory storage.
The evidence for cognitive reserve supports the idea that the brain should be capable of employing multiple paths for memory storage. In this simple schematic, associative input arrives at location A in the brain. To achieve the correct behavioral output, location C must be reached. While the simplest path to C is via B (blue), this linear approach to storing a memory is tenuous. If B becomes damaged, the memory trace is disrupted. However, a flexible mind can circumvent the blue trace if damage occurs and complete the connection to C by utilizing a cognitive reserve pathway (orange). The idea of cognitive enhancers (green path) draws on this same principle by employing molecular modifications to prepare additional locations (e.g. E and F) for participation in the memory trace.
This is consistent with the notion that the educated mind has reached the limit of its cognitive reserve capabilities once symptoms appear. Therefore, if epigenetic mechanisms can be harnessed to further increase flexibility and the mind’s capacity for cognitive reserve, we may have a novel treatment strategy. Preclinical studies with HDACi’s strongly support this possibility (Fischer et al., 2007; Kilgore et al., 2010). Thus, in the spirit of epigenetics (“above the genome”), epigenetic therapies may be epi-etiology; that is to say – “above the cause.” Recognition of this idea broadens the potential neurologic and psychiatric uses of epigenetic-modifying drugs.
What does the ideal epigenetic therapeutic look like?
As we are in the early stages of our neuroepigenetic explorations, now is the time to stop and consider what characteristics we are looking for in epigenetic-modifying drugs. It seems that, in an ideal world, these drugs would be reversible and specific. Reversibility is particularly important for cases of treating unwanted memories, as with post-traumatic stress disorder (PTSD) and relapse in drug addiction. And specificity is best achieved through isoform and gene-target selectivity, as well as the ability to target individual regions of the brain. The notion of using epigenetic intervention to counter cognitive disorders is particularly appealing; as such compounds have the potential to confer greater specificity than the current neurotransmitter-targeting compounds can alone. While the action of neurotransmitter-based pharmaceuticals is limited to receptor availability, epigenetic therapies have the potential to up-regulate the receptor by influencing its transcription rate. This, in turn, would make the neurotransmitter-based pharmaceutical more efficacious. Epigenetic therapies can offer an additional level of influence, as the efficient treatment of memory disorders will likely benefit from up and down-regulating the transcription of memory-related genes (i.e. up-regulation of memory promoters and down-regulation of memory suppressors). Therefore, unraveling the aberrant and complicated epigenetic landscape underlying memory impairments will potentially enable the design of epigenetic modifying drugs that can target the transcription of specific memory genes. This would be accomplished by exerting synergistic effects on chromatin to collectively promote and suppress appropriate targets. Further, epigenetic modifiers provide the same desirable reversibility of traditional drugs because epigenetic marks occur above the level of the genome. This is something that most gene therapies currently under development lack.
Parallels from cancer research can provide us with valuable insight into obstacles we can anticipate in our own field’s pursuit of epigenetic therapies. For example, in cancer, a set of tumor-suppressor genes are hypermethylated within a landscape of global hypomethylation (Jones & Baylin, 2007). Recall that DNA methylation is associated with transcriptional repression. Therefore, global hypomethylation, in conjunction with suppression of tumor-suppressor genes, would support rampant and unchecked cell division. In parallel, effective epigenetic modifying drugs for memory-related disorders must contend with the opposing effects of memory promoter (e.g. reelin, bdnf) and suppressor (e.g. phosphatases [PP1, calcineurin]) genes (Miller & Sweatt, 2007; Miller et al., 2010). This highlights the need for gene- specificity with epigenetic therapies, as a compound that globally elevates transcription would presumably create its own set of problems by pitting memory-promoters and suppressors against one another in a brain that is already struggling to form and maintain memories.
Furthermore, in recent years HDACi’s have received a great deal of attention in the field of cancer as propitious therapeutic drug targets. However, despite their promising potentials for cancer therapy, HDACi’s exhibit toxicity in the clinic that threatens to limit their potential (see Balasubramanian et al., 2009 for review). These adverse effects are likely to arise from the fact that currently available HDACi’s interact with several HDAC isoforms (Kilgore et al., 2010), thus highlighting the importance of isoform-selectivity for the development of epigenetic-modifying therapeutics. HDAC2, for example, would be an excellent target for inhibition (Guan et al., 2007).
Future preclinical efforts will also need to concentrate further on how epigenetic modifications act in concert during the distinct phases of memory formation. Indeed, the biological profile underlying cognitive and behavioral phenotypes may be determined by a collective pattern of epigenetic modifications, rather than individual changes in post-translational histone or DNA modification (see Gräff & Mansuy, 2008, for review). In relation to this, it is important to determine whether an epigenetic treatment for individual memory disorders will require the targeting of one or more epigenetic modifications simultaneously. And more broadly, whether such epigenetic treatments could be useful in combination with other, currently available pharmacologic treatments. It has been proposed in the cancer field that the use of DNMTi’s might be particularly beneficial if used in conjunction with chemotherapy. DNMTi’s could potentially suppress the activation of pro-apoptotic genes in response to cytotoxic agents. This would confer greater resistance to chemotherapy, resulting in less cell death (Kelly et al., 2010). Similarly, epigenetic modifiers might improve the efficacy of the current and developing AD treatments with broad molecular targets (e.g. cholinesterases, mementine, β-secretase inhibitors). Alternatively, epigenetic modifying drugs might prove to be effective sole therapies if they can positively regulate gene targets involved in the degradation and clearance of β-amyloid peptides.
Summary
Epigenetics has a long history in the fields of developmental biology and cancer. Over the past seven or eight years, epigenetics has made its way into the thoughts and experimental plans of neuroscientists. A particularly compelling body of work has accumulated in a surprisingly short period of time that implicates epigenetics in memory processes. In addition, studies are now demonstrating that epigenetic modifying drugs present a promising avenue for the amelioration of memory deficits. The future therapeutic potential of epigenetics in memory relies on both the continued efforts of labs already deeply involved in the research and newcomers providing a fresh perspective. If these research efforts are combined with an historical appreciation of epigenetics and an eye on therapeutics, this could be a decade of enormous advances in cognitive epigenetics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, et al. Chromatin acetylation, memory, LTP are impaired in CBP+/− mice: a model for cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–957. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Anekonda TS. Resveratrol-A boon for treating Alzheimer’s disease. Brain Research Reviews. 2006;52:316–326. doi: 10.1016/j.brainresrev.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straumer O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. Journal of Clinical Oncology. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step. Cancer Letters. 2009;280:211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–83. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Cao X, Südhof TC. A transcriptively active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learning and Memory. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J, Petroze R, Castegna A, Ding Q, Keller JN, Markesbery WR, et al. 4-Hydroxynonenal oxidatively modifies histones: implications for Alzheimer’s disease. Neuroscience Letters. 2004;356:155–158. doi: 10.1016/j.neulet.2003.11.047. [DOI] [PubMed] [Google Scholar]
- Ellinger J, Kahl P, von der Gathen J, Rogenhofer S, Heukamp LC, Gütgemann I, et al. Global levels of histone modifications predict prostate cancer recurrence. Prostate. 2010;70:61–69. doi: 10.1002/pros.21038. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature Neuroscience. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodeling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Gräff J, Mansuy IM. Epigenetic code in cognition and behaviour. Behavioural Brain Research. 2008;192:70–87. doi: 10.1016/j.bbr.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph J, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. Journal of Neuroscience. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–5. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Katzman R, Terry R, DeTeresa R, Brown T, Davies P, Fuld P, et al. Clinical, pathologica, and neurochemical changes in dementia: a subgroup with preserved mental status and numerous neocortical plaques. Annals of Neurology. 1988;23:138–144. doi: 10.1002/ana.410230206. [DOI] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nature Biotechnology. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–12. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- Koshibu K, Gräff J, Beullens M, Heitz FD, Berchtold D, Russig H, et al. Protein phosphatase 1 regulates the histone code for long-term memory. Journal of Neuroscience. 2009;29:13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. The Journal of Biological Chemistry. 2004;279:405450–59. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, et al. Journal of Biological Chemistry. 2006;281:15763–17773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. Journal of Neuroscience. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–7. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Wen PH, Dutt A, Shioi J, Takashima A, Siman R, et al. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutation. Cell. 2003;114:635–645. doi: 10.1016/j.cell.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiology of Aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory function. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiology of Learning and Memory. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, et al. Cortical DNA methylation maintains remote memory. Nature Neuroscience. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen S, Masliah E, Mukherjee C, Meyers D, Cole PA, Ott M. Acetylation of Tau inhibits its degradation and contributes to Tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung NH, Zhu X, Kruman II, Castellani RJ, Petersen RB, Siedlak SL, Perry G, Smith MA, Lee HG. Evidence of DNA damage in Alzheimers disease: phosphorylation of histone H2AX in astroctyes. Age. 2008;30:209–215. doi: 10.1007/s11357-008-9050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficit. Learning and Memory. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro TF, Marques O, Kazantsev A. Therapeutic role of sirtuins in neurodegenerative disease. Biochimica et Biophysica Acta. 2008;1782:363–369. doi: 10.1016/j.bbadis.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Ricobaraza A, Cuadrado-Tejedor M, Pérez-Mediavilla A, Frechilla D, Del Río J, García-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- Roe CM, Mintun MA, D’Angelo G, Xiong C, Grant EA, Morris JC. Alzheimer disease and cognitive reserve: variation of education effect with carbon 11-labeled Pittsburgh Compound B uptake. Archives of Neurology. 2008;65:1467–1471. doi: 10.1001/archneur.65.11.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, et al. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Scarpa S, Fuso A, D’Anselmi F, Cavallaro RA. Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease? FEBS letters. 2003;541:145–148. doi: 10.1016/s0014-5793(03)00277-1. [DOI] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proceedings of the National Academy of Sciences. 2009;106:9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascasde in insular cortex during novel taste learning. Journal of Neuroscience. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohgi H, Utsugisawa K, Nagane Y, Yoshimura M, Genda Y, Ukitsu M. Reduction with age in methylcytosine in the promoter region -224 approximately -101 of the amyloid precursor protein gene in autopsy human cortex. Molecular Brain Research. 1999;70:288–292. doi: 10.1016/s0169-328x(99)00163-1. [DOI] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB: CBP-dependent transcriptional activation. Journal of Neuroscience. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fivecoat H, Ho L, Pan Y, Ling E, Pasinetti GM. The role of Sirt1: at the crossroad between promotion of longevity and protection against Alzheimer’s disease neuropathology. Biochimica et Biophysica Acta. 2010;1804:1690–1694. doi: 10.1016/j.bbapap.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Wenk GL. Neuropathologic changes in Alzheimer’s disease. Journal of Clinical Psychiatry. 2003;64:7–10. [PubMed] [Google Scholar]
- West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. Journal of Molecular Neuroscience. 1995;6:141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- Wilson RS, Herbert LE, Scherr PA, Barnes LL, Mendes de Leon CF, Evans DA. Educational attainment and cognitive decline in old age. Neurology. 2009;72:460–465. doi: 10.1212/01.wnl.0000341782.71418.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, et al. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learning and Memory. 2005;12:111–119. doi: 10.1101/lm.86605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learning and Memory. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]