Abstract
Studies of palladium(II) and platinum(II) binding to well-characterized proteins contribute to understanding the influence of these metals in the environment and the body. The well-characterized apo-protein of azurin has a soft-metal binding site that may be exposed to solvent by mutation of a coordinating histidine-117 residue to glycine. Palladium(II) and platinum(II) form strong 1:1 adducts with the apo form of H117G azurin. A combination of UV-visible, CD, and ICP-MS techniques suggests that the metal binds specifically at His-46 and Cys-112 of the protein.
Platinum-group elements (PGEs) play an integral role in modern chemistry, catalysis, and chemotherapeutic treatments. Because of their heavy use, they have entered our environment, particularly from leaching of catalyst from the catalytic converters in automobiles.1,2 Catalytic converters emit PGEs in inhalable and commutable particle sizes (0.1–20 nm) that are rapidly complexed into soluble and mobile species after deposition.3–5 As a result, PGEs are highly bioavailable and known to bioaccumulate in plant and aquatic life.6,7 Human exposure to PGEs is an increasing concern, specifically following treatment with platinum for battling cancer.8–10 Exposure to cisplatin and its derivates leads to highly elevated and long-term persistent in vivo exposure to bioreactive Pt.11 Thus it is important to understand the chemistry underlying PGE interactions with biomolecules.
Though there has been progress on understanding the interactions of these metals with nucleic acids,12 there remains a need for information on the binding of Pd2+ and Pt2+ to proteins. Interactions of Pd2+ and Pt2+ with proteins have been studied, but the precise location of metal binding is rarely known.13–22 There is also interest in Pd2+-protein adducts as potential scaffolds for catalysis23–25 including hydrolytic cleavage of peptides.26–31 Pd2+ salts also influence amyloid fibril formation.32,33
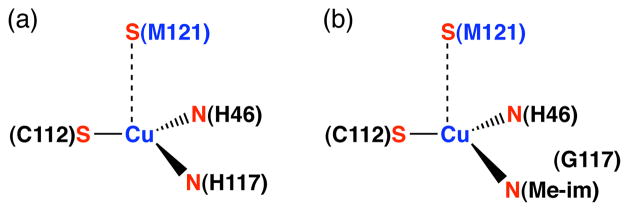
This contribution describes studies utilizing the apo-protein of azurin, a type 1 copper metalloprotein with a characteristic deep blue color. The canonical copper-binding site of Pseudomonas aeruginosa (Pa) azurin consists of four amino acid residues: His-46, Cys-112, His-117, and Met-121 (Figure 1a).34 Pa azurin has been used extensively in the study of protein-metal interactions because it provides a common mononuclear binding site for Zn2+, Ni2+, Co2+, Mn2+, Cd2+, Au+, Ag+, and Hg2+.35–43 Another advantage of azurin is that, as a small protein that is amenable to expression on a large scale from recombinant E. coli, it may easily be mutated to modify the coordination environment of copper44–47 or other metals48,49 in its metal binding site. Among mutations to the primary coordination sphere of the copper site, the H117G mutation is special because it is a small change that allows significant solvent access to the metal, enabling exogenous ligands to bind to copper(II) in place of the missing His-117 residue.46,50,51 For example, addition of copper(II) and an excess of N-methylimidazole to the apo form of H117G apo-azurin results in a characteristic 630 nm absorbance band and a coordination environment similar to the wild type protein.50 Figure 1b illustrates the binding of N-methylimidazole to the H117G copper(II) azurin. We anticipated that the compiled knowledge of H117G azurin would offer a way to unambiguously characterize Pd2+ and Pt2+ binding to native protein residues.
Figure 1.
The copper(II) sites of (a) azurin, (b) H117G azurin with added N-methylimidazole (Me-im).
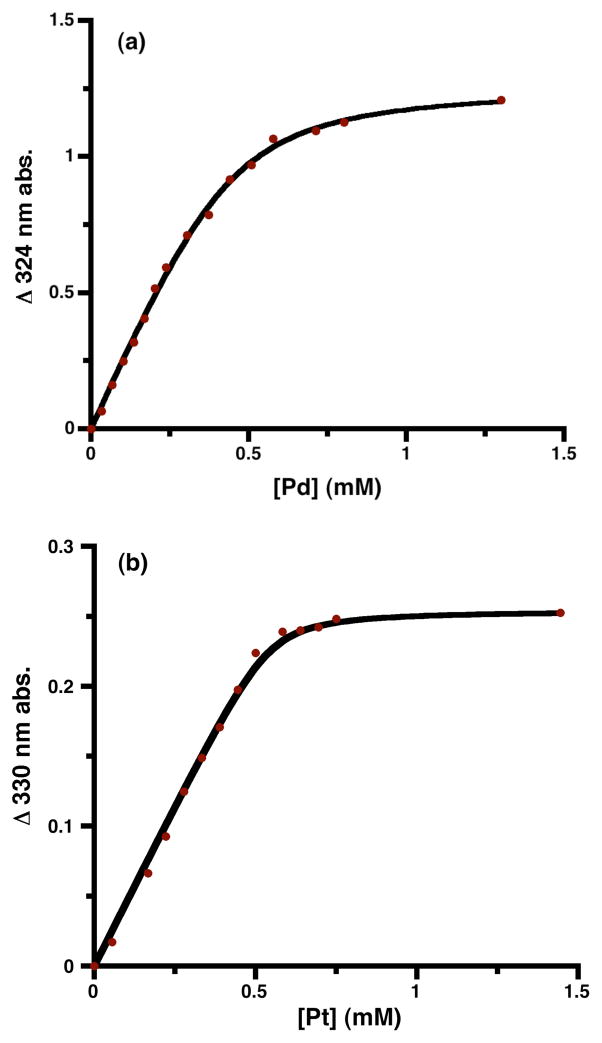
Upon the addition of 1 equiv of MCl42− to a 0.48 mM solution of the apo-form of H117G azurin (buffered at pH 7.0 with 5 mM of 3-(N-morpholino)propanesulfonic acid (MOPS)), a new broad absorbance developed in the UV-vis spectrum for M = Pd (ε = 2.6 mM−1cm−1 at 324 nm) and M = Pt (ε= 0.54 mM−1cm−1 at 330 nm). The spectra are shown in the Supporting Information. Addition of these metal salts to wild type protein does not yield any significant new absorbance, showing that the mutation is essential for PGE binding. The new absorptions in the Pd2+ and Pt2+ adducts of the apo-H117G azurin are reminiscent of those seen in Pd2+ and Pt2+ complexes with anionic sulfur donors, suggesting coordination to cysteinate.52,53 The dependence of the absorbance on added metal concentration (Figure 2) fits to a standard binding curve (eq 1), yielding association constants of 2.1(5) × 104 M−1 for Pd2+ and 1.2(7) × 105 M−1 for Pt2+. Thus, both PGEs bind strongly, and Pt2+ binds roughly 6 times more strongly than Pd2+.
Figure 2.
| (1) |
The development of the new absorbance saturated in each case at approximately 1 equiv of Pd2+ or Pt2+ relative to protein (Figure 2). Exchanging the buffer after addition of the metal salt gave no significant change to the UV-vis spectra, indicating low koff rates. After buffer exchange and dilution, the PGE loaded proteins were digested with acid followed by analysis with inductively coupled plasma spectroscopy (ICP). The metal analysis indicated that the proteins incorporated 95% Pd2+ and 98% Pt2+ into the protein. Since there are no alternative metal binding sites known in azurin,46 and the new UV absorbance is consistent with a thiolate-to-metal charge transfer transition,52–54 these results suggest that the heavy metals bind at the copper site with Cys and His donors.
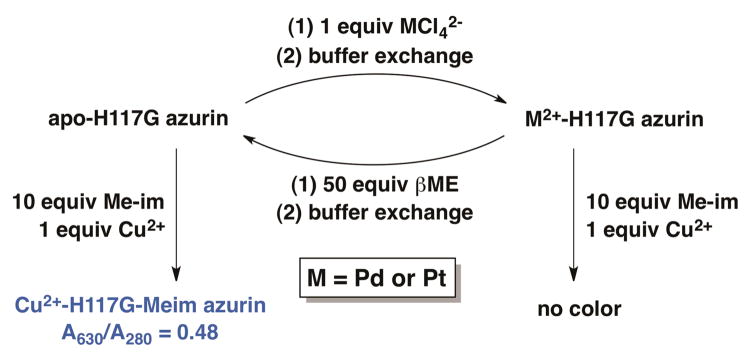
In order to further support the location of metal binding, we took advantage of the fact that Cu2+-bound H117G azurin develops a intense blue color upon binding of N-methylimidazole to regenerate the N2S2 coordination environment of the wild-type copper(II) protein.50 This property enabled us to query the status of the copper binding site as shown in Scheme 1. Addition of copper(II) and an excess of N-methylimidazole to the apo form of H117G azurin resulted in the expected copper-based LMCT absorbance at λmax = 630 nm, with a ratio of absorbances A630/A280 = 0.48. In constrast, addition of Cu2+ and N-methylimidazole to the Pd2+ or the Pt2+ substituted protein did not result in appearance of the 630 nm absorbance band. The inability of the Pd2+ and Pt2+ substituted proteins to bind copper suggests that the heavy metals bind at the copper site, but does not preclude the possibility that the protein is somehow damaged by the heavy metals in a manner that prevents copper binding. We tested for protein damage by incubating the Pd2+ and Pt2+-loaded proteins in an excess of the soft ligand 2-mercaptoethanol for 12 hours to chelate the PGE salt, followed by buffer exchange. After this treatment to remove the heavy metal, the copper binding ability of the protein was completely restored (A630/A280 = 0.48). These results indicate that the protein had undergone no irreversible change, and provides additional support for Pd2+ and Pt2+ binding specifically at the H46/C112 site of the engineered azurin.
Scheme 1.
Using the Cu2+ binding ability of H117G azurin to query M2+ binding at the copper site.
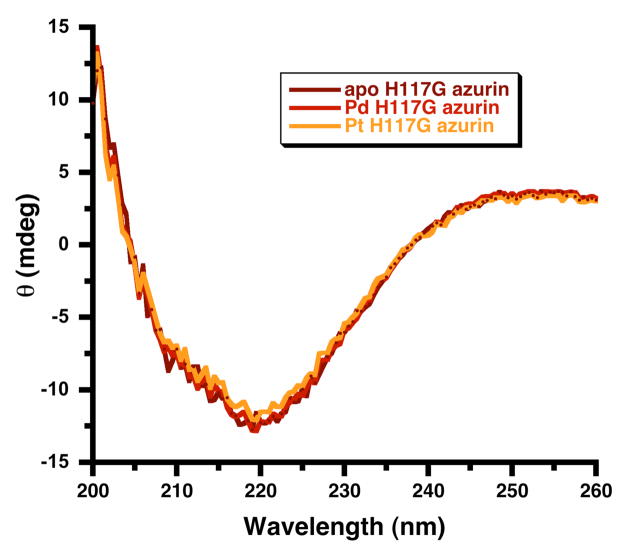
To evaluate whether the addition of Pd2+ and Pt2+ changed the secondary structure of H117G azurin, circular dichroism (CD) experiments were performed after the addition of either Pd2+ or Pt2+ to the apo form of H117G azurin. The CD spectrum, which matches the literature spectrum for the azurin protein,55 was unchanged by the H117G mutation or by the addition of either metal (Figure 3). These results suggest that Pd2+ and Pt2+ binding to the protein does not induce any significant conformational changes. Thus, our experiments argue against a change in the structure of the H117G azurin apo-protein upon PGE binding, and clearly point toward PGE binding at the vacant copper-binding site of H117G apo-azurin.
Figure 3.
Circular dichroism spectra of H117G azurin in the apo form and after the addition of Pd2+ and Pt2+. Samples have 0.22 mM protein in 5 mM MOPS buffer, pH = 7.0.
In conclusion, the copper-binding apo-protein of azurin may be engineered so that it coordinates a single Pd2+ or Pt2+ ion. Each ion binds with Kassoc > 20,000 M−1, and binding to Pt2+ is stronger than to Pd2+. Our evidence indicates that the metal binds to the cysteine at the vacant copper site, and thus we suggest that the metals have square-planar NSCl2 coordination that includes His-46 and Cys-112 of the protein.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (GM-065313). We thank Kara Bren and her group for access to equipment for mutation and protein expression, and we thank Kara Bren and Joseph Wedekind for useful discussions.
Footnotes
Supporting Information. Additional spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Merget R, Rosner G. Sci Total Environ. 2001;270:165. doi: 10.1016/s0048-9697(00)00788-9. [DOI] [PubMed] [Google Scholar]
- 2.Ravindra K, Bencs L, Van Grieken R. Sci Total Environ. 2004;318:1. doi: 10.1016/S0048-9697(03)00372-3. [DOI] [PubMed] [Google Scholar]
- 3.Gomez B, Gomez M, Sanchez JL, Fernandez R, Palacios MA. Sci Total Environ. 2001;269:131. doi: 10.1016/s0048-9697(00)00826-3. [DOI] [PubMed] [Google Scholar]
- 4.Colombo C, Oates CJ, Monhemius AJ, Plant JA. Geochem. 2008;8:91. [Google Scholar]
- 5.Colombo C, Monhemius AJ, Plant JA. Sci Total Environ. 2008;389:46. doi: 10.1016/j.scitotenv.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Sures B. Parasitology. 2003;126:S53. doi: 10.1017/s003118200300372x. [DOI] [PubMed] [Google Scholar]
- 7.Gagnon ZE, Newkirk C, Hicks S. J Environ Sci Health. 2006;A41:397. doi: 10.1080/10934520500423592. [DOI] [PubMed] [Google Scholar]
- 8.Lippert B, editor. Cisplatin - Chemistry and Biochemistry of a Leading Anticancer Drug. Springer-Verlag; Berlin: 1999. [Google Scholar]
- 9.Wang D, Lippard SJ. Nature Rev. 2005;4:307. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 10.Travis LB, Beard C, Allan JM, Dahl AA, Feldman DR, Oldenburg J, Daugaard G, Kelly JL, Dolan ME, Hannigan R, Constine LS, Oeffinger KC, Okunieff P, Armstrong G, Wiljer D, Miller RC, Gietema JA, van Leeuwen FE, Williams JP, Nichols CR, Einhorn LH, Fossa SD. J Natl Cancer Inst. 2010;102:1114. doi: 10.1093/jnci/djq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouwers E, Huitema A, Beijen J, Schellens J. BMC Clinical Pharmacology. 2008;8 doi: 10.1186/1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamieson ER, Lippard SJ. Chem Rev. 1999;99:2467. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 13.Ivanov AI, Christodoulou J, Parkinson JA, Barnham KJ, Tucker A, Woodrow J, Sadler PJ. J Biol Chem. 1998;273:14721. doi: 10.1074/jbc.273.24.14721. [DOI] [PubMed] [Google Scholar]
- 14.Trynda-Lemiesz L, Kozlowski H, Keppler BK. J Inorg Biochem. 1999;77:141. doi: 10.1016/s0162-0134(99)00183-x. [DOI] [PubMed] [Google Scholar]
- 15.Cox MC, Barnham KJ, Frenkiel TA, Hoeschele JD, Mason AB, He QY, Woodworth RC, Sadler PJ. J Biol Inorg Chem. 1999;4:621. doi: 10.1007/s007750050386. [DOI] [PubMed] [Google Scholar]
- 16.Allardyce CS, Dyson PJ, Coffey J, Johnson N. Rapid Commun Mass Spectrom. 2002;16:933. doi: 10.1002/rcm.662. [DOI] [PubMed] [Google Scholar]
- 17.Najajreh Y, Peleg-Shulman T, Moshel O, Farrell N, Gibson D. J Biol Inorg Chem. 2003;8:167. doi: 10.1007/s00775-002-0402-y. [DOI] [PubMed] [Google Scholar]
- 18.Calderone V, Casini A, Mangani S, Messori L, Orioli PL. Angew Chem Int Ed. 2006;45:1267. doi: 10.1002/anie.200502599. [DOI] [PubMed] [Google Scholar]
- 19.Casini A, Mastrobuoni G, Temperini C, Gabbiani C, Francese S, Moneti G, Supuran CT, Scozzafava A, Messori L. Chem Commun. 2007:156. doi: 10.1039/b611122j. [DOI] [PubMed] [Google Scholar]
- 20.Esteban-Fernández D, Montes-Bayon M, Blanco González E, Gómez Gómez MM, Palacios MA, Sanz-Medel A. J Anal At Spectrom. 2008;23:378. [Google Scholar]
- 21.Arnesano F, Natile G. Pure Appl Chem. 2008;80:2715. [Google Scholar]
- 22.Arnesano F, Belviso BD, Caliandro R, Falini G, Fermani S, Natile G, Siliqi D. Chem Eur J. 2011;17:1569. doi: 10.1002/chem.201001617. [DOI] [PubMed] [Google Scholar]
- 23.Ikawa T, Sajiki H, Hirota K. Tetrahedron. 2005;61:2217. [Google Scholar]
- 24.Kiyoyama S, Maruyama T, Kamiya N, Goto M. Ind Eng Chem Res. 2008;47:1527. [Google Scholar]
- 25.Abe S, Hikage T, Watanabe Y, Kitagawa S, Ueno T. Inorg Chem. 2010;49:6967. doi: 10.1021/ic1003758. [DOI] [PubMed] [Google Scholar]
- 26.Karet GB, Kostić NM. Inorg Chem. 1998;37:1021. [Google Scholar]
- 27.Parac TN, Ullmann GM, Kostić NM. J Am Chem Soc. 1999;121:3127. [Google Scholar]
- 28.Milović NM, Kostić NM. J Am Chem Soc. 2002;124:4759. doi: 10.1021/ja012366x. [DOI] [PubMed] [Google Scholar]
- 29.Milović NM, Kostić NM. Inorg Chem. 2002;41:7053. doi: 10.1021/ic025640c. [DOI] [PubMed] [Google Scholar]
- 30.Sun X, Zhang L, Zhang Y, Yang G, Guo Z, Zhu L. New J Chem. 2003;27:818. [Google Scholar]
- 31.Miskevich F, Davis A, Leeprapaiwong P, Giganti V, Kostić NM, Angel LA. J Inorg Biochem. 2011;105:675. doi: 10.1016/j.jinorgbio.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Garnett AP, Jones CE, Viles JH. Dalton Trans. 2006:509. doi: 10.1039/b511553a. [DOI] [PubMed] [Google Scholar]
- 33.Pushie MJ, Ross ARS, Vogel HJ. Anal Chem. 2007;79:5659. doi: 10.1021/ac070312l. [DOI] [PubMed] [Google Scholar]
- 34.Gray HB, Malmström BG, Williams RJP. J Biol Inorg Chem. 2000;5:551. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 35.McMillin DR, Rosenberg RC, Gray HB. Pro Natl Acad Sci. 1974;71:4760. doi: 10.1073/pnas.71.12.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaszak JA, Ulrich EL, Markley JL, McMillin DR. Biochemistry. 1982;21:6253. doi: 10.1021/bi00267a033. [DOI] [PubMed] [Google Scholar]
- 37.Engeseth HR, McMillin DR, Otvos JD. J Biol Chem. 1984;259:4822. [PubMed] [Google Scholar]
- 38.Klemens AS, McMillin DR, Tsang HT, Penner-Hahn JE. J Am Chem Soc. 1989;111:6398. [Google Scholar]
- 39.Nar H, Messerschmidt A, Huber R, Van de Kamp M, Canters GW. FEBS Lett. 1992;306:119. doi: 10.1016/0014-5793(92)80981-l. [DOI] [PubMed] [Google Scholar]
- 40.Moratal JM, Romero A, Salgado J, Perales-Alarcón A, Jiménez HR. Eur J Biochem. 1995;228:653. doi: 10.1111/j.1432-1033.1995.tb20305.x. [DOI] [PubMed] [Google Scholar]
- 41.Jiménez HR, Salgado J, Moratal JM, Morgenstern-Badarau I. Inorg Chem. 1996;35:2737. [Google Scholar]
- 42.McCleskey TM, Mizoguchi TJ, Richards JH, Gray HB. Inorg Chem. 1996;35:3434. doi: 10.1021/ic951055i. [DOI] [PubMed] [Google Scholar]
- 43.Bonander N, Vanngard T, Tsai LC, Langer V, Nar H, Sjoelin L. Proteins. 1997;27:385. doi: 10.1002/(sici)1097-0134(199703)27:3<385::aid-prot6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 44.Tsai LC, Bonander N, Harata K, Karlsson G, Vänngård T, Langer V, Sjölin L. Acta Crystallographica D. 1996;52:950. doi: 10.1107/S0907444996004982. [DOI] [PubMed] [Google Scholar]
- 45.Vidakovic M, Germanas JP. Angew Chem Int Ed. 1995;34:1622. [Google Scholar]
- 46.Kolczak U, Dennison C, Messerschmidt A, Canters GW. Handbook of Metalloproteins. 2001;2:1170. [Google Scholar]
- 47.Marshall NM, Garner DK, Wilson TD, Gao YG, Robinson H, Nilges MJ, Lu Y. Nature. 2009;462:113. doi: 10.1038/nature08551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizoguchi TJ, Di Bilio AJ, Gray HB, Richards JH. J Am Chem Soc. 1992;114:10076. [Google Scholar]
- 49.Fraczkiewicz G, Bonander N, Czernuszewicz RS. J Raman Spect. 1998;29:983. [Google Scholar]
- 50.Den Blaauwen T, Van de Kamp M, Canters GW. J Am Chem Soc. 1991;113:5050. [Google Scholar]
- 51.den Blaauwen T, Hoitink CWG, Canters GW, Han J, Loehr TM, Sanders-Loehr J. Biochemistry. 1993;32:12455. doi: 10.1021/bi00097a025. [DOI] [PubMed] [Google Scholar]
- 52.Isci H, Dag O, Mason WR. Inorg Chem. 1993;32:3909. [Google Scholar]
- 53.Massacesi M, Pinna R, Biddau M, Ponticelli G, Zakharova IA. Inorg Chim Acta. 1983;80:151. [Google Scholar]
- 54.Hansen CN, Kirketerp MBS, Kristensen MB, Nielsen SB, Støchkel K, Wyer JA. Chem Phys Lett. 2011;502:53. [Google Scholar]
- 55.Mei G, Gilardi G, Venanzi M, Rosato N, Canters GW, Finazzi AA. Protein Sci. 1996;5:2248. doi: 10.1002/pro.5560051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.