Abstract
DNA-based nanotechnology is currently being developed as a general assembly method for nanopatterned materials that may find use in electronics, sensors, medicine, and many other fields. Here we present results on the construction and characterization of DNA nanotubes, a self-assembling superstructure composed of DNA tiles. Triple-crossover tiles modified with thiol-containing double-stranded DNA stems projected out of the tile plane were used as the basic building blocks. Triple-crossover nanotubes display a constant diameter of ≈25 nm and have been observed with lengths up to 20 μm. We present high-resolution images of the constructs, experimental evidence of their tube-like nature as well as data on metallization of the nanotubes to form nanowires, and electrical conductivity measurements through the nanowires. DNA nanotubes represent a potential breakthrough in the self-assembly of nanometer-scale circuits for electronics layout because they can be targeted to connect at specific locations on larger-scale structures and can subsequently be metallized to form nanometer-scale wires. The dimensions of these nanotubes are also perfectly suited for applications involving interconnection of molecular-scale devices with macroscale components fabricated by conventional photolithographic methods.
The emerging field of molecular electronics has made considerable recent progress in the development of molecular-scale electronics components and sensors, but the need for templating and patterning at the molecular scale is a major challenge. One approach is the use of DNA-based nanotechnology, which seeks to engineer synthetic DNA polymers to encode information necessary for realization of desired structures or processes on the molecular level (1–6). Properly designed sets of DNA oligonucleotides are able to self-assemble into complex, highly organized structures. The major advantage of DNA as a self-assembling structural material over other currently known chemical systems is DNA's well understood base-pairing rules, which essentially act as a programming language with which to input organizational instructions. In particular, B-form DNA, the structure adopted by double-stranded DNA (dsDNA) under standard solution conditions, consists of the famous antiparallel double helix of complementary strands. Its specifically programmable molecular recognition ability, as well as the huge diversity of available sequences, makes DNA ideal for controlling the assembly of nanostructures. Annealing of complementary strands is achieved by simply heating dissolved oligonucleotide strands above their dissociation temperature then cooling them very slowly, thus allowing strands to find and bind their sequence complements. Unpaired single-strand regions (ssDNA) extending from the ends of double-strand domains can be used to specifically bind other dsDNA displaying a complementary ssDNA sticky end. Regions of ssDNA, therefore, can act as address codes capable of orienting attached substructures to neighboring points in three-space. If DNA is appended to nanoscale objects and chemical moieties, it can act as “smart glue” for organizing such objects and groups in space. An obvious advantage of DNA self-assembly for fabrication is the simultaneous creation of millions or billions of copies of a desired structure due to the massive parallelism inherent in molecular processes.
Hence the use of a programmable, self-assembling polymer such as DNA appears to be one of the most promising avenues available for positioning components for molecule electronics and for targeted connections that can subsequently be metallized to form functional nanowires. The annealing of oligonucleotides has already been used to direct specific association of gold nanospheres into 3D clusters (7–9). Nanometer-scale gold rods have also been bound to one another by hybridization of oligonucleotides (10). The assembly of a silver wire between two prefabricated leads by a DNA hybridization technique has also been reported (11). However, previous studies have used only linear dsDNA; therefore, the available structures were limited.
It remained a challenge to extend these methods to allow for a fine control of diverse associations so as to allow for a wide selection of programmable superstructures.
DNA nanotechnology and some approaches to DNA-based computing rely on unusual structural motifs, including the immobile branched junction (1, 12), which is a stable analogue of the Holliday intermediate in genetic recombination (13). The smallest rigid complexes contain two branched junctions (also known as “crossovers”) connecting two double helices (14) where the helices are designed to be coplanar and are said to be linked by strand exchange at the crossover points. One strand from each helix crosses over and begins base-pairing on the opposite helix. These complexes are known as double-crossover (DX) tiles (15); in the present study we used triple-crossover (TX) tiles, which contain three coplanar double helices linked at each of four crossover points (16). DNA tiles can be programmed to assemble into large 2D lattice sheets by properly designing their sticky ends. Lattices composed of hundreds of thousands of DX or TX tiles and extending up to at least 10 μm on their long edge have been created and examined by atomic force microscopy (AFM) and transmission electron microscopy (TEM) (16, 17). Besides being attractive structural members for simple lattice formation, multihelix tiles have been used as components of self-assembling DNA computers (18–21). One aim of further development of these tile and lattice structures is their use as templates on which to organize desired patterns of materials other than DNA, for example, chemical groups, nanoparticles, nanorods, carbon nanotubes, and proteins. Such DNA templates may be useful as scaffolds for the creation of nanoelectronic circuits and other desired objects and devices requiring nanometer-scale feature resolution.
Here we report the conversion of TX 2D flat lattice super-structures into interesting uniform nanotube superstructures by the addition of thiol groups on a dsDNA stem protruding from the TX tile plane. Nanotube structures are the sole product after annealing the thiol-containing oligonucleotides. The switching of superstructure morphology from 2D flat lattice to tube-like structures has recently been reported in another DNA tiling system by means of the reprogramming of sticky ends, but the previous system resulted in a variety of tube sizes mixed with some fragments of flat lattice (22). Here we present high-resolution images of TX nanotubes from TEM, scanning electron microscopy (SEM), and AFM. DNA has useful molecular recognition and self-assembly characteristics, but its poor electrical properties prevent its direct use in electrical devices (23). We demonstrate here the use of self-assembled TX nanotubes as templates for the construction of highly conductive metallic nanowires. TX-templated nanowire formation is easily reproducible and programmable, and it results in nanowires with dramatically higher conductivity than previously reported dsDNA-templated silver nanowires (11).
Materials and Methods
DNA Sequence Design, Synthesis, Purification, and Annealing. The TAO tiles used here have been extensively characterized and described (16). Briefly, oligonucleotide sequences were designed by using a random hill-climbing algorithm to maximize the likelihood of formation of the desired base-pairing structures while minimizing the chances of spurious pairings; tiles were shown to form as designed by electrophoretic and chemical analyses; and large flat lattice sheets were observed by AFM imaging. Custom oligonucleotides were ordered (Integrated DNA Technology) and gel purified. Sequences of oligonucleotides (in 5′ to 3′ direction) used here are as follows. Tile type A: A1, TCGGCTATCGAGTGGACACCGAAGACCTAACCGCTTTGCGTTCCTGCTCTAC; A2, AGTTAGTGGAGTGGAACGCAAAGCGGTTAGGTCTTCGGACGCTCGTGCAACG; A3, ACGAGCGTGGTAGTTTTCTACCTGTCCTGCGAATGAGATGCCACCACAGTCACGGATGGACTCGAT; and A4, TGCTCGGTAGAGCACCAGATTTTTCTGGACTCCTGGCATCTCATTCGCACCATCCGTGACTGTGGACTAACTCCGCTT. Tile type B: B1, CGAGCAATGAAGTGGTCACCGTTATAGCCTGGTAGTGAGCTTCCTGCTGTAT; B2, ACACAGTGGAGTGGAAGCTCACTACCAGGCTATAACGGACGATGATAAGCGG; B3a, AGCCGAATACAGCACCATCTTTTGATGGACTCCTGAATCGACGTAACTT; B3b, TTGTTACGTCTTTCTACTCGCACCTTCGCTGAGTTTGGACTGTGTCGTTGC; B4, ATCATCGTGGTTCTTTTGAACCTGACCTGCGAGGTATGTGATTTTTCACATACTTTAGAGATTCACCAAACTCAGCGAAGGACTTCAT; B4a, ATCATCGTGGTTCTTTTGAACCTGACCTGCGAGGTATGTGATTT; and B4b, TTCACATACTTTAGAGATTCACCAAACTCAGCGAAGGACTTCAT. Strand B3a was ordered with (B3aSH) and without (B3a) a thiol group attached to its 5′ end, whereas B3b was made with (B3bSH) and without (B3b) a thiol on its 3′ end. The 5′ end of strand B3a and the 3′ end of B3b form a base-paired stem projecting out of the plane of the tile, likewise for B4a and B4b. Thiol-modified oligonucleotides were delivered with–SSH moieties and were reduced to –SH by treatment with dithiothreitol and exchanged into annealing buffer by gel permeation chromatography before use (NAP-5 columns, Amersham Biosciences).
Oligonucleotides were annealed to form tiles and lattices in stoichiometric mixtures by heating to 95°C for 5 min and slowly cooling to room temperature in a thermal cycler by dropping 0.1°C per min (total annealing time was ≈12 h). Exact stoichiometry is determined, if necessary, by titrating pairs of strands designed to hydrogen bond, and visualizing them by nondenaturing gel electrophoresis; absence of monomer is taken to indicate equimolar ratios have been achieved.
TEM Imaging. Transmission electron micrographs were obtained on a Phillips 301 TEM. Direct examination of DNA structures was carried out by pipetting 2–4 μl of annealed DNA solution onto a copper grid that had previously been layered with carbon film. Negative staining involved exposure of the sample on the grid to 10 μl of 2% uranyl acetate solution for 3 min. In all cases excess liquid was blotted from the sample by lightly touching an edge or corner of filter paper to the edge of the grid. Platinum rotary shadow samples were prepared by gently mixing annealed DNA sample with 1 vol of 80% glycerol, then spraying the mixture onto freshly cleaved mica by using a microcapillary and compressed air. The sample was dried on the mica under high vacuum overnight in a Balzer evaporator, tilted to a 4° angle, and rotated 180° (around the normal to the sample plane) during deposition by thermal evaporation of a thin layer of platinum. The sample was then tilted to 90° (flat face toward the electrodes) and coated with a carbon layer. The carbon/platinum film was floated off the mica onto the surface of a domed droplet of buffer and finally captured on a copper grid. Samples were air dried before examination in the TEM.
AFM Imaging. Atomic force micrographs were obtained on a Digital Instruments Nanoscope IIIa with a multimode head by tapping mode in air and tapping mode under buffer. For AFM imaging in air, samples were prepared by pipetting annealed DNA solution (≈2–5 μl) onto freshly cleaved mica (Ted Pella, Redding, CA), allowed to adhere for 2 min, rinsed gently by doming a drop of Milli-Q purified (Millipore) water onto the mica, then air dried either under a stream of nitrogen or by gentle waving. For AFM imaging in buffer, an ≈3- to 5-μl sample was spotted on freshly cleaved mica and left to adsorb to the surface for 3 min. Then, 30 μl of 1× TAE/Mg2+ buffer (40 mM Tris acetate, pH 7.6/2 mM EDTA/10 mM MgCl2) was placed onto the mica and another 30 μl of buffer was pipetted onto the AFM tip. Imaging was performed under 1× TAE/Mg2+ in a tapping mode fluid cell on a Multimode NanoScope IIIa (Digital Instruments), using NP-S tips (Veeco Instruments, Woodbury, NY).
SEM Imaging. All SEM images were taken at 10–15 kV (accelerating voltage) with electron-beam spot size = 3 in a Philips FEI XL30 Thermal Field Emitter SEM. Chamber pressure was between 1.0 × 10–7 and 1.0 × 10–6 mbar (1 mbar = 100 Pa). All samples were prepared on doped n-type silicon substrate with resistivity = 2.0 × 10–5 Ω·m and a 500-nm thermal oxide layer prepared by using the RCA cleaning process without the oxide stripping step. Silicon substrate was treated with aminopropylsilane before DNA sample deposition.
Two-Step Metallization. First, the TX nanotube lattice was seeded with silver by using the glutaraldehyde method (11, 22). Annealed DNA was incubated with 0.2% glutaraldehyde in 1× TAE/Mg2+ buffer on ice for 20 min, then at room temperature for 20 min, then the sample was loaded into a Slide-A-Lyzer mini dialysis unit (Pierce) and dialyzed overnight at 4°C in 1 liter of 1× TAE/Mg2+ buffer. The published method was modified in that the silver seeding was done in aqueous solution for 20 min instead of on substrate. Aldehyde-derivatized DNA was incubated in the dark with an equal volume of 0.1 M AgNO3 in 25% ammonia buffer (pH 10.5) at room temperature for 30–90 min, then 10 μl was deposited onto a silicon substrate and allowed to absorb for 5 min, then excess reagent was rinsed off with distilled water and the sample was dried under a stream of nitrogen. In the second step, HQ Silver electron microscopy formulation (www.nanoprobes.com) was used according to the manufacturer's instructions. One unit of initiator (A) was mixed with one unit of moderator (B) and one unit of activator (C), then 10 μl of this fresh mixture was pipetted onto the sample and left for 5 min. Finally, excess reagent was rinsed off again with distilled water and the sample was dried under a stream of nitrogen.
Results
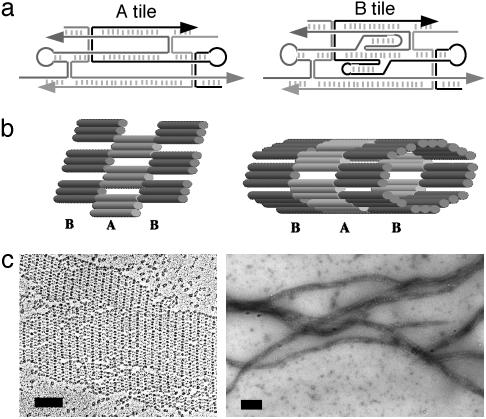
Design and Observation of TAO Nanotubes. The DNA self-assemblies described here were formed from two DNA tile building blocks, a TAO (tile A) and a TAO + 2J (tile B) as shown in Fig. 1a. Both tile types are triple-crossover (TX) tiles referred to as TAO because they contain three coplanar double-helical domains, their strand exchange points are antiparallel (strands reverse direction on crossing-over), and there are an odd number of helical half-turns between crossover points. The B tile contains two extra dsDNA stems, which form junctions with the central helix of the tile such that they project out of the tile plane, with one stem protruding on each side of the tile. The B tiles used here are modified by the replacement of the loop on one protruding stem with two thiol groups, one on a 3′ and the other on a 5′ strand terminus. Sticky ends located at the tile corners have been programmed such that A tiles bind to B tiles and B to A as shown in Fig. 1b. Detailed descriptions of these tiles and the flat lattice superstructure are given in ref. 16.
Fig. 1.
(a) Line drawings showing the strand traces through the two tile types used in the constructions. Oligonucleotides are shown in different shades of gray and with arrowheads marking the 3′ ends. Short vertical hash marks indicate base-pairing within double-helical regions; paired vertical lines represent crossover points. The central helices of both tile types are terminated with–TTTT–loops. Two extra stem-loops (2J) on the central helix of the B tile are designed to protrude, one into and one out of the tile plane. (b) (Left) Cartoon of a section of TAO flat lattice constructed from A and B tiles. Tubes represent double-helical regions; for simplicity, the 2J stem-loops on the B tiles are not shown. (Right) Cartoon model of a section of TAO nanotube shown with eight tiles per layer of tube. (c) (Left) TEM image of platinum rotary-shadowed TAO lattice. The B tiles appear darker than the A tiles, having picked up more platinum on their protruding 2J stem-loops; alternating stripes of A and B tiles are clearly visible with approximately the expected distance of 28 nm. (Right) TEM image of negative-stained TAO nanotubes. (Scale bars = 100 nm.)
Fig. 1b Right shows a section of the proposed structure of the nanotubes with B tile layers alternating with A tile layers, dsDNA helix axes aligned parallel with the tube axis, and thiol groups located inside the tubes. Imaging of TX AB flat lattice sheets (Fig. 1c Left) demonstrates lattice fragments with widely varying dimensions and often with uneven edges. On the other hand, DNA nanotubes (Fig. 1c Right) exhibit uniform widths of ≈25 nm for lengths of up to 20 μm. Negative-stain TEM of the nanotube superstructures provides images of sufficiently high resolution to observe that B stripes (from outwardly projecting dsDNA stem-loops) are perpendicular to the long axis of the nanotubes. This structure provides good evidence for an even number of tiles per ring layer and flat rings rather than a spiral structure. On the basis of experimental measurements and diameters of modeled tubes there appear to be eight tiles per layer. Significantly, the negative-stain TEM shows detailed information of the nanotube with unit periodicity of 27 nm, in good agreement with the modeled length of AB tile units. A recent paper described control of superstructure morphology in another DNA tiling system between flat lattice and elongated tube by changes in base sequence to reprogram the sticky end associations (22). The system described here differs in that the morphology switch is controlled by the presence or absence of thiol modifications on particular oligonucleotides.
Evidence of Tube Structure and Role of Thiol Groups. Fig. 2a is a typical AFM image (tapping mode in air) of the TX AB nanotube. Stripes perpendicular to the long axis of the filaments are clearly visible and indicate closed ring structures in successive layers rather than a spiral structure, which would have given stripes with noticeable diagonal slant. AFM peak–peak width measurement indicated that the individual subunits are separated by ≈27 nm. AFM height measurements (Fig. 2a) give a mean value of ≈2.6 nm, which is twice the height of the 2D flat lattice form. Our interpretation of AFM height data is that the nanotubes have become flattened onto the substrate during sample preparation. Fig. 2b shows a high-resolution AFM image (tapping mode in buffer) of the TX nanotube in which one can discern three gaps and four tiles in some of the layers. The gaps and tiles are arrayed parallel with the axis of the flattened tube. A zoomed-in view of a TEM image (Fig. 2c) of a negative-stained sample shows a section of nanotube that appears to have opened into a flat lattice. The width difference and the outline of the connecting section between the opened (flat) and closed (tube) parts of the imaged structure further suggest its tubular structure.
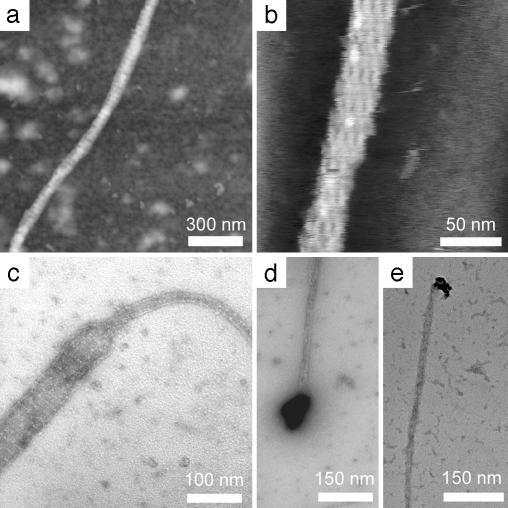
Fig. 2.
(a) AFM image of TAO nanotube taken by tapping mode in air. The B tile layers are visible as brighter stripes (because of increased height) oriented perpendicular to the long axis of the tube. (b) Very-high-resolution image of nanotube taken by tapping mode under buffer. Single helix holes between adjacent tiles are visible as darker patches in several layers. (c) TEM image of negative-stained sample, showing a section of nanotube that appears to have split into flat lattice. (d and e) Negative-stained TEM images of TAO nanotubes with gold nanoparticles attached to the ends, apparently by interaction with thiol sulfurs partially exposed at the tube termini.
Burial of the sulfur moieties within the tubes makes logical sense because disulfide bridges are preferred structures formed by thiol groups under physiologic-like solution conditions such as those used here. The formation of disulfide bonds between neighboring B tiles would cause the lattice to curve and form tubes. The possible existence of free thiols on the outer surface of the nanotubes was probed by using two different gold reagents with reactivity toward sulfur: monomaleimido Nanogold (Nanoprobes, Yaphank, NY) and fresh colloidal gold nanoparticles. The monomaleimido Nanogold failed to react with the tube surfaces (data not shown), whereas the colloidal gold displayed the interesting reactivity shown in Fig. 2 d and e. With very low background levels of unbound gold, the bound gold particles showed a very high probability of attachment to the ends of nanotubes and not to the outer surface anywhere else along the length of the tubes. The indication is that sulfur which is buried within the tubes is exposed to some extent at the open ends of the nanotubes and available for binding with gold. This observation, besides offering evidence of the location of the thiol groups, may also be exploited in future work on targeted binding or formation of electrical contacts with the ends of TX nanotubes.
To further characterize the orientation of TAO tiles in the tubular superstructures, primary amine groups were added to the oligonucleotide termini (B4a and B4b) that base pair to form the dsDNA stem projecting out of the tile plane on the tile face opposite that of the thiol-containing stem. According to our model of the nanotubes, this should place the primary amines on the outside of the tubes and accessible to reaction with sulfo-N-hydroxysuccinimido Nanogold (Nanoprobes). Incorporation of amine-modified oligonucleotide was shown to allow reaction with the monofunctional Nanogold reagent, which successfully attached 1.4-nm gold particles to the outside surface of the assembled tubes (data not shown). Reaction with unmodified tubes resulted in no gold labeling.
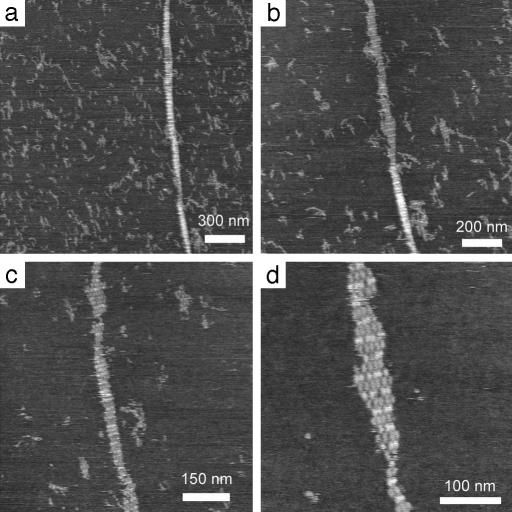
Fig. 3 shows a series of in situ zoomed images showing a TX nanotube being converted to flat lattice by the physical effect of the AFM tip. The series of images was collected by repeatedly reimaging the same nanotube under buffer with an AFM. The high-resolution image (Fig. 3d) shows the detailed nanostructure of the completely torn-open DNA nanotube with alternating layers of A and B tiles clearly discernible. Whole tiles have been scratched away from the opening nanotube by the force of the AFM tip, therefore the tile layers no longer contain all of the tiles that were present in the nanotube. A few layers are seen that still contain six tiles. The combination of the high-resolution AFM images (Figs. 2b and 3d) provides supporting evidence for our model in which individual tile layers in intact nanotubes contain eight tiles.
Fig. 3.
A series of AFM images captured by repeatedly reimaging and zooming in on the same nanotube, which appears mostly tube-like in a, with increasing wear-and-tear through b and c until a section of unfolded tube becomes a single-layer flat lattice (d) displaying stripes composed of lighter (higher) B tiles and darker (shorter) A tiles.
To test the effect of thiol oxidation state on nanotube formation, the nanotube-forming oligonucleotides were annealed in the presence of 20 mM dithiothreitol. AFM examination of this sample (Fig. 4) showed a DNA nanostructure rather different from the nanotubes. It appears to be two sheets of self-assembled DNA nanostructure stacked one on top the other, possibly with interactions between thiol groups from one sheet to another sheet. Further experiments must be completed to clarify a detailed interpretation of this observation; however, because no nanotubes were observed, it is clear that disulfide bridge formation plays an important role in TX nanotube assembly.
Fig. 4.
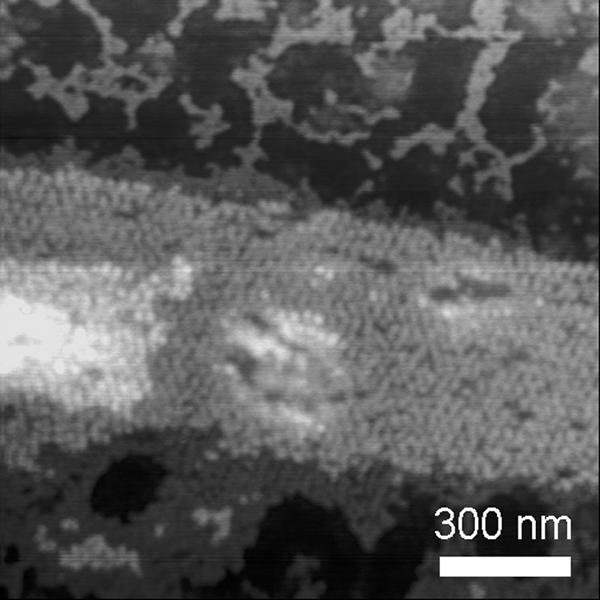
AFM image of self-assembled superstructure formed by annealing the tube-forming thiolated oligonucleotides in the presence of thiol reducing agent (20 mM dithiothreitol). No nanotubes were observed.
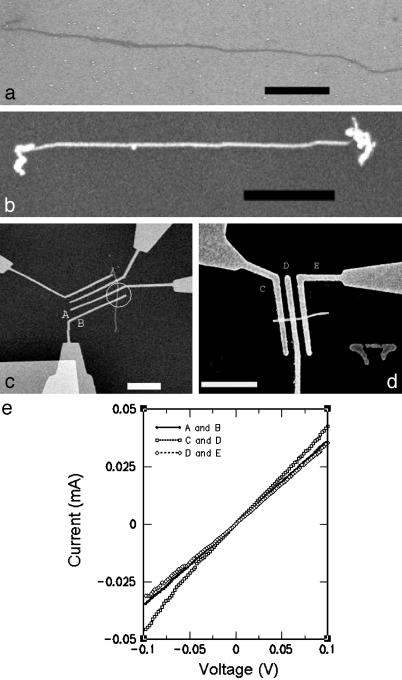
Templating of Metallic Nanowires and Conductivity Measurements. We have metallized the TX nanotubes with silver by using a two-step procedure. The resulting nanowires have been characterized by AFM and SEM (Fig. 5). The metallized nanowires have heights of ≈35 nm, widths of ≈40 nm, and lengths of up to ≈5 μm. We have patterned chromium/gold electrodes onto the wires by electron-beam lithography (5 nm of chromium followed by 30 nm of gold). SEM images of nanowires with attached electrodes are shown in Fig. 5 c and d. Two-terminal current–voltage (I–V) measurements were conducted on these devices (Fig. 5e) with various gap distances: 180, 80, and 100 nm, between electrodes A and B, C and D, and D and E, respectively. The I–V curves show mostly linear (ohmic) behavior and give resistances of 2.80, 2.35, and 2.82 kΩ as measured at 0.1 V. Measurements were done at room temperature (300 K). These numbers correspond to bulk resistivities of ≈1.4, 3.2, and 3.1 × 10–5 Ω·m, respectively. The bulk resistivity of polycrystalline silver is significantly lower (1.6 × 10–8 Ω·m). The higher resistivities of our wires may be due to granularity in the silver structure and/or low-density silver packing within the TX nanotubes. These nanowires demonstrate much higher conductivity than previously reported dsDNA-templated silver nanowires (11), although their conductivity is slightly lower than reported for dsDNA-templated palladium nanowires (24) and slightly lower than a recent self-assembled DNA tile-templated silver nanowire (22). The silver nanowires reported here as well as the other tile-assembly-templated silver wire (22) are of much more uniform width and smooth appearance than the rather bumpy and grainy metallic nanowires templated on dsDNA. Also, although the tile-assembly templates (≈25 nm) are much wider than dsDNA (≈2 nm) the resulting nanowires are generally thinner (≈40 nm). These desirable properties may result from the ability of tile assemblies to bind relatively more glutaraldehyde in the first step of the metallization, which then results in smaller grain size, leading to complete fusion of the wires in the second step.
Fig. 5.
(a) SEM image of TAO nanotube on the silicon substrate before metallization. (b) SEM image of fully metallized TAO nanotube on silicon substrate. The brighter tone of the nanotube in b versus a indicates the increased ability of the metallized nanotube (nanowire) to absorb electrons. (c and d) SEM images showing metallized TAO nanotubes (circled in c), overlaid with chromium/gold electrodes patterned by electron-beam lithography and deposited by thermal vapor deposition. The electrodes are labeled A–E. (Scale bars = 1 μm.) (e) The current–voltage curves measured through the sections of TAO templated nanowire between the indicated electrodes.
Discussion
Our use of a programmable, self-assembling DNA nanostructures appears to be a promising method for positioning components for molecule electronics, providing a diverse set of programmable superstructures for targeted connections that can subsequently be metallized to form functional nanowires. Early DNA-templated metallic nanowires were made of silver and displayed multimegaohm resistance (11). Improved conductivities were observed for gold (25) and palladium nanowires (23). DNA templates have been used for the targeted deposition of other metals, including copper (26) and platinum (27). However, in all previous studies, AFM or EM examination revealed rough wires with significant graininess compared with the smooth, uniform nanowires described here. Previous studies have used only linear dsDNA, in contrast with the DNA nanotube templates used here and with a different DNA tile system we described recently (22). Templates based on simple dsDNA provide structures with limited diversity and less fine control of associations compared with the programmable superstructures described here.
The length scale of the DNA nanotubes is intermediate between the molecular world, where distances are measured in angstroms (10–10 meters), and the macro world, the lower end of which can be measured in micrometers (10–6 meters). One eventual goal of this line of basic research and materials development is the fabrication of reliable interconnects useful in bridging between these disparate length scales. The metallic nanowires reported here may prove to be useful in such future applications.
Acknowledgments
We thank Prof. Jie Liu (Duke University) for access to a Digital Instruments Multimode Nanoscope IIIa AFM, David Anderson (Duke University) for performing the platinum rotary shadowing, and Prof. Gleb Finkelstein (Duke University) for useful discussions of the electrical measurements. This work was supported by National Science Foundation Grants EIA-00-86015 (to J.H.R.) and EIA-0218376 (to T.H.L. and J.H.R.), Defense Advanced Research Projects Agency/Air Force Office of Scientific Research Grant F30602-01-2-0561 (to J.H.R.), and a grant from Taiko Denki, Ltd. (Tokyo) (to T.H.L. and J.H.R.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: dsDNA, double-stranded DNA; TX, triple-crossover; AFM, atomic force microscopy/microscope; TEM, transmission electron microscopy/microscope; SEM, scanning electron microscopy/microscope; TAO, three coplanar double-helical domains, strand exchange points antiparallel (strands reverse direction on crossing-over), and odd number of helical half-turns between crossover points.
References
- 1.Seeman, N. C. (1982) J. Theor. Biol. 99, 237–247. [DOI] [PubMed] [Google Scholar]
- 2.Seeman, N. C. (1999) Trends Biotechnol. 17, 437–443. [DOI] [PubMed] [Google Scholar]
- 3.LaBean, T. H. (2002) in Computational Biology and Genome Informatics, eds. Wang, J. T. L., Wu, C. H. & Wang, P. P. (World Scientific, Singapore), pp. 35–58.
- 4.Reif, J. H. (2002) Comput. Sci. Eng. Mag. 4, 32–41. [Google Scholar]
- 5.Csaki, A., Maubach, G., Born, D., Reichert, J. & Fritzsche, W. (2002) Single Mol. 3, 275–280. [Google Scholar]
- 6.Seeman, N. C. (2003) Nature 421, 427–431. [DOI] [PubMed] [Google Scholar]
- 7.Alivisatos, A. P., Johnsson, K. P., Peng, X., Wilson, T. E., Loweth, C. J., Bruchez, M. P., Jr., & Schultz, P. G. (1996) Nature 382, 609–611. [DOI] [PubMed] [Google Scholar]
- 8.Mirkin, C. A., Letsinger, R. L., Mucic, R. C. & Storhoff, J. J. (1996) Nature 382, 607–609. [DOI] [PubMed] [Google Scholar]
- 9.Mucic, R. C., Storhoff, J. J., Mirkin, C. A. & Letsinger, R. L. (1998) J. Am. Chem. Soc. 120, 12674–12675. [Google Scholar]
- 10.Mbindyo, J. K. N., Reiss, B. D., Martin, B. R., Keating, C. D., Natan, M. J. & Mallouk, T. E. (2001) Adv. Mater. 13, 249–254. [Google Scholar]
- 11.Braun, E., Eichen, Y., Sivan, U. & Ben-Yoseph, G. (1998) Nature 391, 775–778. [DOI] [PubMed] [Google Scholar]
- 12.Seeman, N. C., Chen, J.-H. & Kallenbach, N. R. (1989) Electrophoresis 10, 345–354. [DOI] [PubMed] [Google Scholar]
- 13.Holliday, R. (1964) Genet. Res. 5, 282–304. [Google Scholar]
- 14.Fu, T.-J. & Seeman, N. C. (1993) Biochemistry 32, 3211–3220. [DOI] [PubMed] [Google Scholar]
- 15.Li, X., Yang, X., Qi, J. & Seeman, N. C. (1996) J. Am. Chem. Soc. 118, 6131–6140. [Google Scholar]
- 16.LaBean, T. H., Yan, H., Kopatsch, J., Liu, F., Winfree, E., Reif, J. H. & Seeman, N. C. (2000) J. Am. Chem. Soc. 122, 1848–1860. [Google Scholar]
- 17.Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. (1998) Nature 394, 539–544. [DOI] [PubMed] [Google Scholar]
- 18.LaBean, T. H., Winfree, E. & Reif, J. H. (2000) in Proceedings of the Fifth DIMACS Workshop on DNA Based Computers, DIMACS Series in Discrete Mathematics and Theoretical Computer Science, eds. Winfree. E. & Gifford, D. K. (Am. Math. Soc., Providence, RI), Vol. 54, pp. 123–140. [Google Scholar]
- 19.Winfree, E., Yang, X. & Seeman, N. C. (1998) in Proceedings of the Second DIMACS Workshop on DNA Based Computers, DIMACS Series in Discrete Mathematics and Theoretical Computer Science, eds. Landweber, L. F. & Baum, E. B. (Am. Math. Soc., Providence, RI), Vol. 27, pp. 191–213. [Google Scholar]
- 20.Mao, C., LaBean, T. H., Reif, J. H. & Seeman, N. C. (2000) Nature 407, 493–496. [DOI] [PubMed] [Google Scholar]
- 21.Yan, H., Feng, L., LaBean, T. H. & Reif, J. H. (2003) J. Am. Chem. Soc. 125, 14246–14247. [DOI] [PubMed] [Google Scholar]
- 22.Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. (2003) Science 301, 1882–1884. [DOI] [PubMed] [Google Scholar]
- 23.Storm, A., van Noort, J., de Vries, S. & Dekker, C. (2001) Appl. Phys. Lett. 79, 3881–3883. [Google Scholar]
- 24.Richter, J., Mertig, M., Pompe, W., Monch, I. & Schackert, H. K. (2001) Appl. Phys. Lett. 78, 536–538. [Google Scholar]
- 25.Keren, K., Krueger, M., Gilad, R., Ben-Yoseph, G., Sivan, U. & Braun, E. (2002) Science 297, 72–75. [DOI] [PubMed] [Google Scholar]
- 26.Monson, C. F. & Woolley, A. T. (2003) Nano Lett. 3, 359–363. [Google Scholar]
- 27.Ford, W. E., Harnack, O., Yasuda, A. & Wessels, J. M. (2001) Adv. Mater. 13, 1793–1797. [Google Scholar]