Summary
PGC-1α is an inducible nuclear receptor coactivator with direct functions in both p300-mediated chromatin remodeling and Mediator-dependent transcription in vitro. Here we have employed the PPARγ- and TRα-activated brown adipose tissue-specific UCP-1 enhancer to investigate mechanistic aspects of PGC-1α function. We first demonstrate a cellular role for the PGC-1α interacting MED1 subunit of Mediator in UCP-1 induction, as well as the accumulation of TRα, PPARγ, PGC-1α and MED1 on the UCP-1 enhancer in brown adipocytes. We then use biochemical assays to show (i) that PGC-1α is recruited to the TRα-RXRα-UCP-1 enhancer complex through interaction of an N-terminal LXXLL domain with TRα, (ii) that MED1/Mediator displaces PGC-1α from TRα through LXXLL domain competition and (iii) that, upon loss of PGC-1α-TRα interactions, PGC-1α remains associated with the enhancer complex through an interaction between PGC-1α and MED1 C-terminal domains. These results indicate dynamic MED1-dependent PGC-1α interactions related to functions in both chromatin remodeling and the transition to subsequent transcription initiation.
Introduction
Nuclear receptors are broadly involved in many biological processes that include metabolism, development and reproduction (Mangelsdorf et al., 1995). As conventional DNA-binding transcriptional activators, their function on cognate target genes involves a number of interacting cofactors. These include coactivators, such as the p160 family, that recruit histone modifying factors, and other coactivators, such as the multisubunit Mediator, that act more directly on the general transcription machinery (Glass and Rosenfeld, 2000; Malik and Roeder, 2005; McKenna and O’Malley, 2002). Direct nuclear receptor interactions with these cofactors are generally ligand-dependent and generally involve interactions between nuclear receptor AF-2 domains and LXXLL motifs in the p160 proteins and in the MED1 subunit of the Mediator. The interactions of distinct coactivators, as well as corepressors, with the same nuclear receptor AF-2 domain predicts dynamic cofactor interactions during target gene activation.
Peroxisome proliferator-activated receptor gamma (PPARγ) coactivator-1α (PGC-1α) was originally identified as a PPARγ-interacting protein in brown adipose tissue (BAT) and shown to serve as a coactivator both for PPARγ-activated and for thyroid hormone receptor β (TRβ)-activated transcription of BAT-specific genes such as the uncoupling protein 1 (UCP-1) gene (Puigserver et al., 1998). Since its initial identification, PGC-1α has emerged as a general coactivator for many other nuclear receptors and proved to be an important regulator of a number of metabolic processes that include energy expenditure, mitochondrial biogenesis, and oxidative phosphorylation. In this regard, PGC-1α expression is strongly related to metabolic needs. Thus, it is highly expressed in tissues with high oxidative metabolism such as BAT, skeletal muscle, liver, kidney, heart and brain (Finck and Kelly, 2006; Knutti and Kralli, 2001; Lin et al., 2005; Puigserver and Spiegelman, 2003). It is strongly induced in BAT by cold exposure, which in turn switches on the adaptive thermogenic program through induction of the UCP-1 gene (Puigserver et al., 1998; Ricquier et al., 1986). As further examples, PGC-1α is induced in muscle in response to exercise and in liver and heart in response to fasting (Finck and Kelly, 2006; Knutti and Kralli, 2001; Spiegelman and Heinrich, 2004).
Mechanistically, PGC-1α shows ligand-independent interactions with some nuclear receptors, most notably PPARγ (Puigserver et al., 1998), but interacts with many nuclear receptors such as TRβ (Wu et al., 2002) and estrogen receptor α (ERα) (Tcherepanova et al., 2000) through its LXXLL motifs in a ligand-dependent manner and generally coactivates nuclear receptor-mediated transcription in a ligand-dependent manner. Although PGC-1α also has activator- and promoter-specific functions in RNA processing (Monsalve et al., 2000; Thijssen-Timmer et al., 2006), the main focus of mechanistic studies has been on its primary role as a transcriptional coactivator. In this regard, PGC-1α appears to act in at least two steps of transcriptional activation – an early chromatin remodeling step and a subsequent transcription initiation event. First, consistent with the demonstration of a physical interaction of PGC-1α with the p300 acetyltransferase (Puigserver et al., 1998), our previous studies showed a direct PGC-1α-mediated enhancement of PPARγ- and p300-dependent histone acetylation and transcription from chromatin templates in a reconstituted cell-free system (Wallberg et al., 2003). Second, following the demonstration of a direct interaction of PGC-1α, through a C-terminal domain, with the MED1 subunit of the Mediator, PGC-1α was shown to enhance PPARγ- and Mediator-dependent transcription from DNA templates in the absence of any histone modifying factors (Wallberg et al., 2003). Apart from demonstrating PGC-1α functions in both early chromatin remodeling and subsequent transcription events, these results suggested the possibility that PGC-1α might also facilitate transitions between these steps. Although the mechanism(s) involved in PGC-1α function through the Mediator remains to be elucidated, it must ultimately take into account the seeming paradox that PGC-1α and MED1/Mediator are both recruited to the promoter through interactions of resident LXXLL domains with the ligand-induced AF-2 domains of nuclear receptors.
The dramatic induction of the UCP-1 gene in BAT by PGC-1α provides a good model for further analysis of the mechanism(s) of action of PGC-1α on a specific target gene. Previous studies have implicated a distal 220-bp enhancer in UCP-1 expression in BAT (Kozak et al., 1994), identified functional binding sites for nuclear receptors (including PPARγ and TRα) and other factors in the enhancer (Cassard-Doulcier et al., 1994; del Mar Gonzalez-Barroso et al., 2000; Del Mar Gonzalez-Barroso et al., 2000; Sears et al., 1996) and shown recruitment of nuclear receptors (PPARs and ERRα) to the enhancer (Cao et al., 2004; Debevec et al., 2007). Importantly, and as mentioned above, PPARγ and TRβ were both shown to function synergistically with PGC-1α on the UCP1 enhancer (Puigserver et al., 1998). The physiological importance of the UCP-1/BAT model is underscored by the established role for BAT in thermogenesis and energy expenditure control in rodents (Gesta et al., 2007) and emerging evidence for substantial BAT stores in humans as well (Farmer, 2008; Nedergaard et al., 2007).
To further investigate the mechanisms involved in PGC-1α function on nuclear receptor- activated target genes, especially through the Mediator, we have employed the BAT-expressed UCP-1 gene as a model. Using cell-based assays, we first establish a role for a C-terminal PGC-1α interacting domain of MED1 in UCP-1 induction, as well as the accumulation of TRα, PPARγ, PGC-1α and MED1 on the UCP-1 enhancer in brown adipocytes. We then employ biochemical assays with purified factors to establish dynamic PGC-1α interactions with TRα-RXRα-enhancer complexes – first with TRα through the PGC-1α LXXLL motif and then, following competitive MED1/Mediator interactions with TRα, an enhanced binding that involves interactions of a PGC-1α C-terminal domain with TRα-bound MED1/Mediator. These results emphasize key roles for dynamic MED1 and PGC-1α interactions in the transition from chromatin remodeling to transcription initiation events in nuclear receptor function.
Results
MED1 C-terminus is required for UCP-1 induction in vivo
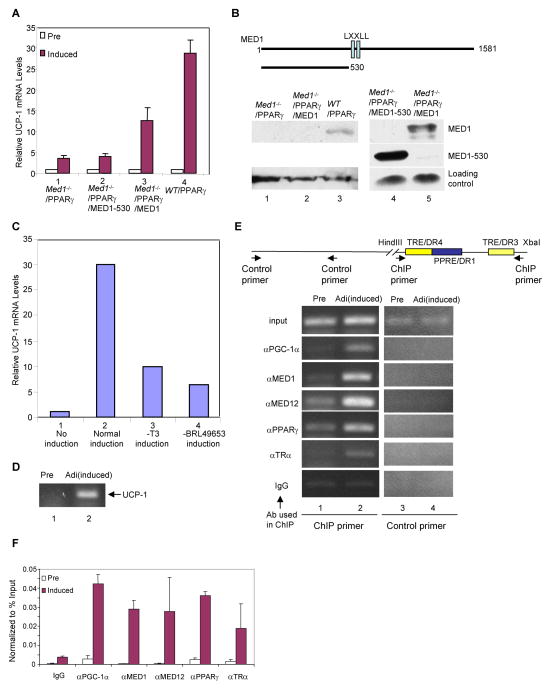
In previous studies with Med1−/− mouse embryonic fibroblasts (MEF), ectopic MED1 and PGC-1α were shown to function synergistically in activation of an ectopic PPARγ-RXRα driven reporter (Wallberg et al., 2003). In order to analyze the role of MED1 in activation of a natural PGC-1α target gene in a more physiological context, we have taken advantage of the ability to induce UCP-1 through the β-adrenergic pathway in in vitro differentiated adipocytes (Puigserver et al., 1998). To this end, we employed Med1−/− MEFs that stably express PPARγ and derived lines that stably express wild type MED1 or MED1 mutants (Ge et al., 2008). These cells were first cultured under conditions leading to differentiation to adipocytes, and then grown in the presence of insulin, a PPARγ agonist and thyroid hormone (T3) before induction of UCP-1 by addition of 9-cis-retinoid acid for 18 hours and 8-Br-cAMP for 6 hours. UCP-1 expression was then monitored by real-time RT-PCR analysis of extracted RNA. As shown in Figure 1A, induction of UCP-1 was greatly impaired in Med1−/−/PPARγ MEFs compared to wild type/PPARγ MEFs (lanes 1 and 4). However, since MED1 is required for MEF differentiation to adipocytes, it was necessary to rule out the possibility that the impaired induction of UCP-1 in Med1−/−/PPARγ MEFs results solely from the failure of MEFs to differentiate to adipocytes. To this end we compared the UCP-1 induction in Med1−/−/PPARγ MEFs that stably express either MED1 or a MED1 mutant (MED1(1–530)) lacking the C-terminus (Figure 1B top). Our previous results showed that both of these cell lines fully differentiate to adipocytes (Ge et al., 2008).
Figure 1. Role of MED1 in UCP-1 induction.
(A). Requirement of the MED1 C-Terminus for UCP-1 induction in differentiated MEFs. WT/PPARγ MEFs, Med1−/−/PPARγ MEFs and Med1−/−/PPARγ MEFs that stably express MED1(1–530) (Med1−/−/PPARγ/MED1–530 MEFs) or full length MED1(Med1−/−/PPARγ/MED1 MEFs) were differentiated to adipocytes, treated to induce UCP-1 gene expression and analyzed for UCP-1 RNA levels by real time RT-PCR as described in Experimental Procedures. Open bars show control non-treated preadipocytes (Pre) and solid bars show UCP-1 induced adipocytes (Induced). Error bars indicate standard deviation of three independent experiments.
(B). Structure and expression levels of MED1 proteins in differentiated MEFs. Top: Schematic representation of Flag-tagged MED1 proteins that were stably expressed in Med1−/−/PPARγ MEFs. Bottom: Analysis of ectopic and endogenous MED1 expression levels by immunoblot with anti-MED1 (lanes 1–3) and anti-Flag (lanes 4 and 5) antibodies.
(C). UCP-1 induction by PPARγ and TR ligands in differentiated MEFs. WT/PPARγ MEFs were first cultured under conditions leading to differentiation to adipocytes and then in complete UCP-1 induction medium (Normal induction, lane 2), induction medium minus T3 (−T3 induction, lane 3) or induction medium minus BRL49653 (−BRL49653 induction, lane 4). UCP-1 expression was monitored by real-time RT-PCR. Non-treated preadipocytes (No induction) are shown in lane 1.
(D) Induction of UCP-1 in brown HIB1B cells detected by RT-PCR.
(E). Induced binding of factors to the UCP-1 enhancer in HIB1B cells. Top: Schematic representation of the UCP-1 enhancer and primers used for ChIP assays. Bottom: ChIP analysis of HIB1B preadipocytes and UCP-1-induced adipocytes with the indicated antibodies.
(F). Induced binding of factors to the UCP-1 enhancer in HIB1B cells. Real time PCR ChIP analyses of preadipocytes and UCP-1 induced adipocytes with the indicated antibodies. Error bars indicate standard deviation of two independent experiments.
As shown in Figure 1A, reintroduction of MED1 to Med1−/−/PPARγ MEFs resulted in significant induction of UCP-1 gene expression (lane 3 versus lane 1). In contrast, reintroduction of MED1(1–530) to Med1−/−/PPARγ MEFs, which leads to normal differentiation to adipocytes, did not rescue UCP-1 induction at all (Figure 1A, lane 2). These results do not reflect effects of differential expression of the ectopic Flag-tagged MED1 proteins as anti-Flag immunoblots revealed that ectopic MED1(1–530) was expressed to a much greater level than ectopic MED1 (Figure 1B lane 4 versus lane 5). In this regard, an anti-MED1 immunoblot revealed that ectopic MED1 was espressed at a very low (nearly undetectable) level relative to that of endogenous MED1 in WT/PPARγ cells (Figure 1B, lane 2 versus 3), which may account for the failure of ectopic MED1 to elicit a level of UCP-1 expression equal to that observed in WT/PPARγ cells. The results also do not reflect differences in the levels of PGC-1α, which was induced to comparable levels in wild type/PPARγ and Med1−/−/PPARγ MEFs (data not shown). Omission of either the TR ligand (T3) or the PPARγ ligand (BRL49653) from the induction medium also resulted in a significant reduction of UCP-1 induction (Figure 1C, compare lanes 3 and 4 to lane 2), indicating roles for both TRα and PPARγ in regulating UCP-1 gene expression.
Overall, these results clearly implicate the MED1 C-terminus (and thus MED1) in UCP-1 induction and, along with prior demonstrations of a role for PGC-1α in UCP-1 induction in adipocytes (Lin et al., 2004), suggest that MED1 and PGC-1α may jointly regulate UCP-1 expression through TRα and/or PPARγ.
MED1 and PGC-1α are recruited to the UCP-1 enhancer in brown adipocytes
As a further test for MED1 and PGC-1α function (through TRα and/or PPARγ) in UCP-1 expression in the natural in vivo context, we employed ChIP assays to monitor factor recruitment to the UCP-1 enhancer in both HIB1B preadipocytes and in differentiated HIB1B adipocytes following UCP-1 induction by a β-adrenergic agonist. As analyzed by RT-PCR, expression of UCP-1 is greatly induced in adipocytes after β-adrenergic agonist treatment (Figure 1D). The 220-bp UCP-1 enhancer contains several nuclear receptor binding sites, including a PPRE/DR1 and two potential TREs (TRE/DR3 and TRE/DR4) (Figure 1E, upper panel). As shown qualitatively in Figure 1E (lower panel) and quantitatively in Figure 1F, differentiated, induced adipocytes show significantly elevated levels of TRα, PPARγ, PGC-1α and components of the Mediator components (MED1, MED12) on the UCP-1 enhancer relative to the levels observed in preadipocytes. These results further indicate that the Mediator and PGC-1α function through TRα and/or PPARγ on the UCP-1 enhancer in brown adipocytes.
PPARγ and TRα are simultaneously recruited to the UCP-1 enhancer
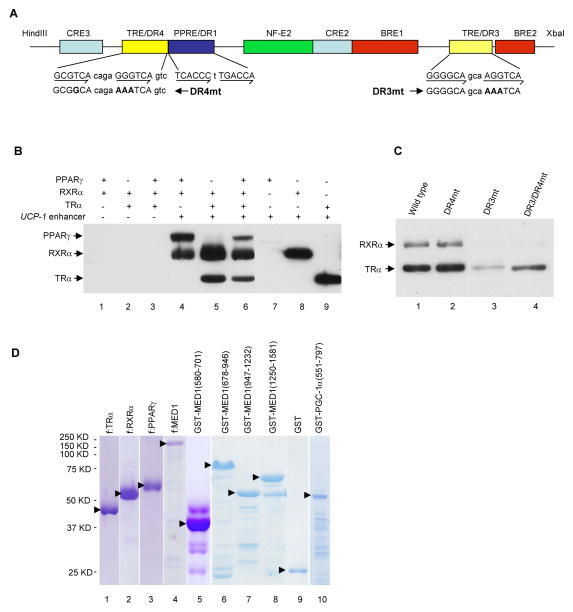
To investigate the biochemical mechanisms of how MED1 and PGC-1α regulate UCP-1 expression, we analyzed their recruitment to the 220-bp UCP-1 enhancer using an immobilized template assay. In this assay, the 220-bp UCP-1 enhancer (Figure 2A) is immobilized on Dynabeads and incubated with purified factors (Figure 2D). After washing, bound proteins are eluted and analyzed by immunoblot. As expected, PPARγ plus RXRα and TRα plus RXRα are independently recruited to the UCP-1 enhancer in a ligand-independent manner (Figure 2B lanes 4 and 5). PPARγ, TRα and RXRα are also jointly recruited to the UCP-1 enhancer (Figure 2B, lane 6). TRα and RXRα, but not PPARγ, also show independent binding, possibly as monomers, to the enhancer in this assay (Figure 2B, lanes 7–9). As controls, none of these receptors bound to control Dynabeads lacking the enhancer (Figure 2B, lanes 1–3). To determine which of the two potential TR binding sites on the UCP-1 enhancer recruits TRα-RXRα, mutations were made in the TRE/DR3 region (DR3mt), in the TRE/DR4 region (DR4mt) or in both regions (DR3/DR4mt) (Figure 2A) and their effects on recruitment of TRα-RXRα were tested. As shown in Figure 2C, DR3mt greatly diminished the recruitment of TRα-RXRα while DR4mt had no effect on TRα-RXRα binding. This indicates that the TRE/DR3 site is responsible for TRα-RXRα recruitment to the UCP-1 enhancer. In light of these results and prior indications (Introduction) of TRα (or TRβ) functions on the UCP-1 enhancer, the following studies have focused on TRα-RXRα-mediated recruitment of cofactors MED1 and PGC-1α.
Figure 2. In Vitro Recruitment of TRα, RXRα and PPARγ to the UCP-1 enhancer.
(A). Schematic representation of the UCP-1 enhancer showing the positions of the different elements and the mutated DR4 and DR3 elements.
(B). Independent and joint recruitment of purified PPARγ, RXRα and TRα to the UCP-1 enhancer. Standard immobilized template recruitment assays with proteins added as indicated were carried out as described in Experimental Procedures. Bound proteins in B and C were detected by immunoblot.
(C). Binding of TRα-RXRα to the DR3 enhancer element. Recruitment assays were carried out with TRα, RXRα and either wild type or mutant enhancers (shown in A) as described in Experimental Procedures.
(D). Purified Flag-tagged proteins from sf9 cells (lanes 1–4) and GST-fusion proteins from bacteria (lanes 5–10) analyzed by SDS-PAGE with Coomassie blue staining. Solid arrowheads indicate the corresponding full length proteins.
Both PGC-1α and MED1 are independently recruited to the UCP-1 enhancer in a ligand-dependent manner
Knowing that TRα-RXRα is efficiently recruited to the UCP-1 enhancer, we then tested if bound TRα-RXRα efficiently recruits purified MED1 or PGC-1α (Figure 2D and Figure 5A). As shown in Figure 3A, PGC-1α was recruited to the UCP-1 enhancer in a TRα-RXRα- and T3-dependent manner. A similar assay with MED1 showed that full length MED1 and a MED1(580–701) fragment containing the two LXXLL motifs are both recruited to the UCP-1 enhancer in a TRα-RXRα- and T3-dependent manner (Figure 3B, lanes 2 and 4; Figure 3C, lanes 2 and 4). These results are consistent with previous demonstrations of direct ligand-enhanced interactions of TRα with MED1 (Yuan et al., 1998) and of TRβ with PGC-1α (Wu et al., 2002; Puigserver et al., 1998) in the absence of DNA. It is noteworthy that whereas these latter studies observed weak interactions of TRβ with PGC-1α in the absence of T3, we observed exclusive ligand-dependent recruitment of PGC-1α to TRα-RXRα on the UCP-1 enhancer. This suggests that our recruitment assays with the natural TRα-RXRα-enhancer complex provide more selective binding conditions for protein-protein interactions than do GST-pull down assays. Moreover, given these results and the ability of TRα alone to bind the UCP-1 enhancer, it also is noteworthy that TRα or RXRα alone fails to recruit either MED1(580–701) or PGC-1α to the enhancer in the presence T3 (Figure 3C, lanes 6–9). This may be due to possible conformational changes of the TRα-RXRα heterodimer upon binding to enhancer sites and suggests that our recruitment assays with the immobilized enhancer more closely approximate the physiological situation.
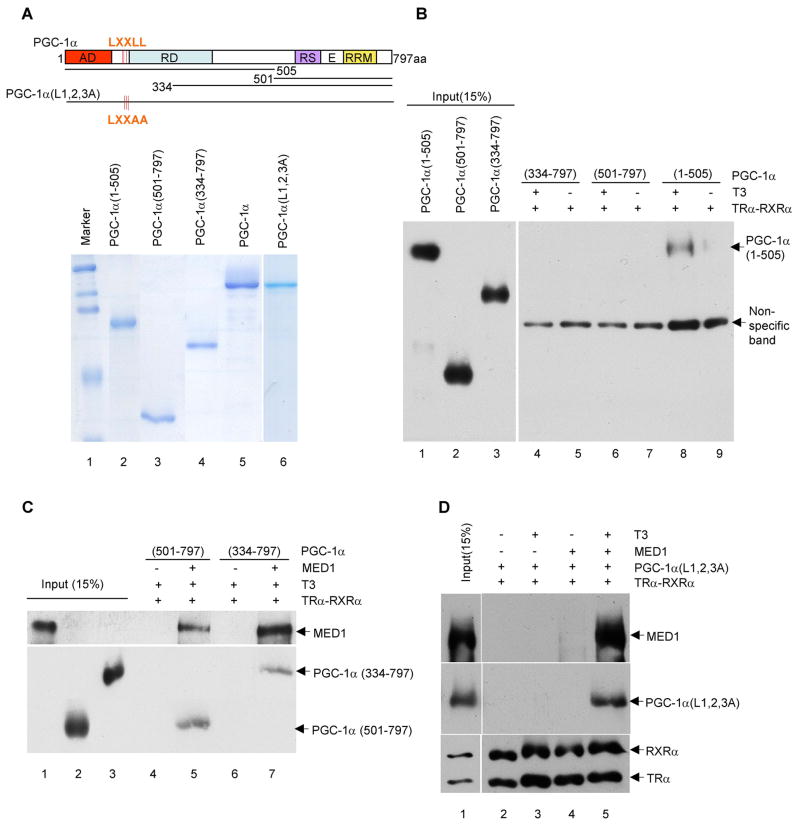
Figure 5. MED1-dependent recruitment of the PGC-1α C-terminus and PGC-1α LXXLL mutants to the TRα-RXRα-UCP-1 enhancer complex.
In panels B, C and D, standard immobilized template recruitment assays with purified proteins and T3 added as indicated were carried out as described in Experimental Procedures and bound proteins were detected by immunoblot.
(A). Wild type and mutant PGC-1α proteins. The upper panel shows a schematic representation of the full length, truncated and triple LXXLL-mutated proteins. The lower panel shows an SDS-PAGE analysis (with Coomassie blue staining) of the purified His-tagged proteins.
(B). Selective T3-dependent binding of an N-terminal PGC-1α fragment to the TRα-RXRα-UCP-1 enhancer complex.
(C). MED1-dependent binding of C-terminal PGC-1α fragments to the TRα-RXRα-UCP-1 enhancer complex.
(D). MED1- and T3-dependent, LXXLL motif-independent binding of full length PGC-1α to the TRα-RXRα-UCP-1 enhancer complex.
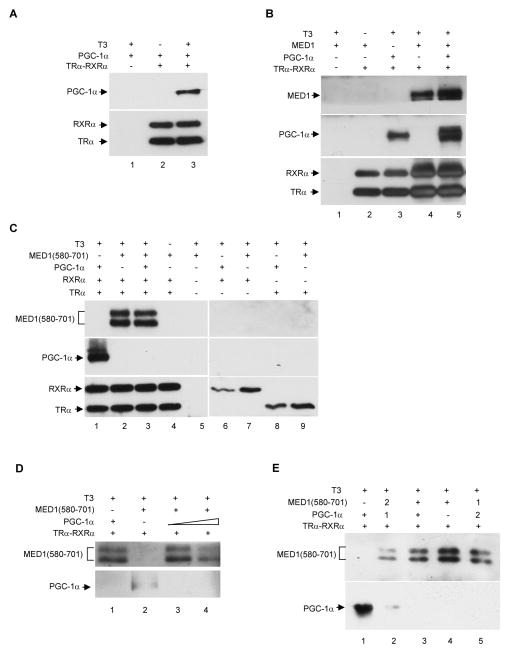
Figure 3. Independent and cooperative binding of PGC-1α and MED1 to the UCP-1 enhancer dependent upon TRα-RXRα and thyroid hormone.
Standard recruitment assays with proteins and T3 added as indicated were carried out as described in Experimental Procedures and bound proteins were detected by immunoblot.
(A). MED1-independent, T3-dependent binding of PGC-1α to the TRα-RXRα-UCP-1 enhancer complex.
(B). PGC-1α independent binding of MED1, as well as cooperative binding of intact MED1 and PGC-1α, to the TRα-RXRα-UCP-1 enhancer complex.
(C) and (D). Competitive binding of MED1(580–701) and PGC-1α to the TRα-RXRα-UCP-1 enhancer complex. All factors were added concommitantly to the immobilized DNA template. as indicated. In panel D the amount of PGC-1α in lane 4 was 10 times the amount in lane 3 and in the analyses in panel C
(E). Active displacement of prebound PGC-1α from the TRα-RXRα-UCP-1 enhancer complex by MED1(580–701). In lane 2 and 5, “1” and “2” indicate the order of addition of the factors to the preformed TRα-RXRα-UCP-1 enhancer complex. Factor 1 was added first and incubated for 30 minutes prior to addition of factor 2 and incubation for another 30 minutes
PGC-1α recruitment to the UCP-1 enhancer is further stabilized through binding to the C-terminus of MED1
As suggested by the ligand-dependent binding of MED1 and PGC-1α to TRα/TRβ, studies with mutant proteins have established that MED1 and PGC-1α interactions with TRs involve interactions of MED1 and PGC-1α LXXLL domains with the TRα or TRβ AF2 domains (Ren et al., 2000; Wu et al., 2002). These results raised the possibility of competitive PGC-1α and MED1 interactions with TRα, although our previous demonstration of direct PGC-1α-MED1 interactions also raised the possibility of a concomitant nuclear receptor-PGC-1α-MED1/Mediator interaction that might contribute to the observed functional synergy between PGC-1α and Mediator.
To further investigate PGC-1α, MED1 and TRα-RXRα interactions on the UCP-1 enhancer, we first tested whether PGC-1α and MED1(580–701), which contains the MED1 LXXLL motifs and previously was found to bind to the PGC-1α C-terminus, can be simultaneously recruited to the TRα-RXRα-enhancer complex. As shown in Figure 3C, and under conditions where PGC-1α and MED1 (580–701) both are independently recruited to the TRα-RXRα-enhancer complex (lanes 1 and 2), the joint addition of PGC-1α and MED1(580–701) to the assay resulted in the exclusive recruitment of MED1(580–701) to the TRα-RXRα-enhancer complex (lane 3). The same result was observed at a 10-fold higher level of PGC-1α (Figure 3D). These results indicate competitive binding of PGC-1α and MED1(580–701) to the same or overlapping sites on TRα. However, and to our surprise, replacement of MED1(580–701) with full length MED1 in the same assay with PGC-1α resulted in a greatly increased recruitment of PGC-1α and a moderately increased recruitment of MED1 to the UCP-1 enhancer (Figure 3B, lane 5 versus lanes 3 and 4). To clearly demonstrate an actual displacement of pre-bound PGC-1α by MED1 LXXLL motifs, competition recruitment assays with reciprocal ordered additions of PGC-1α and MED1(580–701) were performed. As shown in Figure 3E, MED1(580–701) displaced prebound PGC-1α on the TRα-RXRα-enhancer complex (lane 2 versus lane 1), whereas PGC-1α did not displace prebound MED1(580–701) from this complex (lane 5 versus lane 4). Altogether, these results strongly suggest that whereas the ligand-dependent interaction of PGC-1α with TRα-RXRα may be disrupted by a stronger ligand-dependent interaction of the central LXXLL-containing MED1 domain with TRα-RXRα, PGC-1α may still bind to the MED1-TRα-RXRα-enhancer complex through interactions with other domains of MED1. The cooperative binding of PGC-1α and full-length MED1 to the UCP-1 enhancer also suggests that PGC-1α and MED1 form a joint complex with TRα and RXRα on the UCP-1 enhancer.
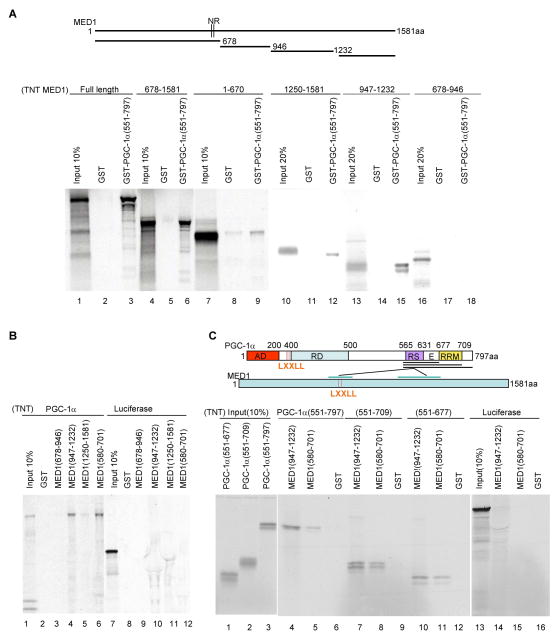
In a previous report, we demonstrated that a C-terminal fragment (residues 551–797) of PGC-1α can interact strongly with MED1 and more weakly with fragments containing the LXXLL motifs but lacking the C-terminal half of MED1 (Wallberg et al., 2003). To further map the MED1 domains that interact with PGC-1α, a purified GST-PGC-1α (551–797) fusion protein (Figure 2D) was immobilized on GST beads and tested for binding to in vitro translated, 35S-methionine-labeled MED1 fragments (Figure 4A upper panel). As shown in Figure 4A (lower panel), full length MED1, MED1(678–1581) and MED1(947–1232) proteins showed strong binding to GST-PGC-1α (551–797) but not to GST alone. Complementary binding studies with in vitro translated PGC-1α and purified GST-MED1 fusion proteins (Figure 2D) confirmed the binding of PGC-1α to MED1(947–1232) and, in agreement with earlier results (Wallberg et al., 2003), to MED1(580–701) (Figure 4B).
Figure 4. Interactions between PGC-1α domains and MED1 domains.
(A). Interactions of MED1 and MED1 fragments with the PGC-1α C-terminus. Bead-immobilized GST or GST-PGC-1α (551–797) fusion proteins were incubated with in vitro translated, 35S-labeled full length MED1 or MED1 fragments and bound proteins were analyzed by SDS-PAGE and autoradiography. The upper panel depicts MED1 and the derived fragments.
(B) Interactions of MED1 and MED1 fragments with full length PGC-1α. Bead-immobilized GST or indicated GST-fusion proteins were incubated with in vitro translated, 35S-labeled PGC-1α (lanes 1–6) or a luciferase control (lanes 7–12) and bound proteins were analyzed by SDS-PAGE and autoradiography.
(C). Interactions of the PGC-1α RS/E domain with MED1. Bead-immobilized GST, GST-MED1(580–701) or GST-MED1(947–1232) fusion proteins were incubated with the indicated in vitro translated, 35S-labeled PGC-1α fragments (lanes 4–12) or a luciferase control (lanes 13–16) and bound proteins were analyzed by SDS-PAGE and autoradiography. The upper panel summarizes the observed interactions of the PGC-1α RS/E domain with an LXXLL-containing MED1 domain (weaker) and a more C-terminal MED1 domain (stronger).
To map the MED1 interaction domains on PGC-1α, purified GST-MED1(580–701) and GST-MED1(947–1232) fusion proteins (Figure 2D) were tested for binding of three successively truncated in vitro translated C-terminal fragments of PGC-1α (Figure 4C, upper panel). As shown in the lower panel of Figure 4C, the smallest tested PGC-1α fragment, comprising residues 551–677 and containing the RS and E domains of the PGC-1α, retained full binding capacity to the two MED1 domains. The analysis in Figure 4C, in agreement with that in Figure 4A, also shows that the PGC-1α fragment binds more strongly to MED1(947–1232) than to MED1(580–701). These results, summarized in the upper panel of Figure 4C, thus establish a strong interaction between C-terminal domains in MED1 and PGC-1α.
To further investigate whether PGC-1α is recruited to the TRα-RXRα-UCP-1 enhancer complex through a MED1 interaction that is independent of the PGC-1α LXXLL-TRα AF2 interaction, His-tagged PGC-1α (1–505), PGC-1α (334–797) and PGC-1α (501–797) fragments were expressed and purified (Figure 5A). PGC-1α (1–505) contains the known TR AF2 interaction domain but not the MED1 interaction domain, whereas PGC-1α (334–797) and PGC-1α (501–797) contain the MED1 interaction domain but not the TR interaction domain (Figure 5A). As expected, only PGC-1α (1–505) was recruited to the UCP-1 enhancer by TRα-RXRα, in a T3-dependent manner, while PGC-1α (334–797) and PGC-1α (501–797) were not (Figure 5B).
However, when full length MED1 was present, both PGC-1α (334–797) and PGC-1α (501–797) were recruited to the TRα-RXRα-UCP-1 enhancer complex (Figure 5C). These results indicate that PGC-1α can be recruited to the UCP-1 enhancer both through binding of its N-terminal LXXLL domains to TRα when MED1 is absent, and through binding of its C-terminal domain to the MED1 C-terminus when MED1 is present. To further confirm this conclusion, a mutant PGC-1α protein (PGC-1α (L1,2,3A)) in which three LXXAA motifs replace three LXXLL motifs was purified (Figure 5A) and tested in the immobilized template assay. In sharp contrast to wild type PGC-1α (Figure 3A), PGC-1α (L1,2,3A) was no longer recruited to the TRα-RXRα-UCP-1 enhancer complex in the absence of MED1 (Figure 5D, lanes 2 and 3). However, in the presence of MED1, PGC-1α (L1,2,3A) and MED1 were jointly recruited to the TRα-RXRα-UCP-1 enhancer in a ligand-dependent manner (Figure 5D, lanes 4 and 5). These results further confirm that, in the absence of MED1, the LXXLL domains of PGC-1α are responsible for its recruitment to the UCP-1 enhancer through binding to TRα, whereas, in the presence of MED1, its C-terminal domain is responsible for its recruitment to the TRα-RXRα-UCP-1 enhancer through binding to MED1. Thus, our results strongly suggest a dynamic recruitment of PGC-1α to the UCP-1 enhancer, first with TRα-RXRα through a PGC-1α N-terminal LXXLL domain and then, following displacement by MED1 LXXLL domains, with receptor-bound MED1 through a PGC-1α C-terminal domain that interacts with the C-terminus of MED1.
PGC-1α and the Mediator bind cooperatively to the TRα-RXRα-UCP-1 enhancer complex through interactions of PGC-1α and MED1 C-terminal domains
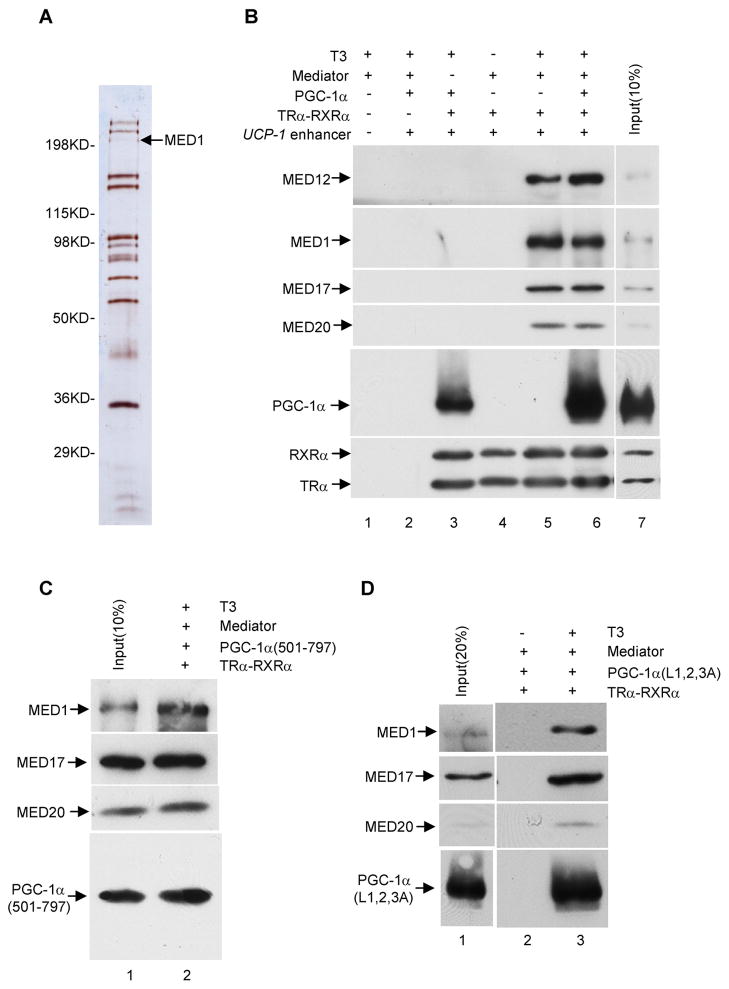
Since MED1 normally functions as part of the multisubunit Mediator complex (Malik et al., 2004), which has been shown to function cooperatively with PGC-1α, it is important to show that the above-described MED1-PGC-1α interactions also occur in the context of the natural Mediator. To this end, the Mediator was purified from Hela cells that stably express a Flag-tagged Nut2 subunit (Figure 6A) and employed in immobilized template assays. Like MED1, the purified Mediator showed ligand-dependent recruitment to the UCP-1 enhancer in the presence of TRα and RXRα (Figure 6B, lanes 4 and 5). As observed for PGC-1α and MED1, co-incubation of PGC-1α and purified Mediator resulted in their joint recruitment to the TRα-RXRα-UCP-1 enhancer complex in a T3-dependent manner; and recruitment of PGC-1α was significantly enhanced by the recruitment of Mediator (Figure 6B, lanes 3 and 6). This result strongly suggests that PGC-1α forms a complex with the Mediator on the UCP-1 enhancer and that this complex further stabilizes PGC-1α binding -- presumably, on the basis of earlier experiments, through interactions of the C-terminus of PGC-1α and the MED1 Mediator subunit. In further support of this hypothesis, PGC-1α (501–797), PGC-1α (334–797) and the PGC-1α (L1,2,3A) LXXLL motif mutant were jointly recruited with the Mediator, in a ligand-dependent manner, to the TRα-RXRα-enhancer complex (Figure 6C and 6D; data not shown). Thus, the PGC-1α-Mediator interactions observed with the TRα-RXRα-enhancer complex parallel the observed PGC-1α-MED1 interactions, and lead to a model for the role of PGC-1α in both chromatin remodeling and transcription.
Figure 6. Mediator-dependent binding of PGC-1α and PGC-1α mutants to the TRα-RXRα-UCP-1 enhancer complex.
In panels B, C and D, standard immobilized template recruitment assays with purified proteins and T3 added as indicated were carried out as described in Experimental Procedures and bound proteins were detected by immunoblot.
(A). Purified Mediator complex analyzed by SDS-PAGE with silver staining.
(B). Mediator-enhanced binding of full length PGC-1α to the TRα-RXRα-UCP-1 enhancer complex.
(C). Joint binding of Mediator and the PGC-1α C-terminus to the TRα-RXRα-UCP-1 enhancer complex.
(D). Joint binding of Mediator and triple LXXLL-mutated PGC-1α to the TRα-RXRα-UCP-1 enhancer complex.
Discussion
PGC-1α and the related PGC-1β and PRC have emerged as remarkably important and versatile transcriptional coactivators, acting primarily through nuclear receptors but also through other activators to mediate a broad range of physiological processes; and a number of target genes, as well as pathways leading to the induction and regulation of PGC-1α, have been identified. (Finck and Kelly, 2006; Lin et al., 2005; Lin, 2009; Yang et al., 2007). However, beyond clear demonstrations of direct PGC-1α-nuclear receptor/activator interactions, there is comparatively less information on the downstream mechanisms of action of PGC-1α on specific target genes and, in most cases, no evidence that PGC-1α acts directly on the identified target genes. Here we have used the thyroid hormone-induced BAT-specific UCP-1 gene as a model to investigate the mechanism of action of PGC-1α. Cell-based assays have established a role for the PGC-1α-interacting MED1 Mediator subunit in UCP-1 induction in BAT. Further biochemical assays have established (i) a strong interaction between C-terminal domains in MED1 and PGC-1α, (ii) a ligand-dependent PGC-1α interaction with a TRα-RXRα enhancer complex that is disrupted by a stronger ligand-dependent MED/Mediator interaction with the TRα-RXRα-enhancer complex and (iii) a persistent, Mediator-enhanced association of PGC-1α with the Mediator-TRα-RXRα-enhancer complex. These results indicate dynamic TRα, PGC-1α, and Mediator interactions on the UCP-1 enhancer that are proposed to facilitate the transition, mediated by PGC-1α functions in each step, between early chromatin remodeling and subsequent transcription initiation steps. It is anticipated that the results reported here will be of general significance for other PGC-1α regulated target genes and possibly for other coactivators.
Joint PGC-1α and MED1 function at the UCP-1 enhancer in adipocytes
UCP-1 is expressed exclusively in BAT and plays a major role in the adaptive thermogenesis response in this tissue (Puigserver et al., 1998; Ricquier et al., 1986). Expression of the UCP-1 gene is regulated through a 220-bp enhancer that has functional binding sites for a number of factors that include TRα/TRβ and PPARγ (del Mar Gonzalez-Barroso et al., 2000; Kozak et al., 1994; Rabelo et al., 1996). In support of earlier studies indicating coactivation of PPARγ and TRβ on the UCP-1 enhancer in reporter-based transfection assays (Puigserver et al., 1998), studies in Pgc-1α−/− mice have clearly shown an involvement of PGC-1α in UCP-1 induction (Lin et al., 2004). Here, we have employed Med1−/− fibroblasts, complemented with MED1 mutants, to show that UCP-1 induction in fibroblast-derived adipocytes (Puigserver et al., 1998) is dependent upon the C-terminus of MED1. Our previous studies showed that this region, in contrast to the conserved N-terminus, is dispensable for PGC-1α independent functions of TRα and PPARγ in vitro, for incorporation of MED1 into the Mediator complex, and for fibroblast differentiation into adipocytes (Ge et al., 2008; Ge et al., 2002; Malik et al., 2004). This requirement for the MED1 C-terminus for PGC-1α function in the UCP-1 enhancer is consistent with our demonstration that it provides a major docking site for PGC-1α and enhances PGC-1α recruitment to the enhancer (discussed below).
As cell-based functional assays do not establish direct effects of factors on target genes, it is important to establish this by other means. In support of direct functions for PGC-1α and MED1/Mediator on the UCP-1 enhancer, our ChIP assays have shown, for the first time, that UCP-1 induction in brown adipocytes is accompanied by an enhanced association of PGC-1α and MED1/Mediator, as well as TRα and PPARγ, with the UCP-1 enhancer. Of note, other recent studies have shown associations of PGC-1α with several different nuclear receptor-activated target genes (Guan et al., 2005; Li et al., 2008).
Ligand-dependent interaction of PGC-1α with the TRα-RXRα-enhancer complex as a basis for recruitment of chromatin remodeling factors
Beyond direct binding of TRα/β to UCP-1 enhancer elements (Rabelo et al., 1996), previous studies have established ligand-dependent binding of PGC-1α to TRβ, as well as PGC-1α coactivation of TRβ on transfected genes, dependent upon PGC-1α LXXLL and TRβ AF2 domains (Wu et al., 2002). As an extension of this work, we show (MED1-sensitive) PGC-1α binding to a natural TRα-RXRα-UCP-1 enhancer complex dependent upon TRα ligand and the PGC-1α LXXLL domains. Given our previous demonstration that the PGC-1α N-terminus, with its three LXXLL domains, is needed for PGC-1α enhancement of p300 function but not for PGC-1α enhancement of Mediator function in vitro (Wallberg et al., 2003), we propose that the ligand-dependent association of PGC-1α with nuclear receptors such as TRα is involved in the recruitment and function of chromatin modifying/remodeling factors in early steps of gene activation. Studies showing PGC-1α interactions with p300 and SRC-1 (Puigserver et al., 1999), the BAF60 subunit of SWI/SNF complex (Li et al., 2008), the HCF1 subunit of SET1/MLL complexes (Lin et al., 2002) and the GCN5 subunit of the SAGA complex (Lerin et al., 2006) are consistent with this proposed PGC-1α function. In limited cases, recruitment of these factors to target genes has been shown to be ligand-dependent, although the cell-based assays do not allow the conclusion that these events are necessarily dependent upon direct ligand-induced nuclear receptor-PGC-1α interactions.
Also of interest is the question of whether PGC-1 and p160 coactivators, which both show ligand-dependent interactions with nuclear receptors and subsequent recruitment of chromatin modifying factors (various histone acetyl- and methyl-transferases), may in some cases have potentially redundant functions in recruiting downstream coactivators such as p300 (discussed in Wallberg et al., 2003). Some studies have indicated synergistic effects of p300 and p160/SRC-1 in coactivating Gal4-PGC-1 functions on reporter genes in transfection assays (Puigserver et al., 1998). However, and relevant to the present studies, a specific (E457A) point mutation on the TRβ AF2 domain was reported to affect p160/SRC-1 interaction and coactivator functions, but not PGC-1α interaction and coactivator functions (Wu et al., 2002).
MED1/Mediator-dependent recruitment of PGC-1α to the TRα-RXRα-UCP-1 enhancer complex
Consistent with our previous demonstration of a functional synergy between Mediator and PGC-1α in PPARγ-dependent transcription in a biochemically defined system (Wallberg et al., 2003), and given MED1 and PGC-1α dependent UCP-1 expression in adipocytes (above), we have demonstrated the ligand-dependent formation of a Mediator-PGC-1α-TRα-RXRα-complex on the native TRE/DR3 site of the UCP-1 enhancer. In this case, however, PGC-1α binding to the TRα-RXRα-enhancer complex is increased relative to the level observed in the absence of Mediator. This appears to be due to an interaction of the PGC-1α C-terminus (RS/E domain) with a C-terminal domain in MED1. This conclusion is supported by the following lines of evidence (i) PGC-1α also shows enhanced binding to a MED1-TRα-RXRα-enhancer complex, (ii) the C-terminus of PGC-1α, as well as a mutant PGC-1α with LXXLL domain mutations that eliminate ligand-dependent binding to TRα, shows both MED1- and Mediator-dependent binding to the TRα-RXRα-enhancer complex, (iii) MED1 and PGC-1α show a strong direct interaction through a C-terminal MED1 domain (947–1232) and a PGC-1α C-terminal domain that contains the RS and E domains, and (iv) a MED1 N-terminal fragment containing two MED1 LXXLL domains shows a strong T3-dependent binding to the PGC-1α-TRα-RXRα-enhancer complex with a concomitant displacement of PGC-1α. The latter result is consistent with previous studies indicating that PGC-1α and MED1 both interact, through LXXLL domains, with the TR AF2 domain (Ren et al., 2000; Wu et al., 2002; Yuan et al., 1998), but is also indicative of a stronger MED1-TRα interaction that is proposed (below) to facilitate the transition from chromatin remodeling to actual transcription. Competitive binding of a different set of coactivators (MED1 and TIF2) to TR has also been observed (Treuter et al., 1999), further indicative of sequential functions of receptor-interacting cofactors in gene activation.
A model for dynamic PGC-1α interactions that facilitate integrated functions in chromatin remodeling and transcription initiation
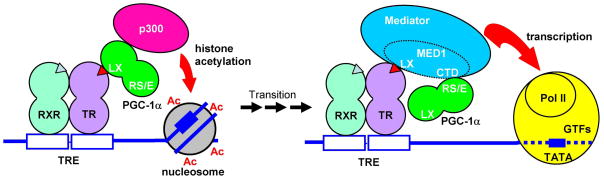
Given the key roles of PGC-1α and its paralogues (PGC-1β and PRC) in the activation of numerous target genes in diverse biological processes, it is increasingly important to understand the actual mechanisms by which PGC-1α functions. On the basis of our current enhancer binding assays, our UCP-1 induction assays in adipocytes, and our previous demonstration of direct PGC-1α functions in both p300-dependent and Mediator-dependent transcription (Wallberg et al., 2003), we propose the model shown in Figure 7 for dual PGC-1α functions, through a TRα-RXRα heterodimer, on the UCP-1 enhancer. The chief features are the following sequence of events: (i) an initial recruitment of PGC-1α through ligand-dependent interactions of LXXLL motifs with the AF2 domain of TRα, (ii) subsequent recruitment of p300, with the possible participation of SRC-1 (Puigserver et al., 1999), to initiate histone acetylation and other chromatin modifications that make the promoter more accessible to the general transcription machinery and the Mediator, (iii) interactions of the Mediator, through the LXXLL motifs in MED1, with the TRα AF2 domain and concomitant displacement of PGC-1α, (iv) a PGC-1α transition involving displacement from TRα followed by retention on the enhancer complex through interactions of the PGC-1α RS/E domains with the C-terminal domain of MED1 and (v) a stimulatory effect of Mediator-bound PGC-1α on formation and/or function of the preinitiation complex at the promoter. How PGC-1α might exert its effect through the Mediator is not yet understood. However, previous studies have shown a T3-mediated juxtaposition of a distal TRE-TRα-RXRα-Mediator complex with the promoter that may facilitate preinitiation complex formation/function (Park et al., 2005); and PGC-1α interactions with the Mediator complex could well influence such functions on the UCP-1 gene.
Figure 7. A model for dynamic PGC-1α interactions that facilitate integrated functions in chromatin remodeling and transcription initiation.
The following sequential steps are proposed. First, PGC-1α is recruited to a target nuclear receptor-enhancer complex through a ligand-dependent interaction of one of its N-terminal LXXLL motifs with the nuclear receptor AF2 domain. Second, binding of p300 (or another chromatin modifying factor) to PGC-1α leads to histone modifications (e.g., acetylation) and/or chromatin remodeling events that make the promoter/enhancer more accessible to Mediator and general transcription factors. Third, MED1/Mediator interactions, through LXXLL motifs, with the liganded nuclear receptor AF2 domain leads to PGC-1α displacement, concomitant with stabilizing interactions between C-terminal domains in PGC-1α and MED1 that result in persistent (or enhanced) PGC-1α recruitment. Fourth, PGC-1α enhances the Mediator-dependent formation and/or function of a preinitiation complex (and may ultimately be transferred to an elongating RNA polymerase II complex for specific RNA processing events). For further details see text.
Another possibility suggested by the model is that PGC-1α, by virtue of its function in both chromatin remodeling and transcription steps through distinct protein-protein interactions, may facilitate the transition between these steps. Although not tested here, but given that PGC-1α interactions with MED1, TRα/β and p300 (Puigserver et al., 1999; Wallberg et al., 2003; Wu et al., 2002) involve different domains in PGC-1α, the transition from chromatin remodeling to transcription could involve formation of a transition complex containing TRα, RXRα, PGC-1α, p300 and Mediator. Another key question, especially in light of the observation that MED1 binds more tightly than PGC-1α to liganded TRα on the enhancer complex, is what determines the order of factor interactions summarized in the model. It is likely that this relates in part to constraints imposed by chromatin structures, such that effective Mediator binding to the TRα-RXRα-enhancer complex depends upon prior PGC-1α-p300 binding and associated histone modifications and chromatin remodeling.
Experimental Procedures
Protein Expression and Purification
Flag-tagged TRα, PPARγ, RXRα, and MED1 were expressed using the Bac-to-Bac Baculovirus Expressing system (Invitrogen) according to the manufacturer’s instructions. Corresponding proteins then were affinity purified on M2-agarose. GST and GST-fusion proteins were expressed in bacteria and purified through binding to Glutathione-agarose beads. His-tagged full length, LXXLL mutant and truncated PGC-1α proteins were expressed in bacteria from the pET23 vector. The expressed PGC-1α proteins are largely insoluble and were solublized, renatured and purified. Briefly, bacteria expressing PGC-1α in pET23 vector were suspended in 20 mM Tris/400 mM NaCl (pH8.0) and disrupted by sonication. After centrifugation of the sonicate, the pellet was washed once with PBST (20 mM NaHPO4/NaH2PO4, pH 7.4, 150 mM NaCl, 0.05% Tween 20) and then dissolved in 6 M Urea in buffer A (20 mM NaHPO4/NaH2PO4, pH 7.4, 200 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol and 0.05% Tween20) by rocking at 4°C overnight. The lysate was then loaded onto a Ni-NTA column. The column was first washed with 6 M urea in buffer A and then with descending concentrations of urea (from 6 M, 3 M, 2 M, 1 M, 0.8 M, 0.6 M, 0.4 M to 0.2 M) in buffer A. The final wash step was performed with BC200 (equivalent to BC500 but with 200 mM KCl) containing 20 mM imidazole, 2 mM β-mercaptoethanol and 0.05% Tween 20. The bound protein was eluted with 400 mM imidazole in BC200 and 0.05% Tween 20. Finally, the purified protein was dialyzed against BC100 (equivalent to BC500 but with 100 mM KCl) with 0.05% Tween 20 and stored at −80°C. The Mediator complex was purified from nuclear extract of a Hela cell line that stably expresses Flag-tagged Nut2 by M2-agarose affinity purification (Malik et al., 2004).
In Vitro Protein Binding Assay
In vitro protein binding was performed with GST-fusion proteins (5 μg) immobilized on glutathione-Sepharose beads (Amersham Pharmacia) and in vitro-translated 35S-methionine-labeled proteins (TNT kit, Promega) as described (Malik et al., 2004).
Immobilized-template Protein Recruitment Assay
A UCP-1 enhancer fragment, containing DNA sequences extending from −2574 to −2374 of the mouse UCP-1 gene (gene bank number, gi:1519064), was obtained by PCR using biotinated oligonucleotides and purified using a Qiagen PCR purification kit. The biotinated DNA fragment was immobilized on Dynabeads M-280 streptavidin (Dynal Biotech) as instructed by the manufacturer. After incubating the beads in blocking buffer (50 mM Tris-HCl, pH 7.3, 100 mM KCl, 0.01% NP-40, 1 mg/ml BSA, 0.5 mM PMSF, 0.01 M DTT, 10 μg/ml of salmon sperm DNA and 1 μg/ml of poly dI-C) for 30 min at room temperature, purified proteins were then added to the reaction and incubated for 30 min at room temperature in the absence or presence of 10 μM T3 with gentle vortexing. The beads were then washed with wash buffer (50 mM Tris-HCl, pH 7.3, 100 mM KCl, 0.01% NP-40, 0.1 mg/ml BSA, 0.5 mM PMSF, 0.5 mM DTT) three times and the bound proteins were eluted by boiling in 2X SDS sample buffer and detected by immunoblot. Standard reactions in 100 μl contained 200 ng enhancer DNA, 3 μg beads and, when present, 20 ng TRα, 20 ng RXRα, 20 ng PPARγ, 200 ng PGC-1α, and 200 ng MED1 or 15 μl Mediator unless otherwise indicated in the Figure Legends.
Cell Culture
Wild type and derivitive MEFs and HIB1B cells were cultured in monolayer in DMEM with 10% fetal bovine serum. Prior to induction of UCP-1 expression, MEFs were differentiated to adipocytes as described (Ge et al., 2004). For subsequent induction of UCP-1 expression, cells were cultured for three days in 10% fetal bovine serum containing 5 μg/ml insulin, 0.5 μM BRL49653 and 100 nM T3. This was followed by addition of 9-cis-retinoic acid (1 μM final concentration) for 18 hr and then by addition of 8-Br-cAMP (1 μM final concentration) for 6 hr before RNA was extracted for real time RT-PCR. To induce UCP-1 expression in HIB1B cells, HIB1B cells were differentiated to adipocytes as described (Puigserver et al., 1998). To induce UCP-1 gene expression, 9-cis-RA (1 μM) was added for 18 hr followed by isoproterenol (1 μM) for 6 hr.
Chromatin Immunoprecipitation Assay
Cells were cross-linked with 1% formaldehyde at 37°C for 10 minutes, washed with cold phosphate-buffered saline and lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1 and 1x protease inhibitor). Lysates were sonicated and, after centrifugation, supernatents were diluted 1:10 in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1 and 1x protease inhibitor), incubated with protein A-Sepharose and 1 μg/ml salmon sperm DNA for 2 hrs at 4°C and then incubated with 2 μg of desired antibodies overnight at 4°C. The immunoprecipitates were collected by incubation with 45 μl protein A-Sepharose and 2 μg/ml salmon sperm DNA for 1 hr. The precipitates were washed sequentially with TSEI (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1 and 150 mM NaCl), TSEII (same as TSEI but with 500 mM NaCl) and buffer III (0.25 mM LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1) and then twice with TE buffer. The precipitates were incubated with elution buffer (1% SDS and 0.1 M NaHCO3) at room temperature for 1 hr and the eluates were then incubated at 65°C for 6 hr for cross-link reversal before being purified with a Qiagen PCR kit. 1/50 of the eluate was used for PCR analysis using the following primer set: forward, 5′-aagcttgctgtcactcctctac-3′; reverse, 5′-tctagagtctgaggaaaggg-3′. The following antibodies were used for ChIP: TRα and PPARγ (Santa Cruz Biotechnology); MED1, MED12 and PGC-1α (from laboratory stocks).
Real-time PCR
Total RNA was collected and equal amounts of total RNA were used for first strand cDNA synthesis using SuperScriptIII First Strand cDNA Synthesis for RT-PCR kit (Invitrogen) following the manufacturer’s instruction. RNA levels were analyzed by quantitative real-time PCR in 25 μl reactions with SYBR Green (Applied Biosystems). 18S RNA was used as an internal control for normalization. Fold-inductions are shown relative to control cells.
Acknowledgments
We thank Dr. Bruce Spiegelman for HIB1B cells and PGC-1α expression vectors and Dr. Anastasis Kralli for PGC-1α mutant expression vectors. This work was supported by National Institutes of Health National Research Service Award 5F32GM68272 to W.C. and by National Institutes of Health grant DK071900 to R.G.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, Collins S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. 2004;24:3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassard-Doulcier AM, Larose M, Matamala JC, Champigny O, Bouillaud F, Ricquier D. In vitro interactions between nuclear proteins and uncoupling protein gene promoter reveal several putative transactivating factors including Ets1, retinoid × receptor, thyroid hormone receptor, and a CACCC box-binding protein. J Biol Chem. 1994;269:24335–24342. [PubMed] [Google Scholar]
- Debevec D, Christian M, Morganstein D, Seth A, Herzog B, Parker M, White R. Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor alpha. Mol Endocrinol. 2007;21:1581–1592. doi: 10.1210/me.2007-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mar Gonzalez-Barroso M, Pecqueur C, Gelly C, Sanchis D, Alves-Guerra MC, Bouillaud F, Ricquier D, Cassard-Doulcier AM. Transcriptional activation of the human ucp1 gene in a rodent cell line. Synergism of retinoids, isoproterenol, and thiazolidinedione is mediated by a multipartite response element. J Biol Chem. 2000;275:31722–31732. doi: 10.1074/jbc.M001678200. [DOI] [PubMed] [Google Scholar]
- Del Mar Gonzalez-Barroso M, Ricquier D, Cassard-Doulcier AM. The human uncoupling protein-1 gene (UCP1): present status and perspectives in obesity research. Obes Rev. 2000;1:61–72. doi: 10.1046/j.1467-789x.2000.00009.x. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Molecular determinants of brown adipocyte formation and function. Genes Dev. 2008;22:1269–1275. doi: 10.1101/gad.1681308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol Cell Biol. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Guan HP, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 2005;19:453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- Kozak UC, Kopecky J, Teisinger J, Enerback S, Boyer B, Kozak LP. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol Cell Biol. 1994;14:59–67. doi: 10.1128/mcb.14.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li S, Liu C, Li N, Hao T, Han T, Hill DE, Vidal M, Lin JD. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta ), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin JD. Minireview: the PGC-1 coactivator networks: chromatin-remodeling and mitochondrial energy metabolism. Mol Endocrinol. 2009;23:2–10. doi: 10.1210/me.2008-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol Cell Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rabelo R, Reyes C, Schifman A, Silva JE. Interactions among receptors, thyroid hormone response elements, and ligands in the regulation of the rat uncoupling protein gene expression by thyroid hormone. Endocrinology. 1996;137:3478–3487. doi: 10.1210/endo.137.8.8754777. [DOI] [PubMed] [Google Scholar]
- Ren Y, Behre E, Ren Z, Zhang J, Wang Q, Fondell JD. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol Cell Biol. 2000;20:5433–5446. doi: 10.1128/mcb.20.15.5433-5446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricquier D, Bouillaud F, Toumelin P, Mory G, Bazin R, Arch J, Penicaud L. Expression of uncoupling protein mRNA in thermogenic or weakly thermogenic brown adipose tissue. Evidence for a rapid beta-adrenoreceptor-mediated and transcriptionally regulated step during activation of thermogenesis. J Biol Chem. 1986;261:13905–13910. [PubMed] [Google Scholar]
- Sears IB, MacGinnitie MA, Kovacs LG, Graves RA. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 1996;16:3410–3419. doi: 10.1128/mcb.16.7.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Tcherepanova I, Puigserver P, Norris JD, Spiegelman BM, McDonnell DP. Modulation of estrogen receptor-alpha transcriptional activity by the coactivator PGC-1. J Biol Chem. 2000;275:16302–16308. doi: 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- Thijssen-Timmer DC, Schiphorst MP, Kwakkel J, Emter R, Kralli A, Wiersinga WM, Bakker O. PGC-1alpha regulates the isoform mRNA ratio of the alternatively spliced thyroid hormone receptor alpha transcript. J Mol Endocrinol. 2006;37:251–257. doi: 10.1677/jme.1.01914. [DOI] [PubMed] [Google Scholar]
- Treuter E, Johansson L, Thomsen JS, Warnmark A, Leers J, Pelto-Huikko M, Sjoberg M, Wright AP, Spyrou G, Gustafsson JA. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- Wu Y, Delerive P, Chin WW, Burris TP. Requirement of helix 1 and the AF-2 domain of the thyroid hormone receptor for coactivation by PGC-1. J Biol Chem. 2002;277:8898–8905. doi: 10.1074/jbc.M110761200. [DOI] [PubMed] [Google Scholar]
- Yang X, Lamia KA, Evans RM. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]