Abstract
MLL5 is a mammalian trithorax group (trx-G) gene identified within chromosome band 7q22, a frequently deleted element found in cytogenetic aberrations of acute myeloid malignancies. MLL5 cDNA was linked with the FLAG and V5 tags at the N and C terminus, respectively, and transfected into 293T cells. Immunofluoresence staining of the expressed tagged MLL5 protein showed localization to the nucleus and exclusion from nucleoli, and no surface staining was detected. Both ectopically introduced and endogenous MLL5 protein displayed a speckled nuclear distribution. By using a series of MLL5-truncated mutants fused with enhanced GFP, a domain (residues 945–1,156) required for foci accumulation was identified, and regions containing functional nuclear localization signals were mapped. Ectopic overexpression of GFP-MLL5 induced cell cycle arrest in G1 phase. This inhibition of cell cycle progression was indicated by delayed progression into nocodazole-induced mitotic arrest and was confirmed by a lack of BrdUrd incorporation. These findings suggest that MLL5 forms intranuclear protein complexes that may play an important role in chromatin remodeling and cellular growth suppression.
The mixed lineage leukemia gene (MLL-1, also known as ALL-1, HRX, and Htrx) (1–4) is located on human chromosome 11q23 and is frequently involved in chromosomal translocations in acute lymphocytic leukemia, myelomonocytic leukemia, and leukemias arising secondary to DNA topoisomerase II inhibitor therapy (5–7). In some patients, MLL undergoes partial tandem duplication (8), but in the majority of cases, the C terminus of MLL is removed by chromosomal translocation and replaced in frame by a variety of different fusion partners.
MLL is homologous to Drosophila Trx and is thus a member of the trithorax group (trx-G) gene family, which together with the Polycomb group (Pc-G) genes control the expression of Hox and other developmental genes through the modulation of chromatin (9–11). MLL harbors a central domain consisting of four plant homeodomain (PHD) zinc fingers with a nested bromodomain and a C-terminal Su(var)3-9, enhancer-of-zeste, trithorax (SET) domain (12–14). Unlike Trx, MLL also contains three AT-hook motifs and a methyltransferase domain but lacks a nuclear receptor zinc finger domain (3). In structural function analysis, the AT-hooks and methyltransferase domain are invariably retained in leukemia-associated fusions and are essential for the full transforming ability of the MLL fusions (15–17).
Trx-G protein function is often associated with the chromatin-remodeling activity of SWI/SNF proteins. In fact, Brahma (BRM), a trx-G family gene, itself, encodes a SWI/SNF ATPase domain. BRM recruits more than seven other SWI/SNF proteins to form a 2-MDa complex that is found to associate with active chromatin (18–20). This complex is cell cycle-regulated, with reduced levels during mitosis and dephosphorylation causing binding to mitotic chromosomes (21, 22). Trithorax (TRX) and its mammalian homolog MLL recruits the BRM complex to specific DNA-binding elements (23, 24). This process is crucial for proper early embryogenesis (25). When MLL activity is altered by chromosomal translocation, the result is infantile leukemia (26). Efficient recruitment of these complexes depends on specific methyltransferase activity on histone H3 lysine 4, which recently has been shown to be an inherent activity of the SET domain of MLL (25, 26).
Four other mammalian genes have been identified in the MLL/TRX gene family. ALR (ALL-1-related gene) maps to chromosome bands 12q12–13, adjacent to a region involved in tumorigenic duplications and translocations (27). MLL2 maps to 19q13.1, a region of frequent rearrangement and amplification in solid tumors. Although MLL2 shares highly conserved domains with MLL, no leukemia-associated MLL2 fusions have been detected (28, 29). Loss of chromosome 7 or deletion of the long arm of chromosome 7 are the most recurrent abnormalities detected in myeloid malignancies. Recently, a scan of these deleted regions resulted in the identification of two unique MLL family genes, MLL3 and MLL5, which mapped to chromosomal bands 7q36 and 7q22, respectively (30, 31).
MLL5 is a relatively small protein (1,858 aa) compared with MLL (3,969 aa). In addition, MLL5 contains only a single PHD rather than a cluster of PHD fingers and its single SET domain is in the center of the protein rather than at the C terminus. Moreover, MLL5 possesses neither AT-hooks nor a methyltransferase homology domain. MLL5 is more distantly related to the human ALR and huASH1 genes and to the Drosophila gene ASH1 (31). MLL5 displays ubiquitous tissue expression and is highly conserved, showing a high degree of homology with its Drosophila counterpart within the PHD (80% identical and 88% similar) and the SET domain (38% identical and 47% similar). No mutations within the MLL5-coding region were detected in all primary leukemias screened (31).
We initially encountered an alternatively spliced version of MLL5 during a screen for genes involved in immune function (V. Vieillard, personal communication) and had studied the localization and function of the smaller protein that it encodes. In the course of this work, the form of MLL5 protein derived from its full-length cDNA was reported (31).
Functional characterization of full-length MLL5 protein by the use of epitope tagging, deletional scanning, and immunofluorescence studies is reported here. We show that MLL5 protein forms intranuclear foci and that overexpression of MLL5 inhibits cell cycle progression, suggesting a role for MLL5 in tumor suppression. The domain required for the formation of foci and the regions important for targeting nuclear localization were also identified.
Methods
DNA Preparation and Plasmid Construction. Total RNA of human peripheral blood lymphocytes, HeLa, Jurkat, COS7, and 721.221 was isolated by using TRIzol reagent (Invitrogen). The first strand of MLL5 cDNA was synthesized by using the SuperScript RT-PCR kit (Invitrogen). The PCR fragments were cloned into the pCRII vector (blunt TOPO TA vector, Invitrogen) and the inserts were subsequently sequenced.
The cDNA sequence encoding the MLL5 protein was fused to FLAG at the N terminus and was cloned into the pEF6-V5 mammalian expression vector (Invitrogen), which provides a V5 C-terminal fusion. To construct full-length and truncated forms of GFP-MLL5 fusion proteins, full-length and various PCR fragments of MLL5 were digested with EcoRI and BamHI and subcloned into the pEGFP-C1 expression vector (Clontech), resulting in N-terminal enhanced GFP fusions.
Cell Culture. The 293T cells were grown in RPMI medium 1640 supplemented with 10% FBS, 2 mM glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, and 400 μg/ml G418 at 37°C in a humidified atmosphere containing 5% CO2/95% air. For mitotic arrest, cells were treated with nocodazole at a final concentration of 400 ng/ml for 16 h, 48 h after transfection. Plasmids were transfected into 293T cells with FuGENE 6 reagent according to the manufacturer's instructions (Roche Molecular Biochemicals) at a 3:2 ratio (fugene/DNA) for full-length MLL5 and 3:1 ratio for truncated MLL5 mutants and mock vectors.
Generation of Anti-MLL5 Polyclonal Abs. Synthetic peptides corresponding to amino acids 227–241 and 801–815 of the MLL5 protein (31) were synthesized (Alpha Diagnostic International, San Antonio, TX) and coupled to keyhole limpet hemocyanin (Pierce) for immunization. Polyclonal sera against these peptides were affinity purified by using a Protein A spin column (Prochem, Acton, MA). Resulting polyclonal Abs were named αMLL5-1 and αMLL5-2.
Immunofluorescence Microscopy. Cells were harvested by trypsinization and attached onto poly(l)-lysine-coated slides (Sigma) by incubation at 37°C for 2 h. Slides were fixed in 100% prechilled methanol for 10 min at –20°C, rehydrated in PBS for 10 min at room temperature (RT), and then permeabilized in 0.1% Triton X-100/PBS for 5 min. Cells were then blocked in PBS/3% BSA at RT for 30 min. This was followed by incubation for 1 h at RT with FITC-conjugated anti-FLAG mAb (1:100, Sigma) or FITC-conjugated anti-V5 mAb (1:100, Invitrogen). For endogenous staining of MLL5, cells were incubated with 2 μg/ml polyclonal Abs αMLL5-1 and αMLL5-2 at RT for 1 h. After extensive washing with PBS, cells were incubated with Alexa Fluor 568-conjugated goat anti-rabbit IgG (1:200, Molecular Probes) at RT for 1 h. In some cases, nuclei were counterstained with 0.1 μg/ml 4′,6-diamidino-2′-phenylindole dihydrochloride.
For BrdUrd incorporation, 48 h after transfection, cells were incubated with 10 μg/ml BrdUrd for 4 or 16 h at 37°C, fixed in 100% methanol at –20°C for 10 min, and permeabilized in 0.1% Triton X-100/PBS for 5 min. BrdUrd-binding sites were exposed by treatment with 2 M hydrochloric acid at RT for 30 min followed by neutralization in 0.1 M borate buffer (pH 8.5). After washing in PBS, cells were incubated for 60 min at RT with Alexa Fluor 594-conjugated anti-BrdUrd mAb (1:100, Molecular Probes) and Alexa Fluor 488-conjugated anti-GFP mAb (1:100, Molecular Probes).
Confocal microscopy was performed on an LSM 510 laser scanning microscope system (Zeiss) equipped with an argon laser at an excitation of 488 nm, a HeNe laser at an excitation of 543 nm, and an Argon UV laser at an excitation of 351 nm. Slides were viewed with ×40 or ×63 microscope objectives. Images were collected and processed by lsm 5 system software (Zeiss).
Western Blotting. Whole-cell lysates of 293T cells transfected with either pEF6-flag/v5-MLL5 or pEF6 vector alone were prepared with the lysis buffer (0.1% SDS/1% sodium deoxycholate/1% Triton X-100/150 mM NaCl) and a mixture of protease inhibitors (Sigma) on ice for 1 h. Lysates (20 μg per lane) were run on an 8% SDS/PAGE gel and transferred to a nitrocellulose membrane filter. After blotting at 4°C overnight with Tris-buffered saline with 0.05% Tween 20, the membrane was incubated with 1 μg/ml polyclonal Abs αMLL5-1 or αMLL5-2 at RT for 1 h. On extensive washing with Tris-buffered saline with 0.05% Tween 20, the membrane was incubated with alkaline phosphatase-conjugated anti rabbit IgG Abs (1:30,000, Sigma). For anti-FLAG (1:1,000, Sigma) and anti-V5 (1:1,000, Invitrogen) mAbs blots, alkaline phosphatase-conjugated antimouse IgG (1:30,000, Sigma) were used as secondary Ab. Signals were developed by incubation with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (BCIP/NBT) tablets according to the manufacturer's instructions (Sigma).
Flow Cytometry and Cell Cycle Analysis. For flow cytometry, 293T cells transfected with pEF6-flag/v5-MLL5 were incubated with FITC-conjugated anti-FLAG mAb (1:100, Sigma) or FITC-conjugated anti-V5 mAb (1:100, Invitrogen). FITC-conjugated mouse IgG1 and IgG2a (Beckton Dickinson) were used as isotype controls. Fluorescence was analyzed by a FACSCalibur flow cytometer (Beckton Dickinson). For cell cycle analysis, 293T cells transfected with GFP-MLL5 constructs were fixed in prechilled 70% ethanol on ice for 2 h. Cells were washed with PBS and resuspended in propidium iodide (PI) staining solution (0.1% Triton X-100/0.2 mg/ml DNase-free RNase A/20 μg/ml PI prepared in PBS) for 15 min at 37°C and analyzed immediately by flow cytometry. The pulse-width/pulse-area signal was used to discriminate between G2/M cells and cell doublets, with the latter gated out. For necrotic cell staining, live cells were treated with 10 μg/ml PI for 5 min and analyzed by flow cytometry. DNA content histograms and other FACS data were analyzed by using flowjo software.
Prediction of Nuclear Localization Signals (NLS). The MLL5 primary sequence was run through the psort ii algorithm (http://psort.nibb.ac.jp), and predicted NLS were obtained based on preset definitions. In brief, NLS were classified as follows: The classical type of NLS uses the following two rules to detect (i) four-residue pattern (“part 4”) composed of four basic amino acids (K or R) or three basic amino acid and either H or P (called “part 4”); (ii) seven residues (“part 7”) starting with P and followed within three residues by a basic segment containing three K/R residues of four. Another type of NLS (“bipartite”) is two basic residues, a 10-residue spacer, and a region consisting of at least three basic residues of five residues.
Results
Nuclear Localization of MLL5 Protein. The MLL5 cDNA sequence obtained from human peripheral blood lymphocytes differed from that originally isolated from bone marrow (31) at residues 1,020, 1,090, 1,099, 1,168, 1,689, and 1,733 (Ala-1020-Val; Pro-1090-Ser, Ser-1099-Phe, Lys-1168-Glu, Ala-1687-Val, and Thr-1733-Ala; the latter amino acid was obtained from this study in each case). The same six residues encoded by cDNA isolated from peripheral blood lymphocytes were found also in the following cell lines: HeLa, Jurkat, 721.221, and COS7. Thus, in this study the MLL5 cDNA amplified from peripheral blood lymphocytes was used for all constructs.
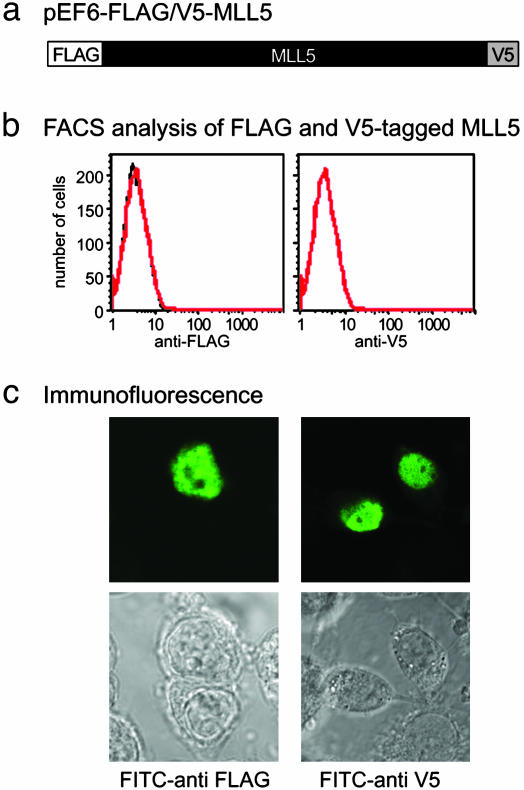
To determine the localization of MLL5, the full-length cDNA was fused to a FLAG epitope tag at the N terminus and a V5 tag at the C terminus (Fig. 1a). This construct (pEF6-flag/v5-MLL5) was transiently transfected into 293T cells, and resulting cell cultures were examined for surface and intracellular expression by using fluorescently conjugated anti-FLAG and anti-V5 mAbs. No surface staining could be detected by flow cytometry analysis (Fig. 1b). Immunofluoresence staining was strictly localized to the nucleus and excluded from nucleoli (Fig. 1c). The same nuclear localization was observed also in other transfected cell lines such as COS and HeLa cells (data not shown). In some cases, discrete intranuclear foci were observed, and the preservation of these foci for staining was sensitive to fixation conditions (I.C., unpublished data).
Fig. 1.
Nuclear localization of MLL5 protein. (a) Full-length MLL5 was tagged with FLAG and V5 epitopes at the N and C termini, respectively, and cloned into the pEF6 vector. (b) The 293T cells were transiently transfected with the pEF6-flag/V5-MLL5 vector for 48 h, stained with 10 μg/ml FITC-conjugated anti-FLAG or FITC-conjugated anti-V5 mAbs for 1 h, and analyzed by flow cytometry (red). Staining for isotype controls is in black. (c) pEF6-flag/v5-MLL5 transfectants were fixed in prechilled 100% methanol for 10 min at –20°C, rehydrated in PBS, and permeabilized in 0.1% Triton X-100/PBS for 5 min. Cells were blocked in PBS containing 3% BSA at RT for 30 min followed by incubation for 1 h at RT with FITC-conjugated anti-FLAG and anti-V5 mAbs.
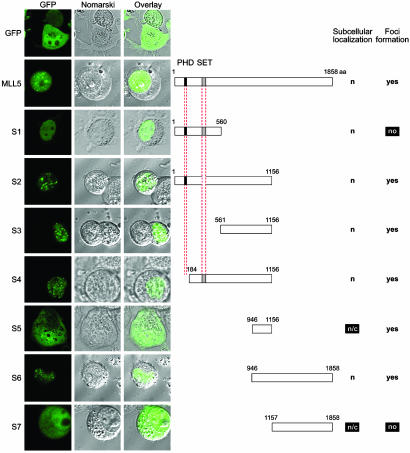
Identification of MLL5 Elements Directing Nuclear Localization and Foci Formation. To investigate its intracellular localization in live cells, MLL5 was fused to GFP and resulting transfectants imaged by confocal microscopy. As seen previously, MLL5 was observed to localize to the nucleus and to form dozens of intensely staining intranuclear foci (Fig. 2). A series of GFP fusion proteins was constructed from various fragments of the MLL5 protein, including several truncations that selectively deleted the PHD and SET domain. Transient transfection of 293T cells with these constructs were examined by confocal microscopy (Fig. 2). Although GFP alone was distributed throughout the cell, nuclear speckled patterns were seen with all constructs used, except those from which codons for amino acids 561–1,156 were deleted (S1 and S7). Because S5, which only encodes amino acids 946–1,156, was able to induce foci formation, this 210-aa region was minimally sufficient for MLL5 foci accumulation.
Fig. 2.
Mapping of MLL5 elements directing nuclear localization and foci formation. Full-length and segments of MLL5 protein were cloned into the pEGFPC1 vector with enhanced GFP fused to their N termini. These constructs were transiently transfected into 293T cells, and live cells were imaged by confocal microscopy. Fluorescence micrographs are shown on the left, with GFP fluorescence followed by Nomarski images and by overlays of Nomarski and fluorescence images. Schematics of the corresponding constructs are displayed on the right. The PHD zinc finger is colored in black, and the SET domain is colored in gray. Amino acid numbers are denoted above each construct. Observed subcellular localization is indicated as nuclear (n) or cytoplasmic (c). The ability to form intranuclear foci is indicated on the right.
At least three separate and functional NLS were found within MLL5: in amino acids 1–560 (S1) and 561–945 (within S3 but not S5), and the region surrounding residue 1,156. A myriad of NLS was predicted in MLL5 by the psort ii algorithm (see Methods). Within the first 560 aa of MLL5 (S1), four part 4 classical NLS, one part 7 classical NLS, and two bipartite type NLS were predicted. A further 13 putative NLS (6 part 4, 3 part 7, and 4 bipartite) were located within amino acids 561–945. At least one functional NLS is within amino acids 1–560 because S1 was exclusively nuclear (Fig. 2). A second NLS maps to amino acids 561–945 because of the nuclear localization of S3 and whole-cell distribution of S5 (Fig. 2). Furthermore, because constructs S5 and S7 yielded both cytoplasmic and nuclear staining (indicating a lack of functional NLS), whereas S6 (encompassing both fragments) was exclusively nuclear, the region surrounding residue 1,156 likely contains a functional NLS. Indeed, two NLS had been predicted between residues 1,151 and 1,158.
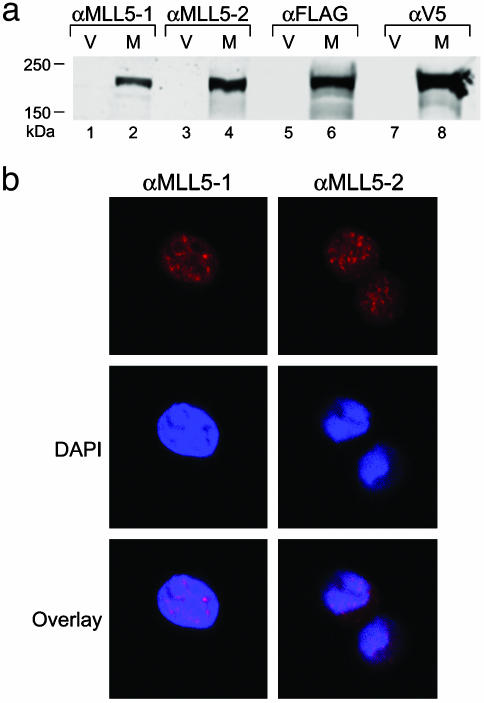
Speckled Distribution of Endogenous MLL5 Protein. To investigate the distribution of endogenous MLL5 protein, rabbit polyclonal Abs were generated against two synthetic peptides comprising residues 227–241 (αMLL5-1) and 801–815 (αMLL5-2) (see Methods). To confirm their reactivity, 293T cells were transfected with a construct that expressed MLL5 protein dually tagged with FLAG and V5 epitopes and subsequently examined by Western blot analysis. Both αMLL5-1 and αMLL5-2 detected protein bands whose migrations were identical to those detected by the anti-FLAG and anti-V5 Abs. Furthermore, these bands were only present in Flag-V5-tagged MLL5 transfected but not mock-transfected cells (Fig. 3a). The polyclonal Abs were also used to stain cells. In each case MLL5 staining showed a nuclear distribution with a speckled pattern (Fig. 3b).
Fig. 3.
(a) Western blot analysis of FLAG-V5-tagged MLL5 expression. Lanes contained whole-cell lysates of 293T cells transfected with either pEF6 vector alone (V) or pEF6-flag-V5-MLL5 (M). Blots were probed with Abs αMLL5-1, αMLL5-2, anti-FLAG mAb, or anti-V5 mAb. The polyclonal Abs αMLL5-1 and αMLL5-2 were raised against synthetic peptides corresponding to amino acids 227–241 and 801–815 of the MLL5 protein, respectively. (b) Endogenous immunofluorescent staining of MLL5 in 293T cells by using rabbit polyclonal Abs αMLL5-1 or αMLL5-2, followed by Alexa 568-conjugated anti-rabbit polyclonal Abs (red). Nuclei were counterstained with 4′,6-diamidino-2′-phenylindole dihydrochloride (DAPI; blue).
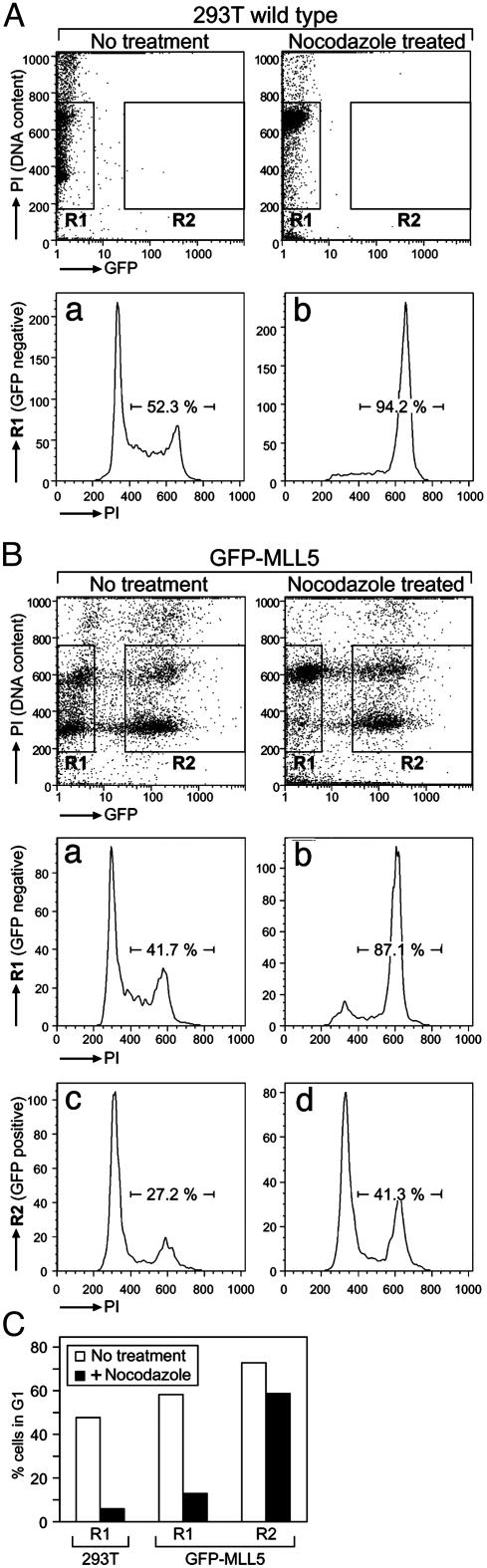
Overexpression of MLL5 Induces Cell Cycle Arrest. Because of its potential function as a tumor suppressor, experiments were carried out to examine the effect of MLL5 gene expression on cell growth. Firstly, cell cycle analysis was carried out on cells transfected with WT or GFP-fused MLL5. Samples were either treated with or without nocodazole to induce arrest in G2/M and then analyzed by flow cytometry. In WT cycling 293T cells, PI staining revealed that ≈50% of cells were in G1 phase (Fig. 4 Aa and C). Almost all of these cells were arrested in G2/M after treatment with nocodazole, as expected (Fig. 4 Ab and C). Transient transfection of GFP-MLL5 allowed examination of two intraculture populations, with GFP expression indicative of transfected cells. The untransfected population displayed nearly identical profiles as WT 293T cells with or without nocodazole treatment (Fig. 4 Ba, Bb, and C). However, in the population in which transfection had been successful, cells did not progress into nocodazole induced arrest in G2/M but mostly remained in G1 phase (Fig. 4 Bc, Bd, and C). The GFP-MLL5-expressing cells were not necrotic as they did not display higher PI staining than negative cells when live populations were stained with PI (data not shown). These data indicate that overexpression of MLL5 prevents cells from progressing past G1 phase.
Fig. 4.
Induction of G1 cell cycle arrest by GFP-MLL5 expression. WT 293T cells or 293T cells transfected with GFP-MLL5 were treated with or without nocodazole for 16 h and then fixed, permeabilized, and stained with PI as described in Methods for cell cycle analysis. (A) WT 293T cells displayed a shift in population to cells with 4N DNA content (G2/M phase) after nocodazole treatment (a and b). (Upper) PI staining (DNA content) was plotted against GFP fluorescence with GFP-negative cells gated on as R1 and positive transfectants gated on as R2. (Lower) DNA content histograms for these respective gates are displayed. (B) The untransfected population (R1) of the GFP-MLL5 transfectant cell culture also displayed a shift to 4N DNA content after nocodazole arrest (a and b). In GFP-MLL5-positive transfectants (R2), however, the profile of cells with 4N content remained constant even after 16-h treatment with nocodazole (c and d). (C) A quantitation of the percentage of cells in G1 phase of the above populations with or without nocodazole treatment indicates an arrest of GFP-MLL5-positive cells (R2) in G1 phase.
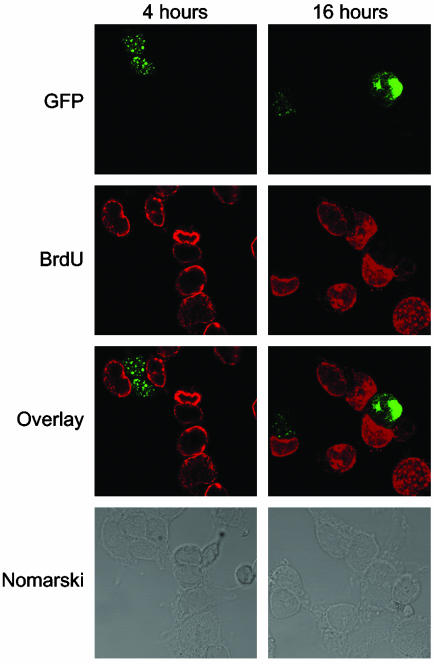
To follow cellular transition into S phase, DNA synthesis was measured by BrdUrd incorporation. After transfection with GFP-MLL5, cells were incubated with BrdUrd for 4 or 16 h to allow the majority of the cells to undergo a complete cell cycle. Cells were fixed, treated with acid to expose BrdUrd sites, and costained with Alexa Fluor 594-conjugated anti-BrdUrd mAb (red) and FITC-conjugated anti-GFP mAb (green). After 16 h, the percentage of BrdUrd-positive and -negative cells among the cells expressing the ectopic protein were analyzed and counted by confocal microscopy (Fig. 5). As expected, only 4% (2 of 50) of cells positive for GFP-MLL5 protein were also BrdUrd-positive. Conversely, almost all (95%) of the GFP-MLL5-negative cells had successfully incorporated BrdUrd. These data confirm that ectopic overexpression of MLL5 inhibits progression of the cell cycle.
Fig. 5.
Overexpression of MLL5 protein in 293T cells inhibits DNA replication. After transfection (48 h), GFP-MLL5 cells were incubated with 20 μg/ml BrdUrd for 4 or 16 h. Resulting cultures were fixed, permeabilized, treated with 1 N HCl to expose BrdUrd sites, and stained with Alexa 568-conjugated anti-BrdUrd mAb (red) and Alexa 488-conjugated anti-GFP mAb (green). The third panel displays an overlay of BrdUrd and GFP-MLL5 staining, and the fourth panel shows the corresponding Nomarski images.
Discussion
In this study, both ectopically introduced and endogenous MLL5 protein were shown to form a nuclear speckled distribution. Overexpression of MLL5 protein prevented cell cycle progression into S phase. Fluorescence imaging of a series of truncated mutants identified at least three functional NLS and a domain required for foci formation.
Overexpression of proteins has been used in many cases as a useful tool in elucidating their physiological role. Indeed, induced overproduction of mammalian tumor-suppressor genes such as p53 and BRCA1 had been used to characterize their biological functions and were shown to induce cell cycle arrest (32, 33). The MLL5 gene was found in a search of candidate myeloid leukemia tumor suppressor genes from an ≈2.5-Mb commonly deleted segment within chromosome band 7q22 (31). Although the molecular mechanism of MLL5 function is unknown, its prohibition of cell cycle entry into S phase could act as a starting point for further functional characterization. Because our experiments were carried out in the HEK 293T cells, which are highly transformed, MLL5 must have a dominant effect on inhibiting cell cycle progression. It would be intriguing to test whether MLL5 expression induces the up-regulation of the signaling proteins that are involved in G1 arrest, such as the cyclin-dependent kinase inhibitor p21 (33, 34). It will also be useful to establish a cell line inducible for MLL5 expression so that modulation of such proteins on MLL5 expression can be monitored more precisely at different stages of the cell cycle.
MLL5 overexpression induced G1 phase arrest, which is also a crucial DNA damage checkpoint that acts as an important safeguard for genomic stability (35). Overexpression of many important proteins associated with DNA damage repair causes the activation of these checkpoints and subsequent growth arrest (32–34, 36). Many of these tumor suppressors such as BRCA1, p53, and Rad51 also form intranuclear foci (34, 37, 38). Such cellular similarities suggest that MLL5 may play a role in DNA repair. Preliminary data indicate that the expression level of endogenous MLL5 protein did not significantly change after DNA damage (e.g., MMS, UV, and γ irradiation), which may not support this role (data not shown). However, this possibility needs to be further investigated by using more accurate quantitative methods to monitor the changes in the protein and RNA levels, e.g., Western blotting or real-time PCR.
In our study, cell lines believed to express endogenous MLL5 (by Northern blot and immunofluorescence analyses) were negative by conventional Western blot analysis by using rabbit polyclonal Abs raised against MLL5 (αMLL5-1 and αMLL5-2; data not shown). Similar results were also described in the detection of WT MLL protein and Drosophila TRX protein (25, 39). It was proposed that MLL and Drosophila TRX protein were highly unstable and generally susceptible to specific proteolytic cleavage. Indeed, a recent study has shown that an N-terminal p320 (N320) and a C-terminal p180 (C180) fragments of MLL1 were generated by the proteolytic cleavage and the two fragments form a stable complex that confers protein stability and correct subnuclear localization (40). When MLL5 was tagged with a FLAG epitope at the N terminus and a V5 epitope at the C terminus, both anti-FLAG and anti-V5 Abs detected the same size of protein bands, suggesting a lack of such proteolytic cleavage. However the endogenous MLL5 may be present at a much lower amount in cells as compared with the ectopically expressed MLL5. Additional care to protect MLL5 from proteolysis may be required for the successful detection of the endogenous protein on Western blot analysis.
The domain required for MLL5 foci formation was identified in this study. The pattern and numbers of foci were sensitive to fixation conditions, and the foci tended to display a more diffuse localization than those imaged on live cells (I.C., unpublished data). Similar speckles are also seen in the MLL-1 and chimeric MLL-1 fusions expressed in the transfected cells (39–41). However, the significance of these foci remains unknown. Many nuclear proteins concentrate into intranuclear speckles or foci, such as cyclin A, PCNA, RPA, Rad51, CAF-1, PML, Mre11, NBS, and BRCA1. These foci represent the functional compartmentalization of many nuclear processes such as DNA replication, repair, transcription, or RNA splicing (37, 42). Thus, it was of great interest to see where MLL5 foci were in relation to other such nuclear proteins. However, none of the proteins tested (BRCA1, Rad51, Bmi-1, and PCNA) colocalized with MLL5 (data not shown).
The absence of either AT-hooks or methyltransferase homology domain in MLL5 suggests that it may not bind to DNA but instead modulates transcription indirectly by protein–protein interaction through PHD and SET domain (31). Consistent with this idea, ALL-1 (-MLL-1) protein self-associates and interacts with the chromatin-remodeling complexes through its SET domain (23, 43). No differences in foci formation and nuclear localization were found in the PHD fingers and SET domain deleted mutants, suggesting that the foci were not large protein complexes formed via these domains. Recently the PHD zinc finger of ING2, a candidate tumor suppressor protein, has been shown to be a nuclear receptor for phosphatidylinositol 5-phosphate, an association that regulates p53-dependent cell cycle arrest (44). Similar experiments addressing an involvement of the PHD zinc finger of MLL5 for signaling and growth arrest would be very intriguing. Additional studies are required to characterize the molecular interactions of the two domains of MLL5. In such a way, the possible biological role of MLL5 could be further explored.
Here, we present a biochemical study on the recently cloned tumor suppressor gene candidate MLL5. We have shown that MLL5 is a nuclear protein that forms distinct foci and that its overexpression induces growth arrest. Much work remains to be done to further elucidate its role in development and leukemogenesis.
Acknowledgments
We thank Drs. Jon Boyson and Derin Keskin for helpful discussion, and Dr. Stanley Korsmeyer and Grace Gill for comments on the manuscript. This work was supported by National Institutes of Health Grants R35 CA47554 and R01 AI50207 and by a fellowship from the Juvenile Diabetes Research Foundation and The Agency for Science, Technology, and Research.
Abbreviations: NLS, nuclear localization signal; PHD, plant homeodomain; PI, propidium iodide; RT, room temperature; SET, Su(var)3-9, enhancer-of-zeste, trithorax.
References
- 1.Ziemin-van der Poel, S., McCabe, N. R., Gill, H. J., Espinosa, R., III, Patel, Y., Harden, A., Rubinelli, P., Smith, S. D., LeBeau, M. M. & Rowley, J. D. (1991) Proc. Natl. Acad. Sci. USA 88, 10735–10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu, Y., Nakamura, T., Alder, H., Prasad, R., Canaani, O., Cimino, G., Croce, C. M. & Canaani, E. (1992) Cell 71, 701–708. [DOI] [PubMed] [Google Scholar]
- 3.Tkachuk, D. C., Kohler, S. & Cleary, M. L. (1992) Cell 71, 691–700. [DOI] [PubMed] [Google Scholar]
- 4.Djabali, M., Selleri, L., Parry, P., Bower, M., Young, B. D. & Evans, G. A. (1992) Nat. Genet. 2, 113–118. [DOI] [PubMed] [Google Scholar]
- 5.Dimartino, J. F. & Cleary, M. L. (1999) Br. J. Haematol. 106, 614–626. [DOI] [PubMed] [Google Scholar]
- 6.Ayton, P. M. & Cleary, M. L. (2001) Oncogene 20, 5695–5707. [DOI] [PubMed] [Google Scholar]
- 7.Ernst, P., Wang, J. & Korsmeyer, S. J. (2002) Curr. Opin. Hematol. 9, 282–287. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher, A. & Magnuson, T. (1997) Trends Genet. 13, 167–170. [PubMed] [Google Scholar]
- 9.Kennison, J. A. (1995) Annu. Rev. Genet. 29, 289–303. [DOI] [PubMed] [Google Scholar]
- 10.Orlando, V. & Paro, R. (1995) Curr. Opin. Genet. Dev. 5, 174–179. [DOI] [PubMed] [Google Scholar]
- 11.Gould, A. (1997) Curr. Opin. Genet. Dev. 7, 488–494. [DOI] [PubMed] [Google Scholar]
- 12.Aasland, R., Gibson, T. & Stewart, A. F. (1995) Trends Biochem. Sci. 20, 56–59. [DOI] [PubMed] [Google Scholar]
- 13.Jeanmougin, F., Wurtz, J., Le Douarin, B., Chambon, P. & Losson, R. (1997) Trends Biochem. Sci. 22, 151–153. [DOI] [PubMed] [Google Scholar]
- 14.Jenuwein, T., Laible, G., Dorn, R. & Reuter, G. (1998) Cell Mol. Life Sci. 54, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeleznik-Le, N. J., Harden, A. & Rowley, J. D. (1994) Proc. Natl. Acad. Sci. USA 91, 10610–10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slany, R. K., Lavau, C. & Cleary, M. L. (1998) Mol. Cell. Biol. 18, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birke, M., Schreiner, S., Garcia-Cuellar, M. P., Mahr, K., Titgemeyer, F. & Slany, R. K. (2002) Nucleic Acids Res. 30, 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khavari, P. A., Peterson, C. L., Tamkun, J. W., Mendel, D. B. & Crabtree, G. R. (1993) Nature 366, 170–174. [DOI] [PubMed] [Google Scholar]
- 19.Cairns, B. (2001) Trends Cell Biol. 11, S15–S21. [DOI] [PubMed] [Google Scholar]
- 20.Wang, W., Cote, J., Xue, Y., Zhou, S., Khavari, P. A., Biggar, S. R., Muchardt, C., Kalpana, G. V., Goff, S. P., Yaniv, M., et al. (1996) EMBO J. 15, 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 21.Muchardt, C., Reyes, J. C., Bourachot, B., Leguoy, E. & Yaniv, M. (1996) EMBO J. 15, 3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 22.Sif, S., Stukenberg, P. T., Kirschner, M. W. & Kingston, R. E. (1998) Genes Dev. 12, 2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozenblatt-Rosen, O., Rozovskaia, T., Burakov, D., Sedkov, Y., Tillib, S., Blechman, J., Nakamura, T., Croce, C. M., Mazo, A. & Canaani, E. (1998) Proc. Natl. Acad. Sci. USA 95, 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoudi, T. & Verrijzer, C. (2001) Oncogene 20, 3055–3066. [DOI] [PubMed] [Google Scholar]
- 25.Kuzin, B., Tillib, S., Sedkov, Y., Mizrokhi, L. & Mazo, A. (1994) Genes Dev. 8, 2478–2490. [DOI] [PubMed] [Google Scholar]
- 26.Armstrong, S. A., Staunton, J. E., Silverman, L. B., Pieters, R., den Boer, M. L., Minden, M. D., Sallan, S. E., Lander, E. S., Golub, T. R. & Korsmeyer, S. J. (2002) Nat. Genet. 30, 41–47. [DOI] [PubMed] [Google Scholar]
- 27.Prasad, R., Zhadanov, A. B., Sedkov, Y., Bullrich, F., Druck, T., Rallapalli, R., Yano, T., Alder, H., Croce, C. M., Huebner, K., Mazo, A. & Canaani, E. (1997) Oncogene 15, 549–560. [DOI] [PubMed] [Google Scholar]
- 28.FitzGerald, K. T. & Diaz, M. D. (1999) Genomics 59, 187–192. [DOI] [PubMed] [Google Scholar]
- 29.Huntsman, D. G., Chin, S. F., Muleris, M., Batley, S. J., Collins, V. P., Wiedemann, L. M., Aparicio, S. & Caldas, C. (1999) Oncogene 18, 7975–7984. [DOI] [PubMed] [Google Scholar]
- 30.Ruault, M., Brun, M. E., Ventura, M., Roizes, G. & De Sario, A. (2002) Gene 284, 73–81. [DOI] [PubMed] [Google Scholar]
- 31.Emerling, B. M., Bonifas, J., Kratz, C. P., Donovan, S., Taylor, B. R., Green, E. D., Le Beau, M. M. & Shannon, K. M. (2002) Oncogene 21, 4849–4854. [DOI] [PubMed] [Google Scholar]
- 32.Braithwaite, A. W., Sturzbecher, H., Addison, C., Palmer, C., Rudge, K. & Jenkins, J. R. (1987) Nature 329, 458–460. [DOI] [PubMed] [Google Scholar]
- 33.Somasundaram, K., Zhang, H., Zeng, Y. X., Houvras, Y., Peng, Y., Zhang, H., Wu, G. S., Licht, J. D., Weber, B. L. & El-Deiry, W. S. (1997) Nature 389, 187–190. [DOI] [PubMed] [Google Scholar]
- 34.Raderschall, E., Bazarov, A., Cao, J., Lurz, R., Smith, A., Mann, W., Ropers, H. H., Sedivy, J. M., Golub, E. I., Fritz, E. & Haaf, T. (2002) J. Cell Sci. 115, 153–164. [DOI] [PubMed] [Google Scholar]
- 35.Bartek, J. & Lukas, J. (2001) Curr. Opin. Cell Biol. 13, 738–747. [DOI] [PubMed] [Google Scholar]
- 36.Haaf, T., Golub, E., Reddy, G., Radding, C. M. & Ward, D. C. (1995) Proc. Natl. Acad. Sci. USA 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scully, R., Chen, J., Ochs, R. L., Keegan, K., Hoekstra, M., Feunteun, J. & Livingston, D. M. (1997) Cell 90, 425–435. [DOI] [PubMed] [Google Scholar]
- 38.Carbone, R., Pearson, M., Minucci, S. & Pelicci, P. G. (2002) Oncogene 21, 1633–1640. [DOI] [PubMed] [Google Scholar]
- 39.Butler, L. H., Slany, R., Cui, X., Cleary, M. L. & Mason, D. Y. (1997) Blood 89, 3361–3370. [PubMed] [Google Scholar]
- 40.Hsieh, J. J., Ernst, P., Erdjument-Bromage, H., Tempst, P. & Korsmeyer, S. J. (2003) Mol. Cell. Biol. 23, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yano, T., Nakamura, T., Blechman, J., Sorio, C., Dang, C. V., Geiger, B. & Canaani, E. (1997) Proc. Natl. Acad. Sci. USA 94, 7286–7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carney, J. P., Maser R. S., Olivares, H., Davis, E. M., Le Beau, M., Yates, J. R., III, Hays, L., Morgan, W. F. & Petrini, J. H. (1998) Cell 93, 477–486. [DOI] [PubMed] [Google Scholar]
- 43.Rozovskaia, T., Rozenblatt-Rosen, O., Sedkov, Y., Burakov, D., Yano, T., Nakamura, T., Petruck, S., Ben-Simchon, L., Croce, C. M., Mazo, A. & Canaani, E. (2000) Oncogene 19, 351–357. [DOI] [PubMed] [Google Scholar]
- 44.Gozani, O., Karuman, P., Jones, D. R., Ivanov, D., Cha, J., Lugovskoy, A. A., Baird, C. L., Zhu, H., Field, S. J., Lessnick, S. L., et al. (2003) Cell 114, 99–111. [DOI] [PubMed] [Google Scholar]