Abstract
We demonstrate the efficacy of a genome-wide protocol in yeast that allows the identification of those gene products that functionally interact with small molecules and result in the inhibition of cellular proliferation. Here we present results from screening 10 diverse compounds in 80 genome-wide experiments against the complete collection of heterozygous yeast deletion strains. These compounds include anticancer and antifungal agents, statins, alverine citrate, and dyclonine. In several cases, we identified previously known interactions; furthermore, in each case, our analysis revealed novel cellular interactions, even when the relationship between a compound and its cellular target had been well established. In addition, we identified a chemical core structure shared among three therapeutically distinct compounds that inhibit the ERG24 heterozygous deletion strain, demonstrating that cells may respond similarly to compounds of related structure. The ability to identify on-and-off target effects in vivo is fundamental to understanding the cellular response to small-molecule perturbants.
The interaction of an organism with its environment is essential to its survival. The currency of this interaction is predominantly small molecules. On a molecular level, small molecules both promote, as in nutrients, and challenge, as in toxins, cell viability. Those gene products that interact with small molecules underlie the organism's ability to adapt to environmental changes and include those that bind, transport, and metabolize small molecules. Specific small molecule–protein interactions are often identified genetically followed by in vitro characterization. Such experiments cannot, however, capture potential interactions with other proteins in the cell. Our experimental protocol allows the identification of all gene products that functionally interact with a small molecule of interest and result in inhibition of cellular proliferation. This chemical genomics assay, haploinsufficiency profiling (HIP), is predicated on our observation that lowering gene dosage from two copies (in a diploid yeast strain) to one copy (in a heterozygous deletion strain) results in a strain that is sensitized to compounds that inhibit the product of the heterozygous locus (1). Functional interactions are then identified genome-wide by competitive growth of a complete collection of molecularly bar-coded heterozygous deletion strains in a single culture, allowing screening of all strains in parallel. Subsequent quantification of relative sensitivities is achieved by using high-density oligonucleotide arrays carrying the bar-code complements (2, 3). Here we highlight the effects of 10 diverse compounds of general interest in 80 genome-wide screens. These compounds include anticancer and antifungal agents, statins, alverine citrate, and dyclonine. For several of the well characterized compounds, we found that some of the most sensitive heterozygous strains often carry a deletion in the gene whose product is known to interact directly with the test molecule; typically, this is the established drug target. A likely explanation for this observation is that the compound inhibits cellular proliferation by reducing the activity of the remaining gene product of the heterozygous locus, thereby mimicking a complete deletion. Thus, in the assay we should primarily identify gene products that are either essential or, when deleted in a homozygous strain, exhibit a slow-growth phenotype. A second class of sensitive heterozygous strains reports nonessential genes that are dosage-limiting for growth only in the presence of compound. These include strains deleted for genes involved in compound transport and/or metabolism. We found that, although most compounds interact primarily with one or a few gene products across the genome, other unexpected effects revealed insights into compound mechanism. These results provide a comprehensive in vivo snapshot of the genome-wide cellular response to small-molecule perturbants.
Materials and Methods
Reagents. Alverine citrate, atorvastatin, methotrexate, 5-fluorouracil (5-FU), miconazole, and amphotericin B were from MicroSource Discovery Systems (Gaylordsville, CT). Lovastatin was the gift of J. Rine (University of California, Berkeley). Cisplatin, itraconazole, and fluconazole were obtained from the Stanford University Pharmacy (Stanford, CA). Dyclonine and fenpropimorph were from Sigma–Aldrich.
Media and Growth Conditions. YPD (yeast extract/peptone/dextrose) was prepared as described (4).
Overexpression Studies. A plasmid overexpressing ERG24 was the gift of C. Mo and M. Bard (Indiana University School of Science, Indianapolis) (5). A plasmid overexpressing the human LBR was the gift of G. Loison (Sanofi-Synthelabo Recherche, Labège, France) (6).
Deletion Pool Construction, Growth, and Chip Experiments. Deletion pool construction and pool growth were as described (3) with the following modifications of growth conditions. After overnight recovery of frozen aliquots of the pools for 10 generations, logarithmically growing cells were diluted in YPD plus compound to an OD600 of 0.0625, and 0.7 ml was pipetted into a well of a 48-well microplate. Cells were grown in a Tecan GENios microplate reader (Tecan U.S., Durham, NC) and every five generations cells were automatically pipetted into 0.7 ml of fresh YPD medium containing the appropriate compound by using a Packard Multiprobe II 4-probe liquid handling system (Perkin–Elmer Life Sciences) controlled by custom labview software (National Instruments, Austin, TX). After 20 generations of growth, cells were saved and frozen at –20°C for subsequent preparation of genomic DNA. Concentrations of compounds screened were based on prescreens against a wild-type strain. The optimal concentration of compound screened was determined empirically. For these concentrations, at least one replicate was generated.
Genomic DNA Preparation, PCR, and Chip Hybridization. Genomic DNA preparation, PCR, and chip hybridization were as described (3).
Data Analysis. Preprocessing of data. Each deletion strain is associated with four hybridization signals on the high-density oligonucleotide array (3). To classify the tags that do not hybridize well enough to the array to yield usable measurements, we estimated the average background intensity from control arrays (for description of control arrays, see Identification of Significantly Sensitive Strains) using a set of 17,964 tags on the chip not represented in any strain. All tags associated with strains that had a measured intensity <3 SD above the mean background intensity in control arrays were eliminated from the analysis. Strains in which all four tags hybridized below this cutoff were therefore removed from the analysis. Lists of these heterozygous deletion strains and homozygous deletion strains that were removed from the analysis can be found in Tables 2 and 3, respectively, which are published as supporting information on the PNAS web site.
Identification of significantly sensitive strains. To best estimate the relative amount of each strain in the population, we used a mixture factor analysis model (7) that combines the intensities of the four tags associated with each strain in each experiment. This factor analysis estimate is a measure of strain abundance. To identify sensitive strains a set of control arrays were used that consists of 36 arrays for the heterozygous analysis and 21 arrays for the homozygous analysis. The control hybridizations are collected from experiments in the standard condition as defined by 20 generations of growth in YPD. Each array was normalized by taking the base 10 logarithm of the intensity values for each position in the array and subtracting the average array intensity. For each strain we fit a Gaussian distribution based on the mean and standard deviation of the factor analysis estimate in the control chips. To quantify the fitness defect (FD) of a strain in question, we calculated the log-likelihood of seeing the experimental factor analysis estimate under the Gaussian for the control chips. The fitness defect score is proportional to the log-likelihood of observing the experimental value given the set of control chips. For convenience we multiply the score by –1, so that large values for scores represent large fitness defects. This definition of fitness defect score quantifies the difference between the abundance of strain in the population in a given experiment to that expected in the standard condition. Strains whose tag intensities fall lower than the mean (implying they were diminishing from the population) are classified as sensitive to treatment, whereas those strains whose tag intensities fall higher than the mean are classified as refractory to treatment. Experimentally, many of the strains that turn up as significantly refractory are slow growers in the standard condition. These strain measurements have tag intensity distributions that are not much above background and are therefore unreliable. Although true resistant strains exist in these lists, they are only a small percentage of the total number of refractory strains. For this analysis, we focus on the sensitive strains and have set the fitness defect score of the refractory strains to zero.
To determine significantly sensitive strains in each experiment, we scored all control chips by using the algorithm described above. For each scored control chip we determined the mean FD score and its standard deviation across all genes. An ANOVA accepts the hypothesis that all control chips come from the same distribution. We next computed the mean and standard deviation of the FD scores in the control chips (mean = 15.6, SD = 0.21). When examining FD scores from experiments, only those strains whose FD score is 3 SD above the mean score of controls was considered significantly sensitive. When replicates for a compound at a particular concentration existed, we computed the average rank for each of the strains that scored significantly sensitive. All raw and analyzed data are presented in Supporting Information, which is published as supporting information on the PNAS web site.
Postprocessing. When a sensitive strain's abundance in the pool is so low that all tags hybridize at background level, it has reached its maximum detectable FD score. Because the actual value of the maximum FD score for each strain depends on the hybridization characteristics of its tags and its relative fitness, we have flagged strains for each experiment that have reached this maximal FD score. The heterozygous strains and the homozygous strains with this maximal FD score are listed in Tables 4 and 5, respectively, which are published as supporting information on the PNAS web site.
Results
Our earlier work demonstrated that drug targets in yeast (encoded by essential or nonessential genes) can be identified by their ability to confer sensitivity when the gene dosage is reduced from two copies in a wild-type strain to one copy in a heterozygous deletion strain (1). We have now scaled the HIP assay to a comprehensive, genome-wide level, taking advantage of the complete collection of molecularly bar-coded heterozygous deletion strains (2, 3). In the assay, pools of ≈6,000 heterozygous deletion strains are grown in the presence of small molecules and cells are collected at specified generation times by using custom robotics. To quantitate the relative abundance of each strain, amplification of the molecular bar codes from resultant genomic DNA is followed by hybridization to high-density oligonucleotide arrays carrying the bar-code complements. Statistical treatment of the resulting signal intensity data allows strain fitness to be quantitatively assessed and ranked in order of sensitivity on a gene-by-gene basis. In this way, each experiment generates a genome-wide profile of functional interactions. The findings of several experiments are confirmed genetically in follow-up assays. Results from 80 experiments profiled at several concentrations of 10 diverse compounds are presented, grouped according to their therapeutic class.
Anticancer Compounds. Because of the extensive homology between yeast and human biochemical pathways and, in particular, that of the cell cycle, we tested the hypothesis that our chemogenomic assay could reveal the mechanism of action of anticancer compounds. We profiled three such compounds: methotrexate, 5-FU, and cisplatin.
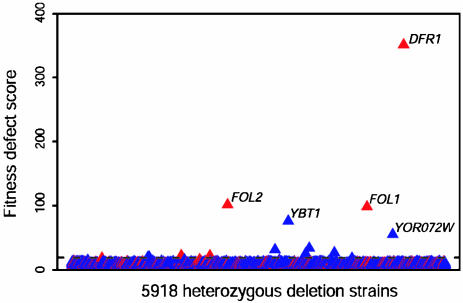
Methotrexate. Dihydrofolate reductase (encoded by DFR1) is the known target of methotrexate (8) and was identified in the HIP assay as a highly sensitive strain at the optimal concentration of 250 μM methotrexate (Fig. 1). Four other strains were identified as significantly sensitive in eight of nine replicate experiments. Two of these strains were heterozygous for the genes FOL1 and FOL2, which act upstream of DFR1 and are required for biosynthesis of folic acid in yeast. Because the readout of the HIP assay is based on growth inhibition, we expect that the only essential gene products identified in our assay will be those that interact directly with compounds and are dosage-limiting for growth. An exception to this may be gene products that are rate-limiting in the drug target pathway. For example, the FOL2 product catalyzes the known rate-limiting step in the biosynthesis of a variety of pterins (9). Although the FOL1 product is not known to be rate-limiting in this pathway, it is possible that under these conditions it may be. Because the HIP assay does not distinguish between gene products that directly interact with a compound from those that become rate-limiting in the presence of a compound, however, it is also possible that FOL1 and FOL2 gene products bind directly to methotrexate. Whatever the case, because FOL1 and FOL2 strains are haploinsufficient in the presence of methotrexate, they are potential candidates for drug targets, because small perturbations in their protein levels lead to growth inhibition. In this case, the human ortholog of FOL2 may be a human therapeutic target because of the high degree of conservation between yeast and humans (blast E value 5 × 10–62). Conversely, FOL1 may be a good antifungal target candidate because it shares significant protein homology to fungal pathogens (Candida albicans blast E value 10–152), but no human homolog exists.
Fig. 1.
A genome-wide readout of heterozygous strain sensitivity profiled at 250 μM methotrexate. The FD score is plotted along the y axis as a function of the 5,918 heterozygous yeast deletion strains ordered by ORF name. The greater the FD score, the more sensitive the strain. Essential ORFs are red and nonessential ORFs are blue. Strains above the dashed line are considered significantly sensitive (see Materials and Methods). Only those genes that scored significantly sensitive in eight of nine replicate experiments are labeled.
The methotrexate results also reveal what became a recurring theme: genes involved in compound availability are often identified by the assay. The YBT1 and YOR072w heterozygous deletion strains are highly sensitive to methotrexate, and both nonessential gene products may be involved in small molecule transport. The human homolog of YBT1 encodes the known methotrexate transporter and up-regulation of this gene in human cancer cells causes methotrexate resistance (10). Although the function of the nonessential gene YOR072w is unknown, homozygous deletants are hypersensitive to wortmannin (11), and the gene encodes a predicted transmembrane domain, indicating that it may play a role in methotrexate transport (12).
5-FU. 5-FU is an antimetabolite used to treat a wide variety of cancers (8). The cytotoxicity of 5-FU is attributed to inhibition of thymidylate synthase and to misincorporation of fluoronucleotides into RNA and DNA (8). Surprisingly, neither the strain carrying the deletion of the yeast thymidylate synthase gene nor any strains deleted for genes involved in DNA-related processes appeared significantly sensitive within a 16-fold concentration range. A functional analysis using the Gene Ontology (GO) term finder [Dolinski, K., Balakrishnan, R., Christie, K. R., Costanzo, M. C., Dwight, S. S., et al. Saccharomyces Genome Database, available at www.yeastgenome.org (accessed September 1, 2003)] revealed that those genes that confer sensitivity to 5-FU when heterozygous are enriched in categories that involve essential RNA processes, notably ribosome biogenesis and assembly (P value 3.9 × 10–9) and rRNA processing (P value 3.6 × 10–9) (Fig. 2 and Table 1). Because the genetic strain background of the deletion library is deleted for URA3, we addressed the possibility that this genotype is responsible for the observed phenotype. Introduction of URA3 on a plasmid into the 10 most sensitive strains confirmed that the auxotrophy was not the cause for their sensitivity (data not shown). Although these results do not rule out inhibition of thymidylate synthase and/or misincorporation into the DNA, they do suggest that the primary mechanism of 5-FU inhibition of yeast cell growth is misincorporation into the RNA leading to the impairment of essential RNA processing functions. Misincorporation into the RNA has also been suggested as the major mechanism of action in human tumor cell lines (13).
Fig. 2.
Distribution of gene ontology (GO) biological process terms for a list of 22 genes that, when heterozygous, confer significant sensitivity in at least two independent experiments in 5-FU at 19.2, 38.5, or 76.9 μM. The percent of genes sensitive to 5-FU is plotted along the y axis in each category relative to that of the whole genome. Only the GO terms with a P value <2.5E-06 are shown.
Table 1.
Gene list used to generate the gene ontology distribution shown in Fig. 2 by using the GO Term Finder*
| Gene | Viability | Biological process |
|---|---|---|
| YPR143W | Essential | Biological process unknown |
| NOP4 | Essential | rRNA processing |
| YPL044C | Essential | Biological process unknown (overlapsNOP4) |
| GLC7 | Essential | Meiosis |
| FUII | Nonessential | Uridine transport |
| MAK5 | Essential | rRNA processing |
| YPR142C | Essential | Biological process unknown (overlaps YPR143W) |
| MAS2 | Essential | Mitochondrial processing |
| MTR4 | Essential | 35S primary transcript processing |
| ITR1 | Nonessential | myo-Inositol transport |
| MAK21 | Essential | Ribosomal large subunit assembly and maintenance |
| DBP7 | Nonessential | 35S primary transcript processing |
| RRP12 | Essential | Processing of 20S pre-rRNA |
| YOR309C | Nonessential | Biological process unknown (overlapsNOP58) |
| MKS1 | Nonessential | Regulation of nitrogen utilization |
| ATP3 | Nonessential | ATP synthesis coupled proton transport |
| PUS7 | Nonessential | Pseudouridine synthesis |
| DIS3 | Essential | 35S primary transcript processing |
| RRP6 | Nonessential | 35S primary transcript processing |
| WHI3 | Nonessential | Regulation of cell size |
| RLR1 | Nonessential | mRNA-nucleus export |
| NOP58 | Essential | rRNA modification |
Genes listed in bold are involved in RNA processing. Two hypothetical ORFs (YPL044c and YOR309c) are also listed in bold because their phenotype is likely due to the disruption of the overlapping ORF.
Dolinski, K., Balakrishnan, R., Christie, K. R., Constanzo, M. C., Dwight, S. S., et al. Saccharomyces Genome Database, available at www.yeastgenome.org.
Cisplatin. Cisplatin is an antineoplastic agent that acts to inhibit cell proliferation by covalently binding to the N-7 position of purines (8). Its antineoplastic activity is thought to result primarily from intrastrand DNA crosslinking. Because cisplatin has no protein target, we expected that the HIP assay would reveal few, if any, sensitive heterozygous strains. In fact, in seven experiments covering a 4-fold concentration range of cisplatin, no strains were consistently sensitive. When (as is the case here) a small molecule has no protein target, it can be more informative to screen the homozygous collection of strains to uncover the compound's mechanism of action. In this case, genes important for cell survival in the presence of compound should be identified, rather than the specific gene products that interact with the small molecule. Indeed, profiling the homozygous pool of strains with cisplatin revealed that the majority of the sensitive strains have homozygous deletions of genes involved in DNA repair (Fig. 3). As expected, these results confirm that the cells cannot survive without repairing their DNA, and, thus, that the DNA itself is likely the primary target of cisplatin.
Fig. 3.
A genome-wide profile of homozygous strain sensitivity to 125 μM cisplatin. Axes shown are equivalent to those in Fig. 1. Only the strains that were scored as significantly sensitive are labeled.
Statin Compounds. Atorvastatin and lovastatin are two commonly used, effective anticholesterol drugs. The target of these drugs is 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase (14). Yeast has two isozymes of HMG-CoA reductase encoded by HMG1 and HMG2. HMG1 contributes the majority (83%) of the enzyme activity in the cell based on in vitro activity assays from extracts of HMG1 and HMG2 homozygous deletion strains (15). Consistent with this observation, only HMG1 heterozygous deletants exhibit sensitivity to atorvastatin and lovastatin in the HIP assay. Individual analysis of the heterozygous and homozygous HMG1 and HMG2 deletion strains confirmed this observation (Fig. 4). As expected, at the concentrations used in the HIP assays, only the HMG1 heterozygous deletion strain exhibits drug-induced haploinsufficiency (Fig. 4B). However, the sensitivity of the HMG1 homozygous deletion strain indicates that a second cellular target must contribute to growth inhibition. Based on predictions from the literature, this target is most likely the gene product of HMG2. Consistent with this observation, we see that only at high concentrations of atorvastatin is HMG2 haploinsufficiency detected (Fig. 4D). This is in agreement with the in vitro activity data and suggests that HMG1 is responsible for the majority of the reductase function in vivo as well. Other strains sensitive to statins include PDR5 (a pleiotropic drug pump) and ERG13. ERG13 encodes HMG-CoA synthase and acts directly upstream of HMG1. The sensitivity of the ERG13 strain to the statins suggests it may be a regulatory step in the mevalonate pathway. Indeed, in cells deprived of ergosterol, the product of ERG13 increases (16).
Fig. 4.
Individual growth analysis of the HMG1 and HMG2 heterozygous and homozygous deletion strains in the presence of atorvastatin. Atorvastatin was 0 μM(A), 62.5 μM(B), 125 μM(C), or 250 μM(D). At the concentrations used in the HIP screens (62.5 and 125 μM), only the HMG1 heterozygous strain is sensitive, not the HMG2 heterozygous strain. At the highest concentration of atorvastatin (250 μM) both heterozygous strains are sensitive, as would be expected if both genes contribute to the HMG-CoA reductase activity of the cell. Comparable analysis in the presence of lovastatin yielded similar results (data not shown).
Antifungal Compounds. Azoles. Antifungals of the azole class primarily target the ERG11 gene product, a cytochrome P450 that catalyzes 14-α-demethylation of lanosterol (17, 18). Data from the HIP assay in the presence of miconazole identify the ERG11 heterozygous strain as one of the most sensitive strains (data are published as supporting information on the PNAS web site). Several other strains likely involved in compound availability, however, do exhibit greater sensitivity than ERG11. We profiled an 8-fold range in concentration. At all concentrations of miconazole, the SET6 heterozygous deletion strain is extremely sensitive. We have observed that the SET6 heterozygote is generally sensitive to compounds that target ergosterol biosynthesis, including fenpropimorph, dyclonine, and alverine citrate (see results below). Because deletion of SET6 in the absence of tested compounds does not have an effect on growth, the function of this gene is more likely to be involved in compound availability. Further experiments are needed to determine if there is a mechanistic link of SET6 to the ergosterol pathway. At 0.05 and 0.1 μM miconazole, the ERG11 deletion strain is significantly sensitive, but the PDR5 strain heterozygous for the known azole pump dominates the profile. At the highest concentration of miconazole tested, 0.2 μM (a dose that inhibits the cells ≈70% with respect to wild type), the ERG11 strain exhibits no significant sensitivity and the profile is composed of dozens of sensitive strains. This loss of target specificity at high concentrations was generally observed for most of the compounds tested.
Although our results with fluconazole (a triazole) across an 8-fold range in concentration were similar to miconazole (an imidazole), the ERG11 target scored as a significantly sensitive strain at only a single concentration (32.6 μM). These results indicate that fluconazole may be less specific for its target, an effect that has also been observed in a previous study (19).
Fenpropimorph. Fenpropimorph is a member of the class of agricultural antifungals known as the morpholines. Fenpropimorph is thought to target both C-8 sterol isomerase (encoded by the ERG2 gene) and C-14 sterol reductase (encoded by the ERG24 gene) based on sterol analysis and in vivo data (5, 20, 21). In the optimal concentration window of 2.3 μM fenpropimorph, the HIP assay detects ERG24 but not ERG2 sensitivity (Fig. 5A). The evidence that the Erg2 protein is the target of fenpropimorph is conflicting; in vitro binding (22) and sterol analysis following compound exposure (20, 21) support Erg2 as a target, whereas the fact that in vivo overexpression of Erg2 does not confer resistance to fenpropimorph [whereas overexpression of Erg24 does (ref. 5 and data not shown)] argues against it. In addition, because the ERG2 homozygous deletion strain is extremely sensitive to fenpropimorph (data not shown) another target in the cell must be causing the observed growth inhibition. One scenario that is consistent with the published data is that Erg2 does indeed interact with fenpropimorph in vivo, but that the interaction with the essential ERG24 gene product is primarily responsible for the observed fungistatic activity of this compound.
Fig. 5.
A comparison of the HIP profiles of fenpropimorph (2.3 μM; A), alverine citrate (500 μM; B), and dyclonine (C; 500 μM). Axes shown are equivalent to those in Fig. 1. In each case, the five most significant strains are labeled. (Insets) Illustration of the compound structure. The chemical core structure shared between the three compounds is shown in red.
Comparison of Compound Structure May Predict Cellular Responses. Alverine citrate, an antispasmodic muscle relaxant, is used to treat irritable bowel syndrome. It is an anticholinergic and binds to the serotonin A1a receptor in cultured human colon cells (23). In the HIP assay, the ERG24 heterozygous strain is most sensitive to this drug, and the profile is very similar to that of fenpropimorph (Fig. 5B). In addition, wild-type yeast carrying a plasmid overexpressing either Erg24 or the human homolog LBR (blast E value × 10–78) shown to complement yeast ERG24 (6) confers resistance to alverine citrate (as is also true for fenpropimorph and dyclonine; data not shown). A third compound with a HIP profile similar to fenpropimorph and alverine citrate is the anesthetic dyclonine (Fig. 5C).
Because the profiles of dyclonine, fenpropimorph, and alverine citrate are very similar (Fig. 5), we compared their chemical structures. The structures revealed a common core structure shared between them (Fig. 5). It is noteworthy that both fenpropimorph and alverine citrate target Erg24 more strongly than dyclonine (Fig. 5) as the dyclonine core structure differs from that of fenpropimorph and alverine citrate. Dyclonine contains a ketone at one of the carbon chain positions. This double-bonded oxygen should limit dyclonine's conformational degrees of freedom and may account for the decreased sensitivity of the ERG24 heterozygous deletion strain to dyclonine compared with either alverine citrate or fenpropimorph. The finding that three such therapeutically distinct compounds with similar profiles share a chemical core structure suggests that the HIP assay may aid in the understanding of structure–activity relationships.
Discussion
Genome-wide profiling of diverse compounds demonstrates that this chemogenomic assay is specific in its ability to identify gene products that functionally interact with small molecules. Based on our results, we primarily identify gene products that (i) interact directly with small molecules and are dosage-limiting for growth and (ii) are involved in bioavailability of small molecules to cells. A key feature of the assay is its ability to assess the consequence of reducing the amount of gene product. Because the HIP assay interrogates heterozygous strains, it differs from most genetic screens that examine the phenotypic consequence of a complete gene deletion in the homozygous (or haploid) condition. By examining only complete deletions, it is difficult to discern the primary effect of compound and impossible to discern the effect of any essential gene. The heterozygous deletion strains allow the study of all ≈1,000 essential gene products simultaneously. Our results support the known small molecule interactions in several cases (methotrexate, azoles, and statins) and revealed previously unknown interactions in each study. For both methotrexate and the statins, pathway-related genes were uncovered that may identify novel drug discovery targets. Our study on 5-FU suggests that the primary mechanism of action is direct incorporation into the RNA. These studies also suggest a previously unknown interaction in one case (alverine citrate). The ability of the HIP assay to uncover subtleties of drug action was evidenced in the statins, where the effect of duplicated genes (HMG1 and HMG2) was realized because one of the genes (HMG1) is responsible for most of the activity in the cell. We also identified several genes involved in transport of small molecules. This set of genes varied widely from compound to compound, suggesting that there is no “best” gene to delete to increase the general drug sensitivity of yeast. Genes involved in transport are important to identify because they may provide clues to mechanisms of drug resistance. As is common in genome-wide experiments, we also identified genes of unknown function in every experiment, providing a rich resource for follow-up experiments. Unlike many genome-wide experiments, where it is difficult to judge which changes in gene activity have biological consequence, the readout of the HIP assay provides a ranking of the relative significance of each gene in the presence of a given compound.
Our results may have a positive impact on the drug discovery process. In the current paradigm, after a lead compound has been identified (typically) in vitro, it is a significant challenge to determine or predict the in vivo effects of such compounds once in the context of all protein targets in a cell. In general, the experimental options to determine in vivo effects are limited to studies involving animal models. A cost-effective, predictive screen such as the HIP assay may be a valuable tool for filtering and prioritizing compounds for further development.
The highly multiplexed nature of our assay permits simultaneous screening of all ≈6,000 gene products for small molecule interactions in a single experiment. By expanding the scope of our chemical libraries and taking advantage of more extensive automation, this in vivo assay may allow the identification of a small molecule inhibitor for every essential gene product in yeast. Because such chemical probes act as reversible conditional “mutants,” their identification would provide powerful chemical genetic tools for functional studies.
Supplementary Material
Acknowledgments
G.G. thanks M. Bard for many helpful discussions. This work was primarily supported by a grant from the National Cancer Institute/National Institute of Biomedical Imaging and Bioengineering and by a National Institutes of Health Alliance for Cellular Signaling GLUE grant (to A.P.A.).
Abbreviations: HIP, haploinsufficiency profiling; 5-FU, 5-fluorouracil; FD, fitness defect; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA.
References
- 1.Giaever, G., Shoemaker, D. D., Jones, T. W., Liang, H., Winzeler, E. A., Astromoff, A. & Davis, R. W. (1999) Nat. Genet. 21, 278–283. [DOI] [PubMed] [Google Scholar]
- 2.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 3.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 4.Guthrie, C. & Fink, G. R. (1991) in Methods in Enzymology, eds. Abelson, J. N. & Simon, M. I. (Academic Press, San Diego), vol. 194.
- 5.Lai, M. H., Bard, M., Pierson, C. A., Alexander, J. F., Goebl, M., Carter, G. T. & Kirsch, D. R. (1994) Gene 140, 41–49. [DOI] [PubMed] [Google Scholar]
- 6.Silve, S., Dupuy, P. H., Ferrara, P. & Loison, G. (1998) Biochim. Biophys. Acta 1392, 233–244. [DOI] [PubMed] [Google Scholar]
- 7.Ghahramani, Z. & Hinton, G. E. (1996) The EM Algorithm for Mixtures of Factor Analyzers, University of Toronto Technical Report CRG-TR-96-1.
- 8.Chabner, B. A., Ryan, D. P., Paz-Ares, L., Garcia-Carbonero, R. & Calabresi, P. (2001) Chemotherapy of Neoplastic Diseases (McGraw-Hill, New York).
- 9.Kaufman, S. & Fisher, D. B. (1972) Pterin-Requiring Aromatic Amino Acid Hydroxylases (Academic, New York).
- 10.Cole, S. P., Bhardwaj, G., Gerlach, J. H., Mackie, J. E., Grant, C. E., Almquist, K. C., Stewart, A. J., Kurz, E. U., Duncan, A. M. & Deeley, R. G. (1992) Science 258, 1650–1654. [DOI] [PubMed] [Google Scholar]
- 11.Zewail, A., Xie, M. W., Xing, Y., Lin, L., Zhang, P. F., Zou, W., Saxe, J. P. & Huang, J. (2003) Proc. Natl. Acad. Sci. USA 100, 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valens, M., Bohn, C., Daignan-Fornier, B., Dang, V. D. & Bolotin-Fukuhara, M. (1997) Yeast 13, 379–390. [DOI] [PubMed] [Google Scholar]
- 13.Scherf, U., Ross, D. T., Waltham, M., Smith, L. H., Lee, J. K., Tanabe, L., Kohn, K. W., Reinhold, W. C., Myers, T. G., Andrews, D. T., et al. (2000) Nat. Genet. 24, 236–244. [DOI] [PubMed] [Google Scholar]
- 14.Alberts, A. W. (1990) Cardiology 77, Suppl. 4, 14–21. [DOI] [PubMed] [Google Scholar]
- 15.Basson, M. E., Thorsness, M. & Rine, J. (1986) Proc. Natl. Acad. Sci. USA 83, 5563–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Servouse, M. & Karst, F. (1986) Biochem. J. 240, 541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truan, G., Epinat, J. C., Rougeulle, C., Cullin, C. & Pompon, D. (1994) Gene 142, 123–127. [DOI] [PubMed] [Google Scholar]
- 18.Turi, T. G. & Loper, J. C. (1992) J. Biol. Chem. 267, 2046–2056. [PubMed] [Google Scholar]
- 19.Dimster-Denk, D., Rine, J., Phillips, J., Scherer, S., Cundiff, P., DeBord, K., Gilliland, D., Hickman, S., Jarvis, A., Tong, L. & Ashby, M. (1999) J. Lipid Res. 40, 850–860. [PubMed] [Google Scholar]
- 20.Marcireau, C., Guilloton, M. & Karst, F. (1990) Antimicrob. Agents Chemother. 34, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baloch, R. I., Mercer, E. I., Wiggins, T. E. & Baldwin, B. C. (1984) Phytochemistry 23, 2219–2226. [Google Scholar]
- 22.Mulholland, J., Preuss, D., Moon, A., Wong, A., Drubin, D. & Botstein, D. (1994) J. Cell Biol. 125, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coelho, A. M., Jacob, L., Fioramonti, J. & Bueno, L. (2001) J. Pharm. Pharmacol. 53, 1419–1426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.