Abstract
The long flexuous virions of the Closteroviridae have a unique bipolar architecture incorporating two coat proteins, with most of the helical nucleocapsid encapsidated by the major coat protein (CP) and a small portion of one end encapsidated by the minor coat protein (CPm). It is not known whether CPm encapsidates the genomic RNA and, if so, which end and what effects transition between the two coat proteins. Two other virus-encoded proteins, an HSP70 homolog (HSP70h) and an ≈61-kDa protein, are required to augment virion assembly. In this work, we examine the in vivo encapsidation of Citrus tristeza virus by its CPm in the absence of CP. In the absence of other assembly-related proteins, CPm protected a family of 5′ coterminal RNAs, apparently because of pausing at different locations along the genomic RNA. Most of the nucleocapsids formed by CPm were short, but a few were full-length and infectious. Mutations within the 5′ nontranslated region demonstrated that the CPm origin of assembly overlaps the previously described conserved stem-and-loop structures that function as a cis-acting element required for RNA synthesis. Thus, in the absence of CP, the CPm encapsidation is initiated from the 5′ end of the genomic RNA. Coexpression of HSP70h and the p61 protein with CPm in protoplasts restricted encapsidation to the 5′ ≈630 nucleotides, which is close to the normal boundary of the bipolar virion, whereas the presence of either HSP70h or the p61 protein alone did not limit encapsidation by CPm.
The principle functions of the virion are to protect the virus genome and to facilitate its transmission from host to host or within the host. Viruses of animals have a general requirement for a precise interaction between the virion and the host cell membrane to allow entry. Thus, the virion might have specific proteins to provide that interaction. A requirement for many plant viruses is a precise interaction with a vector: arthropod, nematode, or fungus. Some viruses have both animal and plant hosts. Virions range from simple, consisting of only one coat protein, which self-assembles with the viral RNA, to extraordinarily complex, involving multiple structural proteins along with numerous accessory proteins, which may be scaffolding proteins or chaperones (1, 2). Although virions exhibit a wide range of structural diversity, size, composition, and complexity, nucleocapsids, in general, are of two basic types of symmetry: helical or icosahedral. Many plant viruses have virions consisting of helical nucleocapsids, which are open structures, usually consisting of a single-coat protein with regular stacking around a nucleic acid in a constant relationship of amplitude and pitch. Helical virions can be rigid or flexible, depending on the coat protein, and virion lengths are determined by the length of the nucleic acid molecule.
The long flexuous virions of Citrus tristeza virus (CTV) and other viruses of the Closteroviridae family have a helical architecture consisting of two coat proteins. The major coat protein (CP) encapsidates most (>95%) of the virion (the “body”) in a manner similar to that of other helical viruses, but the second (minor) coat protein (CPm) encapsidates the other end (the “tail”), resulting in “rattlesnake-like” virions (3–5). It is not known which end of the genome is encapsidated by CPm. The location of the tail was examined previously by shearing virions of the related Beet yellows virus (BYV) and by using antibodies to CPm to enrich broken particles that appeared to have more 5′ sequences (6), but these results were not conclusive. In fact, it is not resolved whether CPm encapsidates any RNA. Alzhanova et al. (7) proposed a model for BYV in which CP encapsidates the entire genomic RNA, whereas the CPm tail portion encapsidates no RNA. If CPm does encapsidate one end of the genomic RNA, no understanding of what defines the transition between CPm and CP exists.
CTV has a positive-stranded RNA genome of ≈20 kb with the ≈5′ half encoding replicase-associated domains that are expressed by a polyprotein. The internally positioned genes, which are not required for replication and are thought to be involved in host interactions, virion assembly, vector specificity, and movement within the plant, are expressed from 3′ coterminal subgenomic mRNAs (8–10). The in vitro assembly of CTV has not been developed. By genetically manipulating the virus in Nicotiana benthamiana protoplasts, we previously demonstrated that two additional virus-encoded proteins, in addition to the two coat proteins, are required for efficient assembly of CTV virions, apparently by functioning together (11). One is a homolog of the ubiquitous HSP70 proteins (HSP70h), which has chaperone-like characteristics, and the other is a 61-kDa protein. Homologs of these two proteins of related closteroviruses have been found to be tightly attached to virions (5, 12, 13), but it is not known whether they augment the assembly process or whether they are minor structural components. Additionally, we found that two conserved stem-and-loop (SL) structures within the 5′ nontranslated region (NTR) (14), which are cis-acting elements necessary for replication, also appear to be involved in virion formation (15).
In this study, we continued dissection of the virion assembly process by examining the encapsidation of a minimal CTV replicon containing the CPm gene in the absence of CP, with or without HSP70h and/or p61. We show that, at least in the absence of CP, the tail covers the 5′ end of the genome and that initiation of assembly by the CPm occurs within the 5′ NTR, in a region that apparently overlaps the previously characterized replication-specific cis-acting element. The CPm in the absence of the other assembly-related proteins was able to encapsidate and protect the entire genomic RNA, albeit at reduced levels. However, the combination of HSP70h, p61, and CPm restricted encapsidation to the 5′ ≈630 nucleotides.
Materials and Methods
Construction of Plasmids. The full-length cDNA clone of CTV, pCTV9R (16, 17), was the basis of all constructs. The nucleotide numbering and sequences of the primers used in this study are according to GenBank (accession no. AY170468) (17). DNA fragments consisting of nucleotides 15115–16058, 13537–16058, 11658–16058, or a DNA fragment comprising nucleotides 11658–13813 and 15115–16058 in tandem, were amplified with forward and reverse primers containing flanking XhoI and PstI restriction sites, digested with XhoI and PstI, and ligated into a similarly digested CTV-p23 replicon (18) to obtain CTV-CPm, CTV-p61-CPm, CTV-HSP70h-p61-CPm, and CTV-HSP70h-CPm, respectively. An additional nonviral cytidylate was introduced at nucleotide 15383 by using overlap extension PCR (19) to disrupt the reading frame of CPm ORF in CTV-CPm to obtain CTV-CPmFS. The CTV-CP replicon containing the CP ORF under its own controller element plus the p23 ORF has been described (18).
The 5′ NTR mutations in the CTV-ΔCla replicon (15) were transferred to CTV-CPm and CTV-CP by removing the SphI (at nucleotide 1660) to NotI (engineered at the 3′ end; ref. 16) fragment, which was ligated into the similarly digested pCTV-ΔCla constructs containing the 5′ NTR mutations (15). The mutations were confirmed by nucleotide sequencing.
In Vitro Transcription, Inoculation of Protoplasts, and Northern Blot Analysis of RNAs. Capped (m7G) in vitro-produced RNA transcripts were used for polyethylene glycol-mediated transfections of N. benthamiana leaf protoplasts (≈1 × 106) as described (16). Protoplasts were harvested at 4 days after inoculation (dpi) to isolate total RNA or to purify nucleocapsids. RNA was analyzed by Northern blot hybridizations by using four different riboprobes, positive- and negative-stranded RNA-specific probes corresponding to the 5′ or 3′ ends of CTV (16, 20), and quantified by scanning and densitometry by using OS-SCAN (Oberlin Scientific, Oberlin, OH). All experiments were repeated three to five times with each replicon.
Isolation and Analysis of Nucleocapsid-Like Particles. Several batches (6–14) of protoplasts were combined; the frozen and thawed protoplast pellets were suspended in 40 mM sodium phosphate buffer, pH 8.2, and clarified by centrifugation at 4,000 rpm for 5 min. The pooled supernatants (10 ml) were layered on a gradient of 1 ml of 70% sucrose and 0.25 ml each of 50% and 20% sucrose in sodium phosphate buffer and were centrifuged at 200,000 × g at 4°C for 75 min in a SW41 rotor. The upper 0.5 ml of 70% sucrose was collected and used to visualize nucleocapsids by electron microscopy or for protein or RNA analyses.
Alternatively, the clarified sap (7 ml) from 4 dpi protoplasts was layered on cesium sulfate-sucrose step gradients (1 ml each of 0%, 15%, 22.5%, and 30% cesium sulfate in 10% sucrose) and was centrifuged at 200,000 × g for 2.5 h at 8°C in a SW41 rotor. The bottom 0.95 ml was discarded, and the next ≈1.5 ml was collected and diluted to 2.5 ml with sodium phosphate buffer, layered on a preformed cesium sulfate-sucrose density gradient (3 ml each of 15%, 22.5%, and 30% sucrose in 10% cesium sulfate), and centrifuged at 200,000 × g for 2.5 h at 8°C in a SW41 rotor. The bottom 3.0 ml were discarded, and the nucleocapsids present in the next 4.0 ml were diluted to 6 ml with sodium phosphate buffer, then precipitated with 4% polyethylene glycol 8000 in the presence of 0.2 N sodium chloride on ice for 1 h, and centrifuged at 15,000 rpm for 15 min at 4°C.
The passage of progeny virions in crude sap between protoplasts, the serological specific electron microscopy, immunogold labeling of CPm virions, and Western immunoblots were described (11, 21, 22).
Mapping the 3′ End of the RNA Encapsidated by CPm. The RNA isolated from nucleocapsids purified by cesium sulfate-sucrose density gradient centrifugation was denatured at 90°C, polyadenylated at the 3′ terminus, cloned as described (23), and sequenced at the Interdisciplinary Center for Biotechnology Research DNA sequencing core facility of the University of Florida (Gainesville).
Results
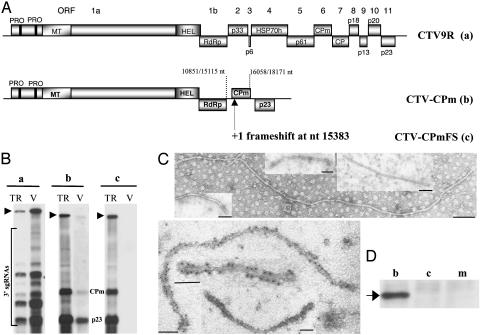
CPm Can Protect the Entire Genomic RNA. In the near-full-length virus, CPm in the presence of both HSP70h and p61 did not assemble detectable amounts of full-length nucleocapsids nor facilitate passage to a new set of protoplasts (11). Here, we examined the ability of CPm to form nucleocapsids in the absence of other assembly-related proteins. For this purpose, we used a minimal replicon, CTV-p23 (18), which contains the 5′ NTR, ORFs 1a and 1b (replicase genes), the p23 gene, which is required to inhibit minus-stranded RNA synthesis so that sufficient single-stranded RNA is produced as substrate for assembly, and finally the 3′ NTR. This replicon was designed to express different combinations of assembly-associated genes to be assayed for nucleocapsid production. CPm was expressed by replicon CTV-CPm that had the CPm gene inserted between ORF 1b and the p23 gene (Fig. 1Ab). CTV-CPmFS, which contained a +1 frameshift mutation in CPm ORF, was a negative control (Fig. 1 Ac). In vitro-produced RNA transcripts of CTV-CPm, CTV-CPmFS, or wild-type CTV (CTV9R) (Fig. 1 Aa; ref. 17) were used to inoculate N. benthamiana protoplasts, and their ability to form nucleocapsids was examined at 4 dpi.
Fig. 1.
(A) Genomic organization of wild-type CTV (CTV9R) (a) and replicons CTV-CPm (b) and CTV-CPmFS (c). The open boxes represent ORFs and their translation products. PRO, papain-like protease; MT, methyltransferase; HEL, helicase; RdRp, RNA-dependent RNA polymerase. (B) Northern blot hybridizations, probed with a 3′ positive-stranded RNA-specific probe, of RNAs from protoplasts inoculated with in vitro produced transcripts (TR) or progeny nucleocapsids (V) in crude sap by CTV9R (a), CTV-CPm (b), or CTV-CPmFS (c). Arrowheads indicate the position of genomic RNA. Wild-type CTV9R 3′ sgRNAs are indicated on the left. The position of CPm and p23 sgRNAs of replicons are shown. (C) Negative-stained electron micrographs of CTV-CPm large and small nucleocapsids (Upper) and nucleocapsids treated with CPm-specific antibodies and decoration with 10-nm gold particles (Lower). (Bar = 100 nm.) (D) Western immunoblot analyses of proteins of partially purified nucleocapsids from protoplasts transfected with CTV-CPm (lane b), CTV-CPmFS (lane c), or mock-inoculated protoplasts (lane m). Arrow indicates the position of CPm-specific protein band.
CTV-CPm nucleocapsids, partially purified by centrifugation through a 70% sucrose cushion and examined by serological specific electron microscopy, were mostly short particles ranging from ≈70 to ≈800 nm in length, plus a few larger particles with a length (≈1,400 nm) expected for that of the complete CTV-CPm genomic RNA (Fig. 1C Upper). We did not find any virus particles from CTV-CPmFS-infected protoplasts, suggesting that the particles observed were produced by the encapsidation of different lengths of the replicon RNA by CPm. Immunogold labeling using a CPm-specific polyclonal antiserum decorated the surface of both the small and the large particles (Fig. 1C Lower), confirming that the particles were constituted by CPm. Protein from the partially purified nucleocapsids from CTV-CPm also strongly reacted with CPm antiserum in Western immunoblots (Fig. 1D, lane b), but not with CP antiserum (data not shown). Nucleocapsid protein extracts from CTV-CPmFS and mock-inoculated protoplasts did not react with CPm antiserum (Fig. 1D, lanes c and m).
We previously developed a rigorous assay for virions (infectious nucleocapsids) by examining their ability to be passaged in crude sap from one set of protoplasts to another set (11). This assay was based on two observations. First, the free unencapsidated RNA was degraded under the conditions of incubation in crude extracts. Second, virions are ≈10,000 times more infectious to protoplasts than free viral RNA, apparently because of their ability to be inserted into protoplasts (22). Incomplete nucleocapsids failed to be passaged under these conditions (11). At 4 dpi, the progeny nucleocapsids were extracted as crude sap from the infected protoplasts and then transfected to a new set of protoplasts. Fig. 1B shows a Northern blot hybridization demonstrating the levels of replication in protoplasts infected by the in vitro produced RNA transcripts compared with their progeny nucleocapsids. The progeny nucleocapsids from the wild-type virus (CTV9R) were efficiently passaged to the next set of protoplasts (Fig. 1Ba). The progeny nucleocapsids from the CTV-CPm replicon also were passaged to the next set of protoplasts, although at much reduced levels (Fig. 1Bb) apparently due to a reduced number of protoplasts infected by the few full-length nucleocapsids formed by CPm. Yet, these data demonstrate that some nucleocapsids formed by only CPm were infectious. No passage of CTV-CPmFS to the next set of protoplasts was detectable (Fig. 1Bc).
Taken together, these data suggest that the CPm, in the absence of other assembly-related proteins, was able to form a few full-length, infectious nucleocapsid (replicon size), but most particles were smaller nucleocapsids of various sizes.
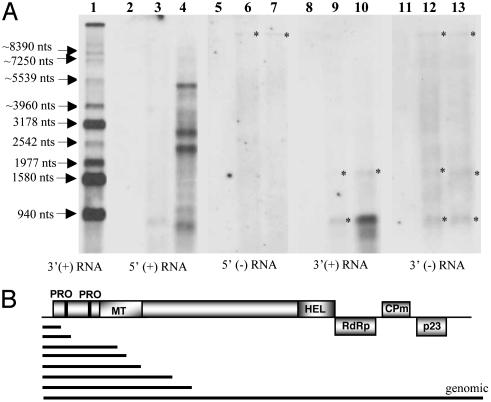
CPm Initiates Encapsidation from the 5′ End of the Genome. CTV produces large quantities of double-stranded RNAs (18) that contaminate nucleocapsid preparations after sucrose density gradient centrifugation, obscuring detection of encapsidated RNAs. To analyze encapsidated RNAs, we had to adapt a system that stringently purifies nucleocapsids from unencapsidated RNAs, particularly double-stranded RNAs. We found that two cycles of cesium sulfate-sucrose density gradient centrifugation effectively eliminated unencapsidated RNAs from nucleocapsids. The nucleocapsid fractions from the cesium sulfate-sucrose gradient centrifugations of CTV-CPm-infected protoplasts were analyzed for encapsidated RNAs by Northern blot hybridization with positive- or negative-stranded RNA-specific probes corresponding to the 5′ or 3′ ends of CTV (Fig. 2A). The 5′- or 3′-specific negative-stranded RNA probe did not hybridize with any small RNAs that would correspond with the observed small nucleocapsids. An RNA of the size of the p23 subgenomic RNA (sgRNA) hybridized with the 3′ positive-stranded RNA-specific probe suggesting that a small amount of this abundant 3′-terminal sgRNA was encapsidated by CPm (Fig. 2 A, lane 10). However, most of the RNA found occurred as a ladder of RNAs that hybridized with the 5′ positive-stranded RNA-specific probe (Fig. 2 A, lane 4). Because these RNAs have a common 5′ end, the data suggest that the CPm, in the absence of other assembly-related proteins, initiated encapsidation from the 5′ end of the replicon RNA but paused at different locations during the assembly process (Fig. 2B). The data also suggest that the 3′ regions of the replicon RNAs remained unencapsidated and were degraded during the extensive purification process.
Fig. 2.
(A) Northern blot hybridization analyses of RNAs from nucleocapsids purified from protoplasts at 4 dpi with no RNA (lanes 2, 5, 8, and 11), CTV-CPmFS (lanes 3, 6, 9, and 12), or CTV-CPm (lanes 4, 7, 10, and 13) hybridized with strand-specific riboprobes. Wild-type CTV 3′ sgRNAs (lane 1) are size markers on the left. Copurified double-stranded RNAs are indicated with asterisks. (B) Schematic diagram of CTV-CPm showing various sized 5′-coterminal RNAs encapsidated by CPm.
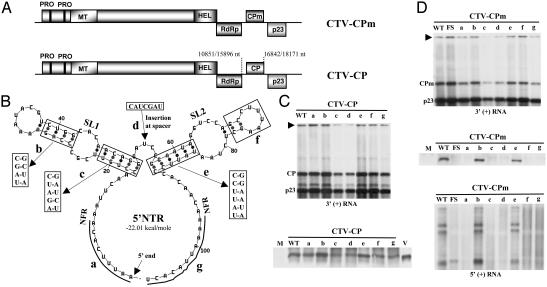
The Origin of Assembly of CPm Is Located in the 5′ NTR. The 107 nucleotides of the 5′ NTR are folded by mfold (24) into two stem-loop structures (SL1 and SL2) separated by a 5-nt spacer region and nonfolding regions on both sides (ref. 15 and Fig. 3B). Using mutational analysis of the 5′ NTR, we found that these secondary structures functioned as a cis-acting element required for replication (15). However, certain mutations in the 5′ NTR that allowed replication did not allow formation of virions capable of being passaged from protoplast to protoplast (15). The previous results plus those presented here, demonstrating that encapsidation involves the 5′ end, led us to examine the effect of these 5′ NTR mutations on initiation of nucleocapsid formation by CPm. We recreated in CTV-CPm seven of the mutations previously found that allowed continued replication at 40–100% of wild-type levels (Fig. 3B; ref. 15). These mutations included deletion of the 5′ 9 nucleotides (mutant a); compensatory mutations in the top (mutant b) and bottom (mutant c) of SL1 and the bottom of SL2 (mutant e); insertion of seven nonviral nucleotides at the spacer (mutant d); deletion of the top portion of SL2 (mutant f); and deletion of nucleotides 97–107 between the structure and the start codon of ORF 1a (mutant g) (Fig. 3B). CTV-CPmFS was the negative control. Mutants a, b, e, and f replicated in N. benthamiana protoplasts, accumulating RNAs at ≈60–100% of the levels of wild-type CTV-CPm, with mutants c, d, and g accumulating at lower levels of ≈40–50% (Fig. 3D Top). Nucleocapsids were partially purified by centrifugation through a 20% sucrose cushion and the Northern hybridization blots demonstrated that only mutants b and e encapsidated the 5′ coterminal RNAs (Fig. 3D Bottom). Also, Western immunoblot analyses using CPm-specific polyclonal antiserum confirmed that CPm nucleocapsids were formed only by mutants b and e (Fig. 3D Middle). Deletions in 5′ and 3′ nonfolding regions (mutants a and g), mutations in the lower part of SL1 (mutant c), insertion at the spacer region (mutant d), and deletion of SL2 top portion (mutant f) prevented encapsidation of the viral RNA by CPm. The compensatory mutations in the top portion of SL1 (mutant b) and bottom portion of SL2 (mutant e) did not prevent encapsidation by CPm. These results suggested that the origin of assembly sequence for the encapsidation of RNA by CPm is located in the 5′ NTR.
Fig. 3.
The effect of 5′ NTR mutations on encapsidation by CPm and CP. (A) Schematic map of CTV-CPm and CTV-CP. (B) The predicted secondary structure of the 5′ NTR of CTV T36 showing the SL structures, SL1 and SL2, separated by a 5-nt spacer and nonfolding regions (NFR) on both sides. Mutations a, f, and g are deletions; mutations b, c, and e are compensatory mutations; and mutation d is an insertion. Accumulation of 3′ positive-stranded RNAs from 5′ NTR mutants (a–g) of CTV-CP (C Upper) or CTV-CPm (D Top). Western immunoblot analyses of proteins from partially purified nucleocapsids from 5′ NTR mutants of CTV-CP (C Lower) or CTV-CPm (D Middle) are shown. Analyses of encapsidated 5′ coterminal RNAs from 5′ NTR mutants of CTV-CPm (D Bottom). Arrowheads indicate the position of genomic RNA. The position of CP, CPm, and p23 sgRNAs are shown in Northern blots. M, mock-inoculated protoplasts; WT, wild-type replicon; V, virions from full-length wild-type CTV; FS, CTV-CPmFS.
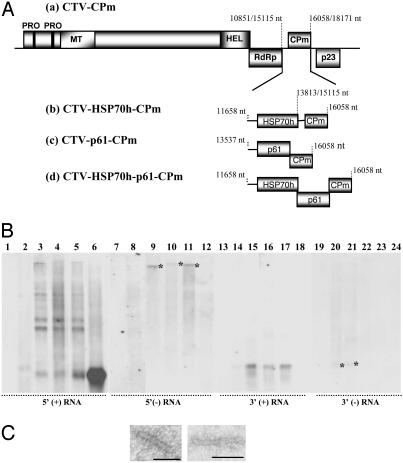
The Combination of HSP70h and p61 Restricts CPm Encapsidation to the 5′ End. We next examined the effects of HSP70h and/or the p61 protein on encapsidation by CPm. The HSP70h and p61 genes, independently (CTV-HSP70h-CPm or CTV-p61-CPm) or together (CTV-HSP70h-p61-CPm), were ligated into CTV-CPm and examined for nucleocapsid production (Fig. 4A). Nucleocapsids from protoplasts at 4 dpi were purified through two cycles of cesium sulfate-sucrose density gradient centrifugation; RNAs were extracted and analyzed by using 5′ or 3′ positive- and negative-stranded RNA-specific probes.
Fig. 4.
The combination of HSP70h and p61 along with CPm restricts encapsidation to 5′≈650 nucleotides of genomic RNA. (A) Schematic diagram of replicons. (B) Northern blot hybridizations of CPm-encapsidated RNAs purified from protoplasts at 4 dpi with no RNA (lanes 1, 7, 13, and 19), CTV-CPmFS (lanes 2, 8, 14, and 20), CTV-CPm (lanes 3, 9, 15, and 21), CTV-HSP70h-CPm (lanes 4, 10, 16, and 22), CTV-p61-CPm (lanes 5, 11, 17, and 23), or CTV-HSP70h-p61-CPm (lanes 6, 12, 18, and 24) by using strand-specific riboprobes. Asterisks indicate double-stranded RNAs copurified with nucleocapsids. (C) Electron micrographs of representative negative-stained nucleocapsid-like particles from CTV-HSP70h-p61-CPm replicon. (Bar = 50 nm.)
The CPm replicons containing either HSP70h (CTV-HSP70h-CPm) or p61 (CTV-p61-CPm) produced particles containing a ladder of positive-stranded RNAs that hybridized with the 5′ positive-stranded RNA-specific probe, similar to that of the CTV-CPm replicon (Fig. 4B, lanes 3–5). A band corresponding to p23 sgRNA also was detected by the 3′ positive-stranded RNA-specific probe (Fig. 4B, lanes 15–17). However, the replicon containing the combination of HSP70h and p61 plus CPm (CTV-HSP70h-p61-CPm) produced particles with a different RNA profile. Only one RNA species of ≈650 nucleotides was found that hybridized with the 5′ positive-stranded RNA-specific probe (Fig. 4B, lane 6). Additionally, the 3′ positive-strand-specific p23 sgRNA was not found (Fig. 4B, lane 18). When visualized by serological specific electron microscopy, CTV-HSP70h-p61-CPm produced small uniform particles of ≈70 nm in length (Fig. 4C). These data suggest that, in the absence of CP and the presence of HSP70h, p61, and CPm, encapsidation was restricted to the 5′ ≈650 nucleotides of genomic RNA.
Western immunoblot analysis of purified nucleocapsids revealed that only a slight increase occurred in the total amount of CPm associated with particles from the replicon containing CPm plus both HSP70h and p61 compared with that of CPm replicons with CPm alone or CPm with either HSP70h or p61 (data not presented). This marginal increase in assembled CPm in the presence of both HSP70h and p61, compared with the ≈50- to 70-fold increase in the amount of the small 5′ RNA (≈650 nucleotides) that was encapsidated, was probably due to CPm encapsidating extended areas of fewer RNAs in the absence of both HSP70h and p61.
CPm Encapsidates the 5′ ≈630 Nucleotides of the Genomic RNA. The replicon CTV-HSP70h-p61-CPm encapsidated large amounts of a relatively uniform RNA of ≈650 nucleotides (Fig. 4B, lane 6). We isolated these encapsidated RNAs from nucleocapsids purified by two cycles of cesium sulfate-sucrose density gradient centrifugation from CTV-HSP70h-p61-CPm-infected protoplasts to determine their 3′ termini. The RNA was poly(A)-tailed, reverse-transcribed, PCR-amplified, and cloned. Restriction enzyme digestion resulted in two uniform sizes of inserts: about 80% large and 20% small. We sequenced 15 clones with the larger insert and 5 clones with the smaller insert. Ambiguities resulted when the apparent terminus was adjacent to a 3′ adenylate residue in the CTV sequence, because we could not distinguish whether the adenylate(s) resulted from the sequence or the poly(A) tail. Nine of the large inserts had identical 3′ termini at nucleotides 627, 628, or 629 with six at nucleotides 630 or 631 (Fig. 5). Three of the small inserts terminated after nucleotide 611 and one each after nucleotide 612 and nucleotide 613 or 614 (Fig. 5). Thus, encapsidation of the genomic RNA in the presence of CPm, HSP70h, and p61 stopped near nucleotides 627–631, occasionally stopping at nucleotides 611–614. It should be noted that both stopping points occur near a series of four cytidylates (CCCC) (Fig. 5).
Fig. 5.
Mapping the 3′ termini of RNAs encapsidated by CPm in the presence of HSP70h and p61 proteins. Arrows show the number of clones terminated at different locations in the genomic RNA sequence. Brackets indicate the possible alternate 3′ termini due to the presence of adenylates after nucleotides 613, 627, and 630.
The 5′ NTR Mutations Do Not Affect Assembly by CP. To examine the effect of 5′ NTR mutations on the translatability of the inserted gene, the stability of the RNAs, and their effect on encapsidation by CP, we introduced the 5′ NTR mutations (a–g) into CTV-CP (Fig. 3A). The CP, in the absence of the other assembly-associated proteins, encapsidates the CTV-CP replicon by forming various sizes of nucleocapsids (T.S. and W.O.D., unpublished data). The 5′ NTR mutants of CTV-CP accumulated levels of progeny RNAs similar to that of the corresponding 5′ NTR mutants of CTV-CPm (Fig. 3C Upper). Mutants a, b, e, f, and g accumulated CP in particles at levels approximately similar to that of wild-type CTV-CP (Fig. 3C Lower), whereas mutants c and d accumulated levels of CP in nucleocapsids at ≈40–50% of the wild-type levels, generally in proportion to their levels of replication (Fig. 3C Lower). Moreover, the progeny nucleocapsids in crude extracts from the 5′ NTR mutants of CTV-CP were successfully passaged to the next set of protoplasts (data not presented), demonstrating that the 5′ NTR mutations did not appreciably affect CP encapsidation.
Discussion
We showed that CPm, at least in the absence of CP, does encapsidate genomic RNA, the tail (perhaps it should be called the “head”) occurs at the 5′ end, and CPm initiates encapsidation within the 5′ NTR. The 5′ halves of different CTV isolates vary considerably in sequence similarity, in particular, within the 5′ NTR where similarities are as low as 42% (25). Yet, it was noticed that all strains were predicted to retain two similar adjacent SL structures (14), in particular, in the positive strands (15). We examined these putative SL structures and found that retention of the structures was important for the ability of the RNA to be replicated, but this cis-acting element tolerated considerable sequence modification (15). Here we found that the element required to initiate assembly of the CPm overlaps this area. Deletions beyond each side of the replication-dependent cis-acting element in nonfolded areas prevented initiation of encapsidation by CPm as did some mutations within the double SL region. Several of the compensatory mutations with sequence changes that allowed replication to continue prevented nucleocapsid assembly. Thus, the CPm origin of assembly appears to be larger and more sequence-specific than the overlapped replication element.
The combination of HSP70h and p61 along with CPm dramatically stopped encapsidation between nucleotides 611 and 631. The BYV HSP70h has been shown to have some in vitro biochemical characteristics of chaperones. The N-terminal domain was shown to have ATPase activity as expected for an HSP70 chaperone, but its C-terminal region did not bind to unfolded proteins (26). Additionally, mutation of the putative ATPase active site inhibited CTV virion assembly (11). Thus, one possible function of the HSP70h would be a chaperone function, perhaps to assist in the proper folding of one or both coat proteins. Cellular HSP70s have been implicated in the assembly of diverse groups of animal viruses (27–31), yet homologs of HSP70h and p61 have been shown to be tightly attached to virions of other members of the Closteroviridae family (5, 12, 13). Does this suggest that they function as minor structural proteins rather than enhancing the assembly process? It is possible that one or both proteins of HSP70h and p61 bind to the transition zone to prevent progression of encapsidation by CPm.
We previously examined assembly of CTV virions by making mutations in 3′ genes and identified four genes needed for efficient production of nucleocapsids (11). Using the full-length virus with the deletion of the CP gene (leaving the CPm, HSP70h, and p61 genes intact) resulted in only a few small particles. That result is consistent with the small nucleocapsids produced by the replicon expressing CPm, HSP70h, and p61 proteins. It is possible that nucleocapsid assembly by these three proteins, in the absence of CP, differs from wild-type virion assembly. However, the efficiency of formation of these short particles and their approximation to the size of the tail of wild-type virions (3, 4) suggest that this subset of encapsidation reflects the normal bipolar virion assembly.
So, what is the advantage of the 5′ end of the genome being encapsidated by a different coat protein? What function does this architecture provide that is not provided by other helical virions composed of only one coat protein? It is common for cell-to-cell and/or long-distance movement of plant viruses to require the ability of virion formation. The tail structure of BYV has been proposed as a “specialized device” required for movement functions (7, 13). However, it is unclear how a minor coat protein on the 5′ end of the genome would affect movement. These bipolar helical virions are transmitted by insect vectors in a semipersistent manner (32). Perhaps the tail has specific affinity for the mouth parts of vectors (5). Another possibility is that the tail assists in disassembly and initiation of infection. The two coat proteins of helical viruses could allow different regions of the virions to have different states of equilibrium: one coat protein could bind more tightly, providing more stability of the virion in different environments, whereas the coat protein at the 5′ end of the genome could be looser, allowing easy initiation of translation after entering a cell. An understanding of the closterovirus virion should allow us to better understand the biology of this unique group of viruses.
Acknowledgments
We thank John Cook for excellent technical assistance, Diann Achor for help with the electron microscope, and Dr. Moshe Bar-Joseph (The Volcani Center, Bet-Dagan, Israel) for suggesting the cesium sulfate-sucrose gradient purification procedure. This research was supported in part by an endowment from the J. R. and Addie S. Graves family and by grants from the Florida Citrus Production Research Advisory Board, the National Citrus Research Council, the U.S.–Israel Binational Agriculture Research and Development Fund, U.S. Department of Agriculture/Agricultural Research Service Cooperative Agreement, the U.S. Department of Agriculture/National Research Initiative Competitive Grants Program, and the Florida Agricultural Experiment Station, and it was approved for publication as Journal Series No. R-09827.
Abbreviations: CP, major coat protein; CPm, minor coat protein; CTV, Citrus tristeza virus; BYV, Beet yellows virus; NTR, nontranslated region; dpi, days after inoculation; SL, stem-and-loop; sgRNA, subgenomic RNA.
References
- 1.Dokland, T. (1999) Cell Mol. Life Sci. 56, 580–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan, C. S. & Pipas, J. M. (2001) Virology 287, 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Agranovsky, A. A., Lesemann, D. E., Maiss, E., Hull, R. & Atabekov, J. G. (1995) Proc. Natl. Acad. Sci. USA 92, 2470–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Febres, V. J., Ashoulin, L., Mawassi, M., Frank, A., Bar-Joseph, M., Manjunath, K. L., Lee, R. F. & Niblett, C. (1996) Phytopathology 86, 1331–1335. [Google Scholar]
- 5.Tian, T., Rubio, L., Yeh, H.-H., Crawford, B. & Falk, B. W. (1999) J. Gen. Virol. 80, 1111–1117. [DOI] [PubMed] [Google Scholar]
- 6.Zinovkin, R. A., Jelkmann, W. & Agranovsky, A. A. (1999) J. Gen. Virol. 80, 269–272. [DOI] [PubMed] [Google Scholar]
- 7.Alzhanova, D. V., Napuli, A. J., Creamer, R. & Dolja, V. V. (2001) EMBO J. 20, 6997–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pappu, H. R., Karasev, A. V., Anderson. E. J., Pappu, S. S., Hilf, M. E., Febres, V. J., Eckloff, R. M. G., McCaffery, M., Boyko, V., Gowda, S., et al. (1994) Virology 199, 35–46. [DOI] [PubMed] [Google Scholar]
- 9.Hilf, M. E., Karasev, A. V., Pappu, H. R., Gumpf, D. J., Niblett, C. L. & Garnsey, S. M. (1995) Virology 208, 576–582. [DOI] [PubMed] [Google Scholar]
- 10.Karasev, A. V., Boyko, V. P., Gowda. S., Nikolaeva, O. V., Hilf, M. E., Koonin, E. V., Niblett, C. L., Cline, K., Gumpf, D. J., Lee, R. F., et al. (1995) Virology 208, 511–520. [DOI] [PubMed] [Google Scholar]
- 11.Satyanarayana, T., Gowda, S., Mawassi, M., Albiach-Martí, M. R., Ayllón, M. A., Robertson, C., Garnsey, S. M. & Dawson, W. O. (2000) Virology 278, 253–265. [DOI] [PubMed] [Google Scholar]
- 12.Napuli, A. J., Falk, B. W. & Dolja, V. V. (2000) Virology 274, 232–239. [DOI] [PubMed] [Google Scholar]
- 13.Napuli, A. J., Alzhanova, D. V., Doneanu, C. E., Barofsky, D. F., Koonin, E. V. & Dolja, V. V. (2003) J. Virol. 77, 2377–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López, C., Ayllón, M. A., Navas-Castillo, J., Guerri, J., Moreno, P. & Flores, R. (1998) Phytopathalogy 88, 685–691. [DOI] [PubMed] [Google Scholar]
- 15.Gowda, S., Satyanarayana, T., Ayllón, M. A., Moreno, P., Flores, R. & Dawson, W. O. (2003) Virology 317, 50–64. [DOI] [PubMed] [Google Scholar]
- 16.Satyanarayana, T., Gowda, S., Boyko, V. P., Albiach-Martí, M. R., Mawassi, M., Navas-Castillo, J., Karasev, A. V., Dolja, V., Hilf, M. E., Lewandowski, D. J., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 7433–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satyanarayana, T., Gowda, S., Ayllón, M. A. & Dawson, W. O. (2003) Virology 313, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satyanarayana, T., Gowda, S., Ayllón, M. A., Albiach-Martí, M. R., Rabindran, S. & Dawson, W. O. (2002) J. Virol. 76, 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 20.Gowda, S., Satyanarayana, T., Ayllón, M. A., Albiach-Martí, M. R., Mawassi, M., Rabindran, S., Garnsey, S. M. & Dawson, W. O. (2001) Virology 286, 134–151. [DOI] [PubMed] [Google Scholar]
- 21.Derrick, K. S. & Brlansky, R. H. (1976) Phytopathology 66, 815–819. [Google Scholar]
- 22.Satyanarayana, T., Bar-Joseph, M., Mawassi, M., Albiach-Martí, M. R., Ayllón, M. A., Gowda, S., Hilf, M. E., Moreno, P., Garnsey, S. M. & Dawson, W. O. (2001) Virology 280, 87–96. [DOI] [PubMed] [Google Scholar]
- 23.Ayllón, M. A., Gowda, S., Satyanarayana, T., Karasev, A. V., Adkins, S., Mawassi, M., Guerri, J., Moreno, P. & Dawson, W. O. (2003) J. Virol. 77, 9232–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 25.Albiach-Martí, M. R., Mawassi, M., Gowda, S., Satyanarayana, T., Hilf, M. E., Shanker, S., Almira, E. C., Vives, M. C., López, C., Guerri, J., et al. (2000) J. Virol. 74, 6856–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agranovsky, A. A., Folimonova, S. Y., Folimonov, A. S., Denisenko, O. N. & Zinovkin, R. A. (1997) J. Gen. Virol. 78, 535–542. [DOI] [PubMed] [Google Scholar]
- 27.Macejak, D. G. & Sarnow, P. (1992) J. Virol. 66, 1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jindal, S. & Young, R. A. (1992) J. Virol. 66, 5357–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cripe, T. P., Delos, S. E., Estes, P. A. & Garcea, R. L. (1995) J. Virol. 69, 7807–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, J., Toft, D. O. & Seeger, C. (1997) EMBO J. 16, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chromy, L. R., Pipas, J. M. & Garcea, R. L. (2003) Proc. Natl. Acad. Sci. USA 100, 10477–10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karasev, A. V. (2000) Annu. Rev. Phytopathol. 38, 293–324. [DOI] [PubMed] [Google Scholar]