Abstract
Cellulase enzymes (endoglucanases, cellobiohydrolases, and β-glucosidases) hydrolyze cellulose into component sugars, which in turn can be converted into fuel alcohols1. The potential for enzymatic hydrolysis of cellulosic biomass to provide renewable energy has intensified efforts to engineer cellulases for economical fuel production2. Of particular interest are fungal cellulases3-8, which are already being used industrially for foods and textiles processing.
Identifying active variants among a library of mutant cellulases is critical to the engineering process; active mutants can be further tested for improved properties and/or subjected to additional mutagenesis. Efficient engineering of fungal cellulases has been hampered by a lack of genetic tools for native organisms and by difficulties in expressing the enzymes in heterologous hosts. Recently, Morikawa and coworkers developed a method for expressing in E. coli the catalytic domains of endoglucanases from H. jecorina3,9, an important industrial fungus with the capacity to secrete cellulases in large quantities. Functional E. coli expression has also been reported for cellulases from other fungi, including Macrophomina phaseolina10 and Phanerochaete chrysosporium11-12.
We present a method for high throughput screening of fungal endoglucanase activity in E. coli. (Fig 1) This method uses the common microbial dye Congo Red (CR) to visualize enzymatic degradation of carboxymethyl cellulose (CMC) by cells growing on solid medium. The activity assay requires inexpensive reagents, minimal manipulation, and gives unambiguous results as zones of degradation (“halos”) at the colony site. Although a quantitative measure of enzymatic activity cannot be determined by this method, we have found that halo size correlates with total enzymatic activity in the cell. Further characterization of individual positive clones will determine , relative protein fitness.
Traditional bacterial whole cell CMC/CR activity assays13 involve pouring agar containing CMC onto colonies, which is subject to cross-contamination, or incubating cultures in CMC agar wells, which is less amenable to large-scale experimentation. Here we report an improved protocol that modifies existing wash methods14 for cellulase activity: cells grown on CMC agar plates are removed prior to CR staining. Our protocol significantly reduces cross-contamination and is highly scalable, allowing the rapid screening of thousands of clones. In addition to H. jecorina enzymes, we have expressed and screened endoglucanase variants from the Thermoascus aurantiacus and Penicillium decumbens (shown in Figure 2), suggesting that this protocol is applicable to enzymes from a range of organisms.
Protocol
1. Screening plate preparation
Add 25g LB growth medium, 15g agar, and 1.5g carboxymethyl cellulose (CMC) substrate to 1L distilled water, and autoclave to sterilize.
After autoclaving, allow medium to cool and add appropriate antibiotics, and IPTG to a final concentration of 100μM.
Pour 200mL medium each into 5 large square petri dishes/bioassay trays (240 x 240 x 20mm or larger). Allow plates to dry completely.
2. Endoglucanase library generation and screen
Generate a library of endoglucanase catalytic domain(s). This can be accomplished in many ways (e.g., site-directed or random mutagenesis, recombination, etc.) and is determined by the researcher.
Clone the library into a vector such that they are under the control of the T7 promoter with the lac operator, and contain a pelB signal sequence for targeting to the periplasm. An example of a suitable vector is pET22b(+) from Novagen.
Transform the endoglucanase library into E. coli strain BL21 (DE3) and plate for single colonies.
Incubate transformation overnight at 37°C.
Using sterile toothpicks, inoculate clones into 96 well deep-well plates containing 400μL LB medium supplemented with appropriate antibiotics. Incubate at 37°C overnight with shaking at 250 rpm.
Add sterile glycerol to 96 well overnight cultures to a final concentration of 10% for long-term storage at -80°C. This will be the master plate.
Using a 96-pin stamp, replicate plate overnight cultures onto the screening plates containing IPTG and CMC. Up to 5 sets of overnight cultures can be stamped onto each screening plate.
Label the screening plates so that the identity and orientation of each 96 well plate is known.
Incubate the stamped libraries at room temperature (17-25°C) overnight or until visible colonies have formed.
Rinse plate with distilled water until all traces of the colonies are removed.
Pour Congo Red (CR; 0.5% in water) onto washed plate. Make sure to add just enough CR to cover the plate.
Incubate 15 minutes at room temperature. Do not exceed 15 minutes for CR incubation. Longer incubation times will result in less distinct halos.
Pour off CR and add a generous amount (more than enough to cover the plate) of 1M NaCl.
Incubate 15 minutes at room temperature.
Pour off NaCl to reveal CMC degradation halos. For better resolution of the halos, longer incubation times with NaCl or multiple NaCl washes may be required.
Using a sterile toothpick, pick active enzymes clones from the master plate for further characterization.
3. Representative Results:
An example of the readout for this high throughput screen is shown in Figure 3. Clones can be identified as inactive, weakly active, and highly active based on the size of the degradation zones. This is especially useful if an enzyme of known activity is included in the library as a reference. As shown here, the identification of active enzymes is robust and reproducible. However, please be aware that adequate labeling of the screening plates is extremely important. The researcher must be able to identify active variants after washing the colonies away, in order to pick them from the master plate stored at -80°C for further characterization. Finally, it is difficult to assess a priori the relationship between activities on artificial vs natural substrates. Therefore, the researcher must test all positive clones on industrially relevant material to determine protein fitness.
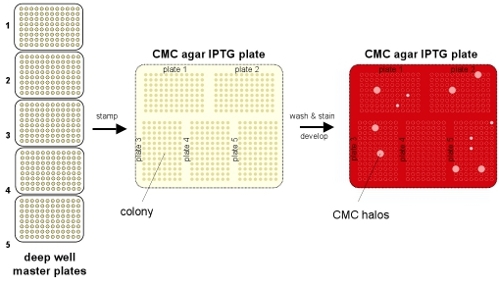 Figure 1. Schematic outline of the screening procedure. Cells are transformed with an endoglucanase library and plated selectively for single colonies. Clones picked and grown overnight in 96 well deep well plates for (1) storage at -80°C and (2) replica plating on plates containing CMC and IPTG to induce endoglucanase expression. Active clones are picked from the master plate stored at -80°C.
Figure 1. Schematic outline of the screening procedure. Cells are transformed with an endoglucanase library and plated selectively for single colonies. Clones picked and grown overnight in 96 well deep well plates for (1) storage at -80°C and (2) replica plating on plates containing CMC and IPTG to induce endoglucanase expression. Active clones are picked from the master plate stored at -80°C.
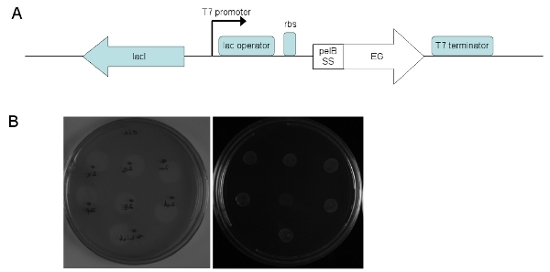 Figure 2. Cloning and expression of fungal catalytic domains in E. coli. (A) Endoglucanase expression vector. Genes were cloned into pET22b(+) (Novagen) under control of the T7 promoter. (B) Right panel: endoglucanase-expressing BL21 (DE3) colonies. Left panel: CMC degradation halos after washing and development with Congo Red.
Figure 2. Cloning and expression of fungal catalytic domains in E. coli. (A) Endoglucanase expression vector. Genes were cloned into pET22b(+) (Novagen) under control of the T7 promoter. (B) Right panel: endoglucanase-expressing BL21 (DE3) colonies. Left panel: CMC degradation halos after washing and development with Congo Red.
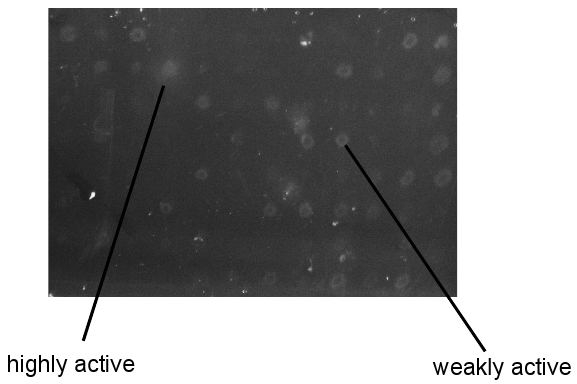 Figure 3. Sample results from CMC/Congo Red screen. Pictures taken of screening plates after washing and Congo Red development. All colonies on screening plates were of similar size.
Figure 3. Sample results from CMC/Congo Red screen. Pictures taken of screening plates after washing and Congo Red development. All colonies on screening plates were of similar size.
Discussion
The protocol described here enables rapid and high throughput identification of active enzymes, with minimal manipulation. Activity detection is fairly sensitive, and qualitatively reflects the amount of activity within the cell. Its ease of use makes this method suitable for a wide range of enzyme libraries, only limited by functional expression in E. coli. Furthermore, activity screening of this kind is not restricted to fungal cellulases, but can be adapted for any endoglucanase activity, or any cellulase activity for which there is a soluble substrate (like CMC).
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was funded by the Gordon and Betty Moore Foundation, and by the UNCF/Merck Science Initiative.
References
- Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- Wilson DB. Cellulases and biofuels. Curr Opin Biotechnol. 2009;20:295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Qin Y. Engineering endoglucanase II from Trichoderma reesei to improve the catalytic efficiency at a higher pH optimum. J Biotechnol. 2008;135:190–195. doi: 10.1016/j.jbiotec.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Nakazawa H. Directed evolution of endoglucanase III (Cel12A) from Trichoderma reesei. Appl Microbiol Biotechnol. 2009;83:649–657. doi: 10.1007/s00253-009-1901-3. [DOI] [PubMed] [Google Scholar]
- Heinzelman P. A family of thermostable fungal cellulases created by structure-guided recombination. Proc Natl Acad Sci. 2009;106:5610–5615. doi: 10.1073/pnas.0901417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzelman P. SCHEMA recombination of a fungal cellulase uncovers a single mutation that contributes markedly to stability. J Biol Chem. 2009;284:26229–26233. doi: 10.1074/jbc.C109.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz SE. Hypocrea jecorina CEL6A protein engineering. Biotechnol Biofuels. 2010;8:3–20. doi: 10.1186/1754-6834-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan SA. Site-directed mutagenesis and CBM engineering of Cel5A (Thermotoga maritima) FEMS Microbiol Lett. 2008;287:205–211. doi: 10.1111/j.1574-6968.2008.01324.x. [DOI] [PubMed] [Google Scholar]
- Nakazawa H. Characterization of the catalytic domains of Trichoderma reesei endoglucanase I, II, and III, expressed in Escherichia coli. Appl Microbiol Biotechnol. 2008;81:681–689. doi: 10.1007/s00253-008-1667-z. [DOI] [PubMed] [Google Scholar]
- Wang H, Jones RW. Properties of the Macrophomina phaseolina endoglucanase (EGL 1) gene product in bacterial and yeast expression systems. Appl Biochem Biotechnol. 1999;81:153–160. doi: 10.1385/abab:81:3:153. [DOI] [PubMed] [Google Scholar]
- Howard RL. Enzyme activity of a Phanerochaete chrysosporium cellobiohydrolase (CBHI.1) expressed as a heterologous protein from Escherichia coli. Afr J Biotechnol. 2003;2:296–300. [Google Scholar]
- Howard Characterisation of a chimeric Phanerochaete chrysosporium cellobiohydrolase expressed from Escherichia coli. Afr J Biotechnol. 2004;3:349–352. [Google Scholar]
- Teather RM, Wood PJ. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Env Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes NR. Mode of action and substrate specificities of cellulases from cloned bacterial genes. J Bio Chem. 1984;259:10455–10459. [PubMed] [Google Scholar]


