Abstract
We present evidence that, in Escherichia coli, polar positional information is present at midcell independent of known cell division factors. In filamented cells, IcsA, which is normally polar, localizes at or near potential cell division sites. Because the cell pole is derived from the septum, the sites to which IcsA localizes in filaments correspond to future poles. IcsA localization to these sites is independent of FtsZ, MinCDE, septation, and nucleoid occlusion, indicating that positional information for the future pole is independent of cell division and chromosome positioning. Upon IcsA localization to these sites, septation is inhibited, suggesting that IcsA recognition of this polar positional information may influence cell division.
The new pole of a bacterial cell is derived from the septum formed during the previous cell division, and the old pole is derived from the septum formed at an earlier cell division. Bacterial cell division occurs by recruitment of the tubulin-like protein FtsZ to midcell, polymerization of FtsZ into a cytokinetic ring, and contraction of that ring (1, 2). Polymerization and contraction of the FtsZ ring normally involves at least 10 associated proteins, each of which directly or indirectly depends on FtsZ for localization to midcell (3, 4).
Two factors known to be important in placement of the FtsZ ring at midcell are the Min proteins (MinCDE) and positioning of the daughter chromosomes (5–9). The choice of division site in Escherichia coli depends in part on MinCDE, because in their absence an FtsZ ring can form at any of three potential division sites located at midcell or at either pole. MinCDE prevent division at the two polar potential division sites, thereby restricting FtsZ ring localization to midcell (5). The molecular mechanism of nucleoid–mediated occlusion of cell division is less clearly understood. Experimental evidence for an effect of nucleoid occlusion is based on the observation that, in mutants in which DNA replication and/or segregation is perturbed, FtsZ rings and septa tend to form in nucleoid-free regions (7). Moreover, in anucleate cells, localization of FtsZ rings to midcell is less precise (8).
Several bacterial proteins, including proteins essential to virulence of bacterial pathogens, are known to localize to one or both cell poles (10). Shigella move through the cytosol of infected mammalian cells by assembly of an actin tail on one pole of the bacterium (11). The Shigella outer membrane protein IcsA (VirG) localizes to the pole of the bacterium (12), where it mediates assembly of the actin tail. Targeting of IcsA to the pole occurs in the bacterial cytoplasm or at the cytoplasmic face of the inner membrane (13, 14). Each of two regions within IcsA (IcsA1–104 and IcsA507–620) is sufficient for polar localization (14). When expressed in E. coli and other Enterobacteriacae, IcsA, IcsA1–104, and IcsA507–620, localize to the pole (14, 15), indicating that the polar targeting mechanism is widely conserved. The positional information that enables IcsA and other polarly localized proteins to recognize one or both poles is incompletely understood.
Because the pole is derived from a prior cell division event, we examined whether positional information necessary for proper localization of IcsA depends on factors known to be involved in septation. We found that IcsA localizes to the sites of future poles independent of FtsZ, MinCDE, and nucleoid occlusion. These data indicate that polar positional information required for IcsA localization is independent of both known components of the cell division machinery and nucleoid occlusion.
Materials and Methods
Bacterial Strains and Growth Conditions. Bacterial strains used in this study are listed in Table 1. Strains were grown in LB or M9 minimal media supplemented with 0.2% glycerol. WT strains were maintained at 37°C. ftsZ84 (Ts) (gift of J. Beckwith, Harvard Medical School), ftsA12 (Ts) dnaA5 (Ts) (gift of L. I. Rothfield, University of Connecticut Health Center, Farmington), and ΔminCDE dnaA5 (Ts) strains were maintained at 30°C. ftsZ84 (Ts) ΔminCDE and mukB strains were maintained at 25°C. When required, antibiotics were used at the following concentrations: ampicillin, 100 μg·ml–1; chloramphenicol, 25 μg·ml–1; kanamycin, 50 μg·ml–1; and tetracycline, 10 μg·ml–1. Standard techniques were used for cloning and analysis of DNA, PCR, electroporation, transformation, and P1 phage transduction (16, 17).
Table 1. Bacterial strains and plasmids used in this study.
| Strains or plasmids | Genotype | Source or ref. |
|---|---|---|
| E. coli strains | ||
| MC4100 | F-araD139 ΔlacU169 relA1 rpsL150 thi mot flb5301 deoC7 ptsF25 rbsR | 33 |
| PB103 | dadR trpE trpA tna purB+ | 34 |
| EC307 | MC4100 leu::Tn10 ftsZ84 (Ts) | 26 |
| PB114 | PB103 ΔminCDE::kan | 5 |
| AJ35 | PB114 leu::Tn10 ftsZ84 (Ts) | This study |
| WM949 | MG1655 ΔmukB::kan | W. Margolin, University of Texas Medical School, Houston |
| AJ41 | PB103 ΔmukB::kan | This study |
| WC1016 | PB103 ftsA12 (Ts) zic501::Tn10 dnaA5 (Ts) | 35 |
| AJ46 | PB114 dnaA5 (Ts) zic501::Tn10 | This study |
| Shigella strain | ||
| MBG341 | icsP | 36 |
| Plasmids | ||
| pBAD24-gfp | PBAD-gfp | 14 |
| pBAD24-icsA1-104::gfp | PBAD-icsA1-104-gfp | 14 |
| pBAD24-icsA507-620::gfp | PBAD-icsA507-620-gfp | 14 |
| pTRI56 | pBAD33-icsA507-620-gfp | Unpublished data |
| pMBG270 | pBR322-icsA | 18 |
Expression of IcsA-GFP. Induction of IcsA-GFP fusions was by addition of l-arabinose to the culture medium to 0.2%. For IcsA localization in cells filamented by inhibition of FtsI, aztreonam, which inhibits FtsI, was added to cells to a final concentration of 1 μg·ml–1 at OD600 0.2, and growth was continued to OD600 0.4–0.6, at which point IcsA-GFP expression was induced for 40 min at room temperature. For IcsA-GFP localization studies in ftsZ84 (Ts), ftsZ84 (Ts) ΔminCDE, ftsA12 (Ts) dnaA5 (Ts), and ΔminCDE dnaA5 (Ts) strains, cells were grown to OD600 0.3–0.4 and then shifted to 42°C (the restrictive temperature) for 60 min, at which point IcsA-GFP expression was induced for an additional 10 min at 42°C; for IcsA-GFP localization to midcell in the WT strain, identical growth conditions were used.
Protein Analysis and Quantification of IcsA Expression. Whole-cell proteins were prepared from bacterial cultures immediately before microscopy. For quantification of levels of IcsA expression, protein loading was normalized to cell density, and Western blot analysis was performed by using antisera to either IcsA (12) or GFP (Molecular Probes). Band densitometry was performed by using quantity one quantitation software (Bio-Rad). Comparison of the level of expression of IcsA-GFP to that of native IcsA in exponential phase Shigella was performed in a similar manner. The level of native IcsA was determined for a Shigella strain that carries a disruption in the gene encoding the protease that cleaves IcsA at the bacterial surface (icsP), because in this strain, essentially all translated IcsA is cell associated and therefore detected in whole-cell protein preparations. This approach differs from an approach we used previously for making such comparisons (14). In the previous approach, we compared the level of IcsA-GFP to the level of native IcsA in WT Shigella, for which a significant amount of translated IcsA fractionates to the culture supernatant and not to the pellet used for the whole-cell proteins that were analyzed. This previous approach undoubtedly underestimated the level of expression of native IcsA (18).
Localization of IcsA-GFP in the Presence of Full-Length IcsA. To test the localization of IcsA507–620-GFP in the presence of full-length IcsA, full-length icsA was expressed under the control of the native icsA promoter in pBR322 (18). To maintain plasmid compatibility, the IcsA507–620-GFP fusion plasmid was expressed under the control of the arabinose promoter in pBAD33. Cells were filamented by addition of aztreonam as described above. To attain levels of expression of IcsA-GFP that were comparable to that attained in the pBAD24 expression vector, expression of the pBAD33 derivative was induced for up to 80 min at room temperature. The intensity of the diffuse signal from delocalized IcsA-GFP in cells that were also expressing full-length IcsA was measured by using iplab scientific imaging software (Scanalytics, Fairfax, VA). Mean pixel intensity was determined for each of three 2-by-2-pixel squares spaced along the length of each of 20 filaments for each strain.
Localization of IcsA-GFP and FtsZ. Cells were filamented by treatment with aztreonam, and IcsA-GFP expression was induced as described above. The cells were subsequently fixed with 80% methanol (19, 20) and immunostained with antiserum to FtsZ (1:20,000 dilution, gift of D. RayChaudhuri, Tufts University School of Medicine, Boston) and a Texas red-conjugated secondary antibody. Cells were then stained with 10 μl of 4′,6-diamidino-2-phenylindole (DAPI, 10 μg·ml–1) for 1 min to visualize the nucleoids.
Microscopy. Bacteria were mounted onto a 1% agarose or LB-agar pad containing 0.4% glucose on a 15-well slide (ICN) and were visualized and imaged as described (14). For time-lapse experiments, bacterial samples immobilized on slides were maintained in a 30°C incubator (that encompasses the microscope stage). Unless otherwise indicated, staining for nucleoids was by incubation in the presence of 0.2 μg·ml–1 DAPI for 5 min immediately before mounting.
Tabulation of Targeting Patterns. The interval and regularity of spacing of IcsA-GFP foci was determined by measuring and tabulating the distance from one end of the cell to each of >40 individual GFP foci per strain. For the ΔminCDE mutant, only filaments greater than or equal to two normal cell lengths were tabulated. For tabulation of IcsA-GFP foci within nucleate or anucleate regions of chromosome partitioning mutants, the spacing between two foci was included in the tabulation only when both foci were located within the same nucleate or anucleate region. Tabulation of IcsA-GFP targeting patterns in WT cells was performed on 100 bacteria that had been previously randomly selected on phase images as described (14).
Inhibition of Cell Division Assay. For determining the effect of expression of IcsA-GFP or GFP on cell division of the ftsZ84 (Ts) mutant upon restoration of functional FtsZ, cells were grown for 60 min at 42°C (the restrictive temperature), at which point growth was continued for an additional 10 min at 42°C with or without induction of IcsA-GFP or GFP. Cultures were then shifted to the permissive temperature and plated for colony counts at each of two times: (i) at the time of shift to the permissive temperature, and (ii) 30 min after growth at the permissive temperature. In each case, immediately before the shift to the permissive temperature, cells were washed to remove the inducer.
For determining the effect of expression of IcsA-GFP or GFP on cell division of the ftsZ84 (Ts) strain at the permissive temperature or the WT strain, cells were grown after subculturing for 2 h at 30°C, at which point growth was continued for an additional 30 min at 30°C, with or without induction of IcsA-GFP or GFP. Samples for colony counts were taken and plated at each of two times: (i) at the end of the 2-h growth at 30°C, immediately before induction, and (ii) at the end of the 30-min period of additional growth with or without induction. In each case, immediately after induction, cells were washed to remove the inducer.
The effect of expression of IcsA-GFP or GFP on cell division in each of these strains and conditions was determined from colony counts as follows. For each experiment, the ratio of the colony-forming units (CFUs) from the sample taken at the second plating (time ii) to the CFUs from the sample taken at the first plating (time i) was calculated. The numbers seen in Table 3 represent the mean of three or more independent experiments. The efficiency of cell division was calculated as the extent of inhibition of cell division normalized to that of GFP alone in the same strain background under the same conditions.
Table 3. Effect of IcsA-GFP constructs on cell division.
| Ratio of CFUs†
|
|||||
|---|---|---|---|---|---|
| Growth temperature* | GFP construct | - | + | Efficiency of cell division‡ | |
| ftsZ84 (Ts) | 42°C/30°C | IcsA1-104-GFP | 14.4 | 1.6 | 0.2 |
| IcsA507-620-GFP | 11.2 | 2.0 | 0.3 | ||
| GFP | 6.0 | 3.2 | 1.0 | ||
| ftsZ84 (Ts) | 30°C/30°C | IcsA1-104-GFP | 1.6 | 2.8 | 1.2 |
| IcsA507-620-GFP | 1.2 | 1.5 | 0.9 | ||
| GFP | 2.6 | 3.5 | 1.0 | ||
| WT | 42°C/30°C | IcsA1-104-GFP | 1.0 | 0.9 | 0.7 |
| IcsA507-620-GFP | 0.8 | 0.9 | 0.9 | ||
| GFP | 0.9 | 1.2 | 1.0 | ||
| WT | 30°C/30°C | IcsA1-104-GFP | 2.7 | 4.5 | 0.9 |
| IcsA507-620-GFP | 4.2 | 3.0 | 0.9 | ||
| GFP | 4.8 | 8.8 | 1.0 | ||
Before first plating (time i)/before second plating (time ii) (see Materials and Methods).
Ratio of CFUs from the sample taken at the second plating (time ii) to CFUs from the sample taken at the first plating (time i) (see Materials and Methods).
The extent of inhibition of cell division normalized to that of GFP alone in the same strain background under the same conditions (see Materials and Methods).
Results and Discussion
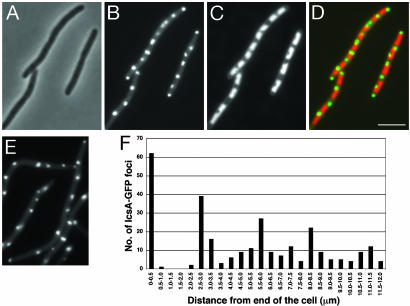
Upon Inhibition of Septation, IcsA Localized at or near Cell Division Sites. We analyzed localization of IcsA507–620-GFP and IcsA1–104-GFP, which remain cytoplasmic, in E. coli treated with aztreonam, which blocks FtsI (penicillin-binding protein 3, PBP3), a cell division protein that is recruited toward the end of the cell division dependency cascade (21, 22). As expected, treatment with aztreonam induced the formation of filaments (Fig. 1A). GFP fluorescent foci from IcsA507–620-GFP and IcsA1–104-GFP localized at regularly spaced intervals along the length of the filaments and at the extreme ends of the filaments (Fig. 1B and data not shown). The IcsA-GFP foci were localized to gaps between the nucleoids (Fig. 1 C and D), which correspond to potential cell division sites, suggesting that IcsA-GFP was positioned at or near the cell division site.
Fig. 1.
Localization of IcsA507–620-GFP at intervals along the lengths of aztreonam-treated and ftsZ84 (Ts) E. coli filaments. (A–D) WT (MC4100) treated with aztreonam: phase (A), GFP (B), DAPI-stained nucleoid (C), and overlay of GFP (green) and nucleoid (red) (D). (E and F) ftsZ84 (Ts) mutant (EC307) grown at the restrictive temperature: GFP (E) and periodicity and spacing of IcsA507–620-GFP fluorescent dots (F)(n = 349). Distribution of dots is shown as a function of distance from the end of the cell. Localization and distribution of IcsA1–104-GFP fluorescent dots were similar (data not shown). (Scale bar: 5 μm.)
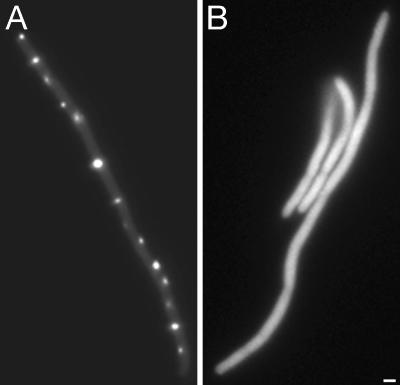
Interference of Full-Length IcsA with IcsA507–620-GFP Localization. To determine whether the localization of IcsA507–620-GFP foci at regularly spaced intervals along the length of filaments was specific, we examined the ability of full-length IcsA to interfere with this localization. Upon expression of full-length IcsA in trans, the GFP signal from IcsA507–620-GFP was diffuse (Fig. 2), suggesting that full-length IcsA interferes with interactions of IcsA507–620-GFP with a putative target. In these studies, to maintain plasmid compatibility, IcsA507–620-GFP was expressed from the arabinose promoter in pBAD33, whose copy number is lower than that of pBAD24, which was used in the other experiments presented in this study. To adjust for this, we also examined the interference of full-length IcsA with IcsA507–620-GFP expressed for a prolonged period (80 min) from pBAD33, under which conditions the level of expression of IcsA507–620-GFP was comparable to that attained from pBAD24 under experimental conditions used elsewhere in this study, as determined by Western blot analysis (data not shown). Under these conditions, full-length IcsA continued to interfere with IcsA507–620-GFP, as determined by a significant 2-fold increase in the intensity of the diffuse delocalized GFP signal in these cells compared to cells that expressed only IcsA507–620-GFP. Of note, under these conditions, the level of IcsA-GFP in the cells was ≈2- to 6-fold greater than that of native IcsA in Shigella. These results strongly suggest that localization of IcsA507–620-GFP at intervals in filaments is specific and not a result of the formation of inclusion bodies or protein aggregates.
Fig. 2.
Localization of IcsA507–620-GFP in the presence of full-length IcsA in aztreonam-treated WT (MC4100) E. coli filaments. IcsA507–620-GFP alone (A) and IcsA507–620-GFP in the presence of full-length IcsA (B). (Scale bar: 1 μm.)
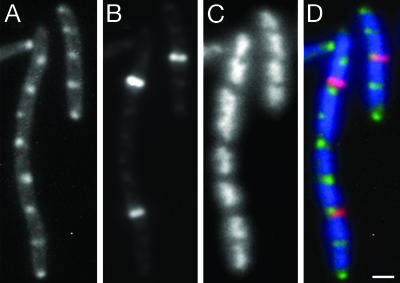
IcsA Was Indeed at or near Cell Division Sites. We examined localization of IcsA and FtsZ rings in E. coli cells filamented by treatment with aztreonam. In these filaments, FtsZ rings localize with or adjacent to IcsA507–620-GFP foci (Fig. 3). In general, FtsZ rings formed only at every other or at every fourth potential division site along the length of the filament, as determined by the spacing of the nucleoids. In contrast, IcsA-GFP localized not only at or adjacent to those potential cell division sites at which FtsZ rings were found, but also at or adjacent to essentially all potential cell division sites between FtsZ rings. A similar pattern of FtsZ ring localization has been observed (23). The localization of some of the IcsA foci at or adjacent to FtsZ rings indicates that at least some, and perhaps all, of the sites to which IcsA localizes in filaments are at or near the potential cell division sites. The localization of IcsA to positions between the nucleoids that correspond to potential cell division sites, but at which FtsZ rings are not present, suggests that IcsA positional information may be independent of FtsZ.
Fig. 3.
Localization of IcsA507–620-GFP and FtsZ at intervals along the lengths of aztreonam-treated WT (MC4100) E. coli filaments. IcsA507–620-GFP (A), FtsZ by immunolabeling (B), DAPI-stained nucleoid (C), and overlay of the IcsA-GFP (green), FtsZ (red), and nucleoid (blue) images (D). (Scale bar: 1 μm.)
IcsA Localization at or near Cell Division Sites Is Independent of FtsZ. Many proteins are known to colocalize with the cell division site; each protein that has been shown to localize to cell division sites depends on FtsZ for its localization. P1 and F plasmids also localize to midcell (24); localization of these plasmids may be independent of FtsZ, because upon FtsZ ring depletion with cephalexin (which inhibits FtsI), the plasmids localize at regular intervals along the length of filaments (23, 24).
In contrast to other proteins known to colocalize with the cell division site, we found that IcsA is independent of FtsZ for its localization. We analyzed localization of IcsA507–620-GFP and IcsA1–104-GFP in an ftsZ84 (Ts) background. As expected, at the restrictive temperature, ftsZ84 (Ts) cells formed filaments with no visible constrictions, because of their inability to assemble an FtsZ ring at the cell division site (25–27). After growth at the restrictive temperature, IcsA507–620-GFP and IcsA1–104-GFP each formed fluorescent foci at regularly spaced intervals along the length of the filaments and at the extreme ends of the filaments (Fig. 1E and data not shown).
The spacing of the IcsA-GFP foci was similar to the spacing of cell division sites. We calculated the interval and regularity of spacing of the foci. The distance from one end of the filament of each IcsA507–620-GFP focus was plotted as a function of cell length (Fig. 1F). In vegetative E. coli, one bacterial cell length is ≈2–3 μm (28), and in filamentous cells, FtsZ rings localize at similar intervals of 2–3 μm (29). In ftsZ84 (Ts) filaments, IcsA507–620-GFP foci were spaced at a distance of 2.6 ± 0.5 μm (mean ± SD) (Fig. 1F and Table 2), which is similar to the spacing of foci in WT cells that were filamented by treatment with aztreonam (Table 2). This spacing places approximately one focus per cell length. IcsA1–104-GFP foci showed similar spacing intervals. Taken together, these results indicate that IcsA localizes at or near potential division sites. Furthermore, unlike all other proteins that colocalize with FtsZ, IcsA localization occurs independently of the recruitment of FtsZ and downstream cell division proteins to the cell division site.
Table 2. Interval and regularity of spacing of IcsA507–620-GFP foci.
| Strain background | Relevant genotype | Region of filaments analyzed | Distance between IcsA507-620-GFP foci, mean ± SD in μm (N) |
|---|---|---|---|
| MC4100 | WT* | All | 2.1 ± 0.5 (109) |
| PB103 | WT* | All | 2.7 ± 1.2 (40) |
| EC307 | MC4100 ftsZ84 (Ts) | All | 2.6 ± 0.5 (349) |
| PB114 | PB103 ΔminCDE | All | 2.6 ± 0.7 (209) |
| AJ35 | PB103 ftsZ84 (Ts) ΔminCDE | All | 3.3 ± 1.2 (246) |
| AJ41 | PB103 mukB | All | 2.6 ± 1.1 (243) |
| Nucleate | 2.4 ± 1.1 (103) | ||
| Anucleate | 2.3 ± 1.2 (39) | ||
| WC1016 | PB103 ftsA12 (Ts) dnaA5 (Ts) | All | 2.9 ± 1.2 (307) |
| Nucleate | 2.5 ± 0.8 (44) | ||
| Anucleate | 2.2 ± 1.1 (80) | ||
| AJ46 | PB103 ΔminCDE dnaA5 (Ts) | All | 3.9 ± 1.7 (216) |
| Nucleate | 3.6 ± 1.3 (69) | ||
| Anucleate | 3.2 ± 1.2 (73) |
WT strains were filamented by treatment with aztreonam (see Materials and Methods).
IcsA Positioning in ftsZ84 (Ts) Filaments Is Independent of Min Proteins. The Min system functions as a negative regulator of FtsZ ring placement (30). In the absence of Min proteins, FtsZ is capable of forming rings at each of the three potential division sites, leading to a mix of short filaments, anucleate mini cells, and normal-sized cells. We analyzed IcsA1–104-GFP and IcsA507–620-GFP localization in a ΔminCDE mutant and a ftsZ84 (Ts) ΔminCDE double mutant. As for the ftsZ84 (Ts) filaments, in both the ΔminCDE and ftsZ84 (Ts) ΔminCDE filaments, IcsA-GFP fluorescent foci localized at or near potential division sites. In ΔminCDE filaments, IcsA507–620-GFP foci were spaced at 2.6 ± 0.7 μm (mean ± SD, Table 2), which places approximately one focus per normal cell length. The spacing of foci in the ftsZ84 (Ts) ΔminCDE filaments was slightly, albeit not significantly wider (3.3 ± 1.2 μm, Table 2). The significance of this finding is uncertain, as foci were generally located between nucleoids, suggesting that the sites to which the foci located nevertheless represented potential cell division sites and potential poles. These results indicate that IcsA recognition of polar positional information at or near potential cell division sites occurs independently of both FtsZ and the Min proteins.
Localization of IcsA-GFP Foci Is Independent of Nucleoid Occlusion. The observed colocalization of IcsA507–620-GFP dots with nucleoid-free gaps (Fig. 1D) could be consistent with a role of nucleoid occlusion in its positioning. If nucleoid occlusion played a role in IcsA targeting, we predicted that IcsA localization would be random in anucleate cells or anucleate segments of filaments.
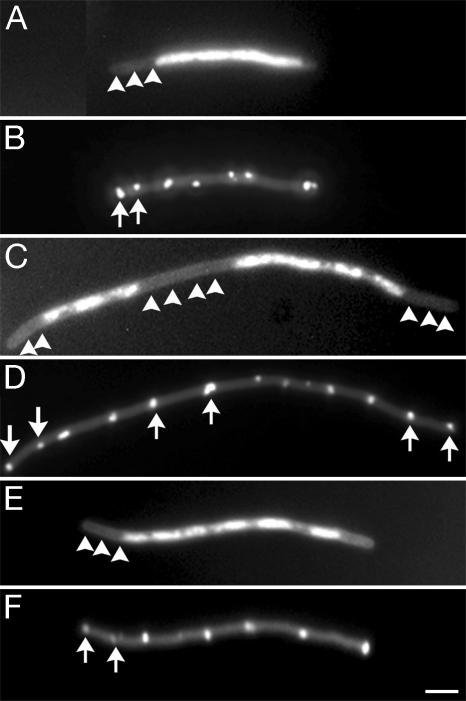
We examined positioning of IcsA1–104-GFP and IcsA507–620-GFP in anucleate segments of filamented cells. Null mutants in mukB display defects in nucleoid structure and segregation, leading to ≈5–13% of cells being anucleate (31). In mukB mutants, FtsZ rings appear to form on top of nucleoids in some instances and lack precision in localization to midcell (32). We examined the localization of IcsA507–620-GFP foci in mukB cells that were filamented with aztreonam, quantifying only those cells that, on DAPI staining, clearly contained anucleate segments. IcsA507–620-GFP foci were localized at fairly regular intervals along the lengths of the filaments, including within anucleate segments (Fig. 4 A and B and Table 2). The dots were spaced at 2.6 ± 1.1 μm (mean ± SD), which again places approximately one focus per cell length; the mean of 2.6 μm is similar to those observed for foci in WT cells filamented with aztreonam and ftsZ84 (Ts) filaments (Table 2). The anucleate segments comprised 22.1% of the total length of the cells that were quantified. IcsA507–620-GFP foci were spaced similarly in nucleoid and nucleoid-free regions of the filaments (Table 2). These data indicate that IcsA localization at or near potential cell division sites is independent of nucleoid occlusion.
Fig. 4.
IcsA507–620-GFP localization at intervals in anucleate and nucleate segments of chromosome partitioning mutants and chromosome partitioning and cell division double mutants. A mukB mutant (AJ41) filamented by growth in the presence of aztreonam (A and B); an ftsA12 (Ts) dnaA5 (Ts) strain (WC1016) after growth at the restrictive temperature (C and D); and a ΔminCDE dnaA5 (Ts) strain (AJ46) after growth at the restrictive temperature (E and F). (A, C, and E) DAPI-stained nucleoids, showing anucleate segments (arrowheads). (B, D, and F) GFP image of the same microscopic fields, showing spacing of IcsA507–620-GFP foci in the anucleate segments (arrows). (Scale bar: 3 μm.)
The Min proteins and nucleoid occlusion have redundant effects on the position of the cell division septum (8). To address whether the two might have a redundant role in IcsA localization, we examined the positioning of IcsA-GFP in mutants in which both cell division and chromosome positioning were defective. The spacing of IcsA507–620-GFP foci was determined in filaments of an ftsA12 (Ts) dnaA5 (Ts) strain and a ΔminCDE dnaA5 (Ts) strain. When grown at the restrictive temperature (42°C), each of these strains forms filaments with extended chromosome-free regions at the filament ends (Fig. 4 C–F). Fifty-two percent of the total length of the ftsA12 (Ts) dnaA5 (Ts) filaments and 46% of the total length of the ΔminCDE dnaA5 (Ts) filaments were a nucleate. In each strain, IcsA507–620-GFP foci localized at periodic intervals within anucleate and nucleate regions of the filaments (Fig. 4 C–F). The spacing of IcsA-GFP foci in the ftsA12 (Ts) dnaA5 (Ts) filaments was similar to that of the WT strains filamented with aztreonam, the ftsZ84 (Ts) strain, and the ΔminCDE strain. Moreover, the spacing of foci was similar within the anucleate and nucleate regions of these filaments (Table 2). The spacing of IcsA-GFP foci in the ΔminCDE dnaA5 (Ts) filaments was slightly, albeit not significantly, wider (3.9 ± 1.7 μm, Table 2). Of note, this strain lacks the Min proteins, whereas the ftsA12 (Ts) dnaA5 (Ts) strain contains them. The wider spacing observed in these filaments appears to not be determined by the positioning of the nucleoid, because foci were spaced similarly in the anucleate and nucleate regions of the filaments (Table 2). The SD on the mean distance between foci was slightly larger for the ΔminCDE dnaA5 (Ts) filaments (1.7 μm) than for other strains (0.5–1.3 μm, Table 2), indicating that the regularity of spacing of IcsA positional information was also altered slightly. Thus, in the absence of both proper chromosome partitioning and the Min proteins, IcsA positional information is spaced at slightly wider than normal intervals and with slightly less precision, suggesting that the redundant effects of the two systems contributes to the precise spacing of polar positional information. Nevertheless, in the absence of these two systems, IcsA localization was not markedly perturbed, indicating that IcsA recognition of polar positional information is largely independent of the redundant effects of the cell division apparatus and nucleoid occlusion.
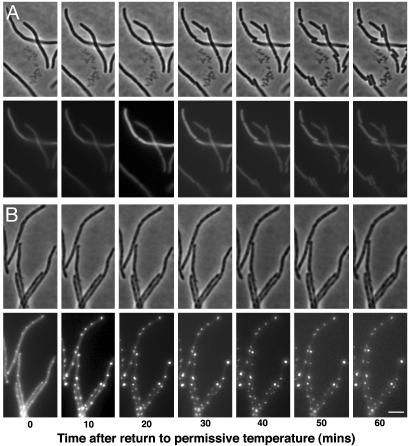
Expression of IcsA-GFP Constructs Inhibits Cell Division of ftsZ84 (Ts) Filaments upon Return to the Permissive Temperature. Given that IcsA localized at or near the potential cell division sites in filaments, we explored whether IcsA might be recognizing a structure involved in the cell division pathway. We reasoned that if IcsA was recognizing such a structure, its localization at or near potential division sites might block subsequent cell division. Therefore, we tested whether IcsA localization in filaments lacking localized FtsZ rings inhibited cell division upon restoration of functional FtsZ. We monitored cell division patterns of ftsZ84 (Ts) filaments, in which either IcsA was expressed or not expressed, upon subsequent return to the permissive temperature. When IcsA507–620-GFP was expressed, septation appeared blocked, with cells remaining filamentous for up to 60 min at the permissive temperature (Fig. 5B). In contrast, when IcsA507–620-GFP was not expressed, within 20 min, septa began to form, and by 60 min, most filaments had divided into multiple individual cells (Fig. 5A). IcsA expression did not alter FtsZ stability, and the level of expression of IcsA507–620-GFP was comparable to that in other experiments (data not shown). In a small percentage of filaments in which IcsA507–620-GFP had been induced, division occurred, but these filaments lacked fluorescence, indicating minimal or absent IcsA-GFP expression in this subpopulation of cells (data not shown).
Fig. 5.
IcsA507–620-GFP and IcsA1–104-GFP inhibit cell division of ftsZ84 (Ts) filaments upon return to the permissive temperature. Time-lapse sequence of phase and GFP images of ftsZ84 (Ts) (EC307) filaments after return to the permissive temperature in the absence of expression of IcsA507–620-GFP (A) or after expression of IcsA507–620-GFP (B). Time lapse of filaments in which IcsA1–104-GFP was expressed appeared to be similar (data not shown). (Scale bar: 5 μm.)
We then determined whether, upon FtsZ release, the number of individual cells in the population increased significantly less in the presence of IcsA than in the absence of IcsA. The relative increase in CFUs that occurred upon FtsZ release (expressed as the ratio of CFUs 30 min after FtsZ release to CFUs immediately before FtsZ release) was significantly less when IcsA1–104-GFP or IcsA507–620-GFP was expressed than when the IcsA-GFP fusion was not expressed (Table 3). Expression of GFP alone had only a minor effect on the increase in CFUs (Table 3). In parallel, we examined cell division upon IcsA-GFP expression in the ftsZ84 (Ts) strain grown at the permissive temperature; in this case, expression of the IcsA-GFP fusions had no effect on cell division (Table 3). However, filaments do not form under these conditions, and IcsA does not localize at or near the cell division sites (data not shown and see below). These results suggest that IcsA-GFP localization at or near potential division sites in filaments may inhibit subsequent cell division. IcsA-GFP may inhibit cell division by blocking proper localization or function of FtsZ; however, our results do not exclude the possibility that IcsA-GFP blocks FtsZ renaturation or that IcsA-GFP competes with FtsZ for a chaperone. Taken together, IcsA recognition of polar positional information appears to influence cell division, yet this positional information is independent of known cell division proteins.
In WT cells, IcsA is found at essentially all old poles, but only ≈50% of new poles (14), leading to the hypothesis that IcsA positional information either depends on maturation of the pole or is transiently masked at new poles. Consistent with this hypothesis is the absence, in WT cells, of IcsA at midcell: <1% of cells expressing IcsA1–104-GFP and ≈1% of cells expressing IcsA507–620-GFP had fluorescent foci at midcell. Additionally, expression of IcsA-GFP fusions had no effect on cell division of the WT strain at 42°C, 37°C, or 30°C (Table 3 and data not shown). Thus, examination of IcsA localization in filaments has permitted the uncoupling of the known cell division pathway from the generation of polar positional information.
Uncoupling of the known cell division pathway from the generation of polar positional information has enabled us to demonstrate that polar positional information is independent of the known cell division pathway. This polar positional information is present in the absence of septation, and therefore is set up independently of the cell division site becoming a pole. Furthermore, nucleoid occlusion is not the basis of this positional information, although nucleoid positioning and the Min proteins appear to have some redundant effect on the precision of the spacing of IcsA foci. The structure recognized at the cell pole by IcsA, and perhaps by other polarly localized proteins, is present in some form in the cytoplasm or at the cytoplasmic face of the inner membrane and at or near the cell division site. IcsA binding to this site inhibits cell division, suggesting that the two may be linked.
Acknowledgments
We thank D. RayChaudhuri, P. A. J. de Boer, W. Margolin, S. Hiraga, L. I. Rothfield, and J. Beckwith for providing antibody and strains and A. D. Grossman and S. B. Calderwood for critical reading of the manuscript. M.B.G. was supported by National Institutes of Health Grant AI35817, and A.J. was supported by a postdoctoral fellowship award from the American Heart Association.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CFU, colony-forming unit; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Addinall, S. G. & Holland, B. (2002) J. Mol. Biol. 318, 219–236. [DOI] [PubMed] [Google Scholar]
- 2.Margolin, W. (2001) Curr. Opin. Microbiol. 4, 647–652. [DOI] [PubMed] [Google Scholar]
- 3.Buddelmeijer, N. & Beckwith, J. (2002) Curr. Opin. Microbiol. 5, 553–557. [DOI] [PubMed] [Google Scholar]
- 4.Errington, J., Daniel, R. A. & Scheffers, D. J. (2003) Microbiol. Mol. Biol. Rev. 67, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer, P. A., Crossley, R. E. & Rothfield, L. I. (1989) Cell 56, 641–649. [DOI] [PubMed] [Google Scholar]
- 6.Bi, E. & Lutkenhaus, J. (1993) J. Bacteriol. 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulder, E. & Woldringh, C. L. (1989) J. Bacteriol. 171, 4303–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu, X. C. & Margolin, W. (1999) Mol. Microbiol. 32, 315–326. [DOI] [PubMed] [Google Scholar]
- 9.Woldringh, C. L., Mulder, E., Huls, P. G. & Vischer, N. (1991) Res. Microbiol. 142, 309–320. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro, L., McAdams, H. H. & Losick, R. (2002) Science 298, 1942–1946. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg, M. B. (2001) Microbiol. Mol. Biol. Rev. 65, 595–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg, M. B., Barzu, O., Parsot, C. & Sansonetti, P. J. (1993) J. Bacteriol. 175, 2189–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandon, L. D., Goehring, N., Janakiraman, A., Yan, A. W., Wu, T., Beckwith, J. & Goldberg, M. B. (2003) Mol. Microbiol. 50, 45–60. [DOI] [PubMed] [Google Scholar]
- 14.Charles, M., Perez, M., Kobil, J. H. & Goldberg, M. B. (2001) Proc. Natl. Acad. Sci. USA 98, 9871–9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandlin, R. C. & Maurelli, A. T. (1999) Infect. Immun. 67, 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 17.Miller, J. H. (1992) A Short Course in Bacterial Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 18.Magdalena, J. & Goldberg, M. B. (2002) Cell Motil. Cytoskeleton 51, 187–196. [DOI] [PubMed] [Google Scholar]
- 19.RayChaudhuri, D. (1999) EMBO J. 18, 2372–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiraga, S., Ichinose, C., Niki, H. & Yamazoe, M. (1998) Mol. Cell 1, 381–387. [DOI] [PubMed] [Google Scholar]
- 21.de Pedro, M. A., Quintela, J. C., Holtje, J. V. & Schwarz, H. (1997) J. Bacteriol. 179, 2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss, D. S., Chen, J. C., Ghigo, J. M., Boyd, D. & Beckwith, J. (1999) J. Bacteriol. 181, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pogliano, J., Pogliano, K., Weiss, D. S., Losick, R. & Beckwith, J. (1997) Proc. Natl. Acad. Sci. USA 94, 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon, G. S., Sitnikov, D., Webb, C. D., Teleman, A., Straight, A., Losick, R., Murray, A. W. & Wright, A. (1997) Cell 90, 1113–1121. [DOI] [PubMed] [Google Scholar]
- 25.Addinall, S. G., Bi, E. & Lutkenhaus, J. (1996) J. Bacteriol. 178, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.RayChaudhuri, D. & Park, J. T. (1994) J. Biol. Chem. 269, 22941–22944. [PubMed] [Google Scholar]
- 27.Begg, K. J. & Donachie, W. D. (1985) J. Bacteriol. 163, 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madigan, M. T., Martinko, J. M. & Parker, J. (2000) Brock Biology of Microorganisms (Prentice–Hall, Upper Saddle River, NJ).
- 29.Sun, Q. & Margolin, W. (2001) J. Bacteriol. 183, 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothfield, L. I., Shih, Y. L. & King, G. (2001) Cell 106, 13–16. [DOI] [PubMed] [Google Scholar]
- 31.Sun, Q., Yu, X. C. & Margolin, W. (1998) Mol. Microbiol. 29, 491–503. [DOI] [PubMed] [Google Scholar]
- 32.Yu, X. C., Sun, Q. & Margolin, W. (2001) Biochimie 83, 125–129. [DOI] [PubMed] [Google Scholar]
- 33.Chen, J. C., Weiss, D. S., Ghigo, J. M. & Beckwith, J. (1999) J. Bacteriol. 181, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Boer, P. A., Crossley, R. E. & Rothfield, L. I. (1988) J. Bacteriol. 170, 2106–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook, W. R. & Rothfield, L. I. (1999) J. Bacteriol. 181, 1900–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shere, K. D., Sallustio, S., Manessis, A., D'Aversa, T. G. & Goldberg, M. B. (1997) Mol. Microbiol. 25, 451–462. [DOI] [PubMed] [Google Scholar]