Abstract
Primary dystonia is a disease characterized by involuntary twisting movements caused by CNS dysfunction without underlying histopathology. DYT1 dystonia is a form of primary dystonia caused by an in-frame GAG deletion (ΔE302/3) in the TOR1A gene that encodes the endoplasmic reticulum luminal protein torsinA. We show that torsinA is also present in the nuclear envelope (NE), where it appears to interact with substrate, and that the ΔE302/3 mutation causes a striking redistribution of torsinA from the endoplasmic reticulum to the NE. In addition, ΔE302/3-torsinA recruits WT torsinA to the NE, potentially providing insight into an understanding of the dominant inheritance of the disease. DYT1 dystonia appears to be a previously uncharacterized NE disease and the first, to our knowledge, to selectively affect CNS function.
DYT1 dystonia is a childhood-onset CNS disorder characterized by dramatic motor dysfunction due to abnormal interneuronal signaling rather than neurodegeneration (1, 2). Therefore, understanding the molecular determinants of this disease might shed light on basic mechanisms that regulate neuronal function and plasticity.
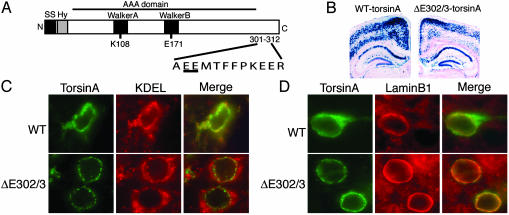
TorsinA is a widely expressed member of the AAA (ATPase associated with different cellular activities) protein family (3) (Fig. 1A). AAA proteins participate in a diverse range of biological functions, including membrane trafficking, powering cellular motors, organelle biogenesis, and protein chaperone activities (4–7). These proteins use ATP binding and hydrolysis to induce conformational changes in substrate molecules via a repetitive cycle of substrate binding and release; they tend to multimerize and typically unfold proteins or disassemble protein complexes (e.g., N-ethylmaleimide-sensitive factor; NSF) (5, 6). The ΔE302/3-torsinA mutation lies within a C-terminal domain that is unique to a number of torsin-related proteins (8) but is not part of the AAA domain (Fig. 1 A). Biochemical and cellular studies of torsinA in overexpressing cellular systems suggest that the ΔE302/3 mutation does not alter the stability or solubility of torsinA or its ability to multimerize (9, 10).
Fig. 1.
ΔE302/3-torsinA is abnormally localized in the neuronal NE. (A) Schematic representation of the torsinA protein. The primary sequence surrounding the DYT1 mutation is shown with the site of the glutamic acid deletion underlined. SS, signal sequence; Hy, hydrophobic domain; K108 and E171, site of key Walker box residues mutated for functional studies of torsinA. (B) β-Galactosidase histochemistry of brain sections from WT- and ΔE302/3-torsinA-expressing transgenic mice. (C and D) Frontal cortex sections from transgenic mice expressing either WT-torsinA or ΔE302/3-torsinA and double stained with antibodies against torsinA and KDEL (C) or laminB1 (D). No torsinA immunolabeling was observed in nonbitransgenic littermates (data not shown).
To elucidate the molecular mechanisms of primary dystonia, we examined the effect of the ΔE302/3 mutation on the behavior of torsinA. We find that the ΔE302/3 mutation causes torsinA to relocalize from the endoplasmic reticulum (ER) to the nuclear envelope (NE) in both patient tissue and neurons from ΔE302/3-torsinA transgenic mice. Further, we show that disease-associated torsinA recruits WT protein to the NE. Additional experiments suggest that torsinA is normally present in the NE, where it appears to interact with substrate(s).
Materials and Methods
TorsinA cDNA Plasmid Construction and Generation of Transgenic Mice. Human torsinA cDNA was cloned into pcDNA3.1 (Invitrogen). Mutations were introduced by using QuikChange (Stratagene). Enhanced GFP (Clontech) was cloned into an NheI site introduced between residues 21 and 22 of torsinA [immediately following the N-terminal signal sequence cleavage site (11)]. WT-, ΔE302/3-, and E171Q-torsinA cDNAs were also cloned into the pCMV multiple cloning site of a modified pBudCE4.1 vector (Invitrogen) that contained the lacZ reporter in the pEF-1α promoter multiple cloning site. The constructs for transgenic mice were made by cloning either human WT- or ΔE302/3-torsinA cDNA into the multiple cloning site of the pBi3 plasmid (12) (a kind gift H. Bujard, University of Heidelberg, Heidelberg), which contains a bidirectional tetracycline operator motif with minimal cytomegalovirus promoters (tetO) and the β-galactosidase gene fused to a nuclear localization signal sequence (NLS-lacZ). WT- and ΔE302/3-torsinA cDNA containing pBi3 plasmids were injected by the Columbia University Transgenic Facility, and founder mice were mated with the previously described prion promoter–tetracycline transactivator mice (13). Mice were used in accordance with National Institutes of Health guidelines for the use of live animals, and the animal protocol was approved by the Institutional Animal Care and Use Committee of Columbia University.
Cell Culture. BHK21 cells were grown in DMEM containing 10% FBS. Cells were transfected by using Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions and examined 48 h posttransfection. Immunofluorescent staining was performed on cells grown on collagen-coated glass coverslips (Carolina Biological Supply) and fixed by 5-min immersion in methanol prechilled to –20°C. Protein was extracted by homogenizing cells with 5 vol of 1% Triton X-100 lysis buffer including 2 mM EDTA, 2 mM DTT, and protease inhibitor cocktail (Sigma). Lysates were gently rotated at 4°C for 45 min and centrifuged at 13,000 rpm for 10 min, and then the supernatant was used in SDS/PAGE. BHK21 cells stably transfected with GFP-tagged torsinA were selected by using 1 mg/ml geneticin, and individual clones were picked and screened for observable GFP fluorescence after 4% formaldehyde fixation. Human fibroblasts were grown in MEM supplemented with 15% FBS and 1% BME vitamins (Sigma).
Immunofluorescent Labeling. Mouse brains were removed after 4% formaldehyde perfusion fixation, postfixed at 4°C overnight, and transferred into 30% sucrose/PBS for 36 h at 4°C. Tissue was subsequently frozen, and 30-μm floating sections were cut and mounted onto Fisher Plus slides. Sections were postfixed for 10 min in 4% formaldehyde, washed and permeabilized in PBS including 0.25% Triton X-100 (PBS-Triton), and treated with NaBH4 in PBS for 10 min. Labeling of mouse brain sections and tissue culture cells was subsequently performed identically. Samples were blocked for 1 h at room temperature (PBS-Triton; 50 mM NH4Cl; 10% normal donkey serum), then incubated at 4°C overnight in blocking solution including primary antibodies. Rabbit polyclonal anti-torsinA antibody was raised against a peptide of amino acids 319–332 of human torsinA and was a kind gift from B. Lauring (Columbia University). Mouse monoclonal anti-torsinA “DM28A” was a kind gift from X. Breakefield (Harvard University, Boston). Other primary antibodies used were anti-KDEL (StressGen Biotechnologies, Victoria, Canada), anti-laminB1 (Santa Cruz Biotechnology), anti-βactin (Sigma), anti-GFP (AbCam), and anti-β-galactosidase (Sigma). The following day samples were washed, incubated for 2 h with secondary antibodies (Jackson ImmunoResearch) diluted in blocking solution, washed a final time, and prepared for fluorescent microscopy by mounting with VECTASHIELD with DAPI (Vector Laboratories). Immunofluorescent samples were examined by using a Zeiss Axioskop fluorescent microscope. Images were acquired by using a Zeiss LSM 510 multiphoton confocal microscope with the Columbia Optical Microscope Facility. Quantification of transiently transfected cells was performed by a blinded observer assessing 120 torsinA immunoreactive cells from each cDNA transfection for concentrated NE labeling and the presence of spheroid bodies. Each cDNA construct was transfected on three separate occasions, and differences between mutated and WT torsinA were assessed by using the unpaired, two-tailed Student's t test.
ImmunoGold Electron Microscopy. Confluent BHK21 cells stably expressing GFP-ΔE302/3-torsinA were fixed with 4% paraformaldehyde/0.1% glutaraldehyde in 0.1 M cacodylate (pH 7.4) for 30 min, washed with 0.05 M ammonium chloride in the same buffer for 15 min, treated with 0.5% Triton X-100 in PBS for 2 min, washed with PBS, blocked with 1% BSA-PBS, and incubated with anti-GFP in 0.5% BSA-PBS overnight at 4°C. After washing with PBS to remove primary antibody, the cells were probed with goat anti-rabbit conjugated to 5-nm gold particles. Labeled cells were then fixed with 2.5% glutaraldehyde in 0.1 M cacodylate (pH 7.4) for 2 h, postfixed with 1% osmium tetroxide in the same buffer for 1.5 h, treated with 0.5% aqueous uranyl acetate, dehydrated in graded alcohol, treated with propylene oxide, and embedded in Embed 812 (Electron Microscopy Sciences, Fort Washington, PA). Ultrathin sections were viewed by using a JEOL 100cx electron microscope operated at 80 kV. The distance of gold particles from heterochromatin was measured on electron micrographs of four randomly selected, separate cells.
Results
We began our studies of the mechanism of the ΔE302/3 mutation by determining the subcellular distribution of torsinA in neurons, the dysfunctional cell type in DYT1 dystonia. We examined torsinA immunoreactivity in the brains of transgenic mice that express comparable levels of either WT or ΔE302/3-torsinA as assessed by β-galactosidase reporter activity (Fig. 1B) and fluorescent intensity of torsinA immunoreactivity (Fig. 1 C and D). WT-torsinA predominantly colocalized with an ER marker (KDEL epitope) in all neuronal types examined, including frontal cortex neurons (Fig. 1C). In contrast, ΔE302/3-torsinA was strikingly colocalized with laminB1 in the neuronal NE (Fig. 1D) and almost entirely excluded from the bulk ER (Fig. 1C).
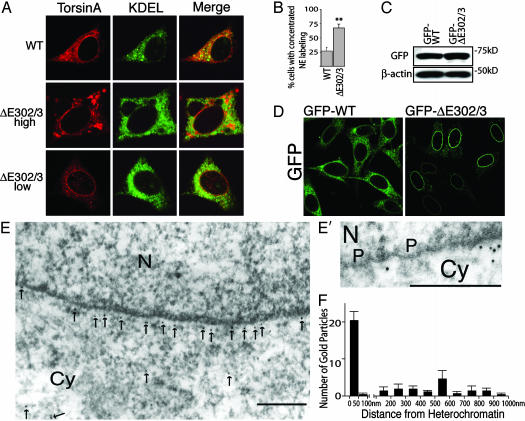
To explore the structural and functional features of torsinA that mediate its interaction with the NE, we studied the behavior of torsinA in transfected BHK21 cells (Fig. 2). Highly expressed WT and ΔE302/3-torsinA remained fully sensitive to digestion by endoglycosidase H (data not shown), indicating that they both enter and are normally retained within the ER (9, 10). Similar to our observations in neurons, WT-torsinA largely colocalized with KDEL, although it also displayed greater NE immunoreactivity than is typically observed of ER proteins (Fig. 2 A). ΔE302/3-torsinA displayed marked NE labeling alongside the previously reported spheroid bodies (Fig. 2 A) (9, 10, 14), which were only observed in the highest-expressing cells (Fig. 2 A Middle). Notably, we observed selective NE torsinA immunoreactivity in cells expressing low to moderate amounts of ΔE302/3-torsinA (Fig. 2 A Bottom); selective NE immunoreactivity was never observed in cells transfected with WT-torsinA. These spheroid bodies appear similar to the cytoplasmic structures that are frequently observed when NE proteins are expressed at high levels (15–17).
Fig. 2.
ΔE302/3-torsinA is concentrated in the NE in vitro.(A) Immunofluorescent labeling of BHK21 cells transiently transfected with WT- or ΔE302/3-torsinA. (Middle and Bottom) NE concentration of ΔE302/3-torsinA in both high and low ΔE302/3-torsinA-expressing cells. Only high ΔE302/3-torsinA-expressing cells displayed spheroid body accumulation of torsinA. No differences were observed between high and low WT-torsinA-expressing cells. (B) Percentage of transfected cells with concentrated NE torsinA labeling. Each cDNA transfection was performed three times; error bars represent SEM. **, Significant (P < 0.01) difference from WT. (C) SDS/PAGE of Triton X-100-extracted protein illustrating that GFP-tagged torsinA proteins were expressed at similar levels. (D) Direct GFP fluorescence from BHK cells stably expressing GFP-tagged WT-torsinA or ΔE302/3-torsinA. (E and E′) ImmunoGold electron microscopic localization of GFP in GFP-ΔE302/3-torsinA cell line. Gold particles are highlighted with arrows. N, nucleus; Cy, cytoplasm; P, nuclear pore complex. (Scale bars, 200 nm.) (F) Distance of gold particles from electron dense heterochromatin was measured in images from four randomly selected cells. Error bars represent SEM.
To rigorously test the significance of the differences observed between transfected WT- and ΔE302/3-torsinA, we quantified the percentage of cells displaying concentrated NE torsinA immunoreactivity. These experiments confirmed that a significantly higher percentage of ΔE302/3-torsinA-transfected cells display concentrated NE immunoreactivity than WT-torsinA-transfected cells (Fig. 2B; P < 0.01). Nevertheless, it was still possible that these findings resulted from a difference between the expression levels of WT and ΔE302/3-torsinA. Therefore, we generated clonal cell lines that stably express similar amounts of either GFP-tagged WT- or ΔE302/3-torsinA to determine whether the differences in subcellular distribution persisted (Fig. 2 C and D). The GFP fluorescence of GFP-WT-expressing cells was distributed throughout the ER, whereas GFP-ΔE302/3 was again specifically concentrated in the NE (Fig. 2D), consistent with an intrinsic, expression level-independent difference between WT- and ΔE302/3-torsinA.
The NE is a double-membrane structure (separated by a 20- to 50-μm perinuclear space) in continuity with the ER and is interrupted by nuclear pore complexes (18). To further verify that ΔE302/3-torsinA is present in the perinuclear space, we examined the subcellular distribution of GFP-ΔE302/3-torsinA in the stably expressing cell line by using ImmunoGold electron microscopy (Fig. 2 E and E′). We found that the majority of gold labeling was located close to the nucleus (Fig. 2 E and F). Because it was necessary to solubilize intracellular membranes to obtain immunolabeling, the inner and outer nuclear membranes rarely remained in labeled cells. Consequently, to further investigate the localization of gold particles, we determined the thickness of the NE in nonpermeabilized cells by measuring the distance between heterochromatin and the outer nuclear membrane. This distance was 35 nm, consistent with previous measurements of the NE (18). We found that perinuclear gold particles averaged 21 ± 1 nm from heterochromatin, and we did not observe labeling of nuclear pores (Fig. 2E′). Thus, the ultrastructural localization of ΔE302/3-torsinA appears consistent with the perinuclear immunofluorescent pattern; these data, together with the fact that torsinA has a signal peptide and is resident within the pre-Golgi secretory pathway, suggest that ΔE302/3-torsinA is concentrated within the perinuclear space (Fig. 2E). Transmembrane inner nuclear membrane proteins are selectively retained in this specialized membrane by interacting with nuclear elements (19–21). Because torsinA is not a transmembrane protein (10), our observations in neurons and BHK21 cells may reflect the fact that ΔE302/3-torsinA interacts with the ER luminal domain of a NE protein.
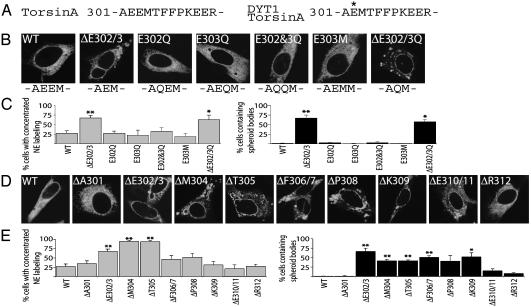
The DYT1 mutation removes one of a pair of negative charges (Fig. 3A); so, we examined whether a charge alteration was responsible for phenotypic differences between WT- and ΔE302/3-torsinA. However, the E302Q and E303Q mutations did not recapitulate the redistribution of ΔE302/3 from the ER into the NE (Fig. 3 B and C). The DYT1 mutation also results in the closer apposition of A301 and M304, but mimicking this feature (E303M) also failed to replicate the DYT1 cellular phenotype (Fig. 3 A–C). Furthermore, removal of the negative charge that remains in ΔE302/3-torsinA (ΔE302/3Q) did not prevent the DYT1-like redistribution (Fig. 3 B and C). These observations suggested to us that removal of an amino acid in this region is key to the redistribution phenotype; so, we tested this possibility by producing a series of mutant proteins containing deletions between positions 301(A) and 312(R) (Fig. 3 A, D, and E). Single amino acid deletions at positions 304 and 305 were indistinguishable from ΔE302/3-torsinA. Deletions at positions 306–309 caused a phenotype intermediate between WT and DYT1 proteins, and deletion of E310/311 or R312 closely resembled WT (Fig. 3 D and E). These data support the idea that a deletion per se, rather than loss of negative charge, underlies the DYT1 cellular phenotype. The fact that several, but not all, deletions in this region produce redistribution to the NE suggests that a specific structural disruption underlies this phenotype and that some of these other deletions might also be capable of producing DYT1 dystonia. This raises the question of why only the ΔE302/3-torsinA mutation has been identified in patients with DYT1 dystonia, and why it has arisen de novo in multiple populations (22). A possible explanation for the selective occurrence of this mutation may be that the sequence surrounding the site of the ΔE302/3 mutation contains an imperfect 24-bp tandem repeat, which is stabilized by the GAG deletion (22).
Fig. 3.
Multiple single amino acid deletions in torsinA mimic ΔE302/3-torsinA mislocalization to the NE. (A) Primary amino acid sequence of the C-terminal region surrounding the DYT1 mutation. (B) Immunofluorescent torsinA labeling of transiently transfected BHK21 cells. (C) Percentage of transfected cells with concentrated NE torsinA labeling or spheroid bodies. (D) Immunofluorescent torsinA labeling of BHK21 cells transiently transfected with versions of torsinA containing single amino acid deletions. (E) Percentage of transfected cells with concentrated NE torsinA labeling or spheroid bodies. All cDNA transfections were performed three times; error bars represent SEM. * and **, Significant (P < 0.05 and P < 0.01, respectively) difference from WT.
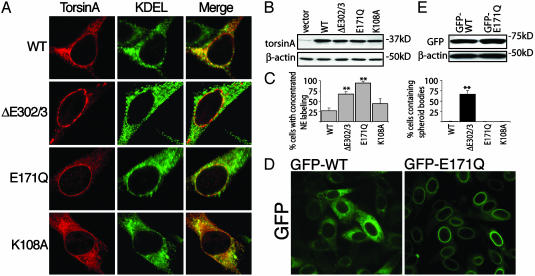
We exploited the presence of the AAA Walker A and B domains in torsinA (Fig. 1 A) to explore whether torsinA substrate(s) reside in the NE. A well characterized mutation in the Walker B domain (E to Q at position 171 in torsinA) leaves ATP binding intact but inhibits ATP hydrolysis by Walker box-containing proteins (23–25); this mutation results in a permanently ATP-bound protein, locking it in a high-affinity substrate-associated transition state (7). We found that E171Q-torsinA localized exclusively to the NE (Fig. 4 A and C). However, we also observed diffuse labeling of the ER in cells expressing high levels of E171Q-torsinA, as is characteristic of highly expressed NE proteins (16). In contrast, a mutation in the Walker A domain that inhibits binding of ATP (K to A at position 108 in torsinA) did not significantly alter the normal subcellular localization of the protein (Fig. 4 A and C). To confirm that these results were not confounded by differences in levels of expression between constructs, we generated a clonal cell line that expressed GFP-tagged E171Q-torsinA at a similar level to the GFP-WT-torsinA line (Fig. 4 D and E). The GFP fluorescence in the GFP-E171Q-torsinA cell line was specifically concentrated in the NE, similar to that observed with the GFP-ΔE302/3-torsinA line (Figs. 2D and 4D). Considering that preventing ATP hydrolysis causes high-affinity interactions between a wide range of AAA proteins and their respective protein targets (23, 24, 26–29), the specific localization of E171Q-torsinA to the perinuclear space is consistent with the possibility that a torsinA substrate resides in the NE.
Fig. 4.
ATP-bound torsinA is selectively localized at the NE. (A) Immunofluorescent labeling of transiently transfected BHK21 cells. (B) SDS/PAGE of Triton X-100-extracted protein shows that the torsinA constructs were expressed at similar levels. (C) Percentage of transfected cells with concentrated NE torsinA labeling or spheroid bodies. Each cDNA transfection was performed three times; error bars represent SEM. **, Significant (P < 0.01) difference from WT. (D) Direct GFP fluorescence from BHK21 cells stably expressing GFP-tagged WT- or E171Q-torsinA. (E) SDS/PAGE of Triton X-100-extracted protein demonstrating that GFP-tagged torsinA proteins were expressed at similar levels.
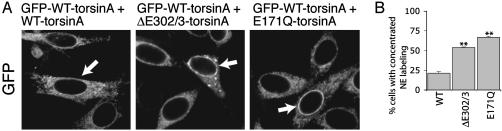
Most AAA proteins function as multimeric protein complexes (5, 7), raising the possibility that ΔE302/3-torsinA exerts a dominant effect on WT-torsinA. Our findings, together with the dominant inheritance of the human disease, suggest that ΔE302/3-torsinA may associate with and abnormally concentrate WT protein at the NE. To further assess this possibility, we transfected the GFP-WT-torsinA cell line with untagged WT-, ΔE302/3-, or E171Q-torsinA. Transfection of unlabeled WT-torsinA did not affect the distribution of GFP immunoreactivity (Fig. 5A; arrow indicates transfected cell). In contrast, transfection of either unlabeled ΔE302/3- or E171Q-torsinA into GFP-WT-torsinA cells clearly altered the pattern of GFP immunoreactivity. Transfection of unlabeled ΔE302/3-torsinA caused GFP-WT-torsinA to relocate to the NE and spheroid bodies (Fig. 5 A and B; arrow indicates transfected cell). Similarly, transfection of E171Q-torsinA caused a dramatic relocalization of GFP immunoreactivity to the NE (Fig. 5 A and B; arrow indicates transfected cell). The fluorescent intensity of the NE (and spheroid bodies) of GFP-WT-torsinA cells transfected with ΔE302/3-torsinA was greater than any region of the surrounding untransfected cells, presumably because of the recruitment and concentration of GFP-WT-torsinA in these structures.
Fig. 5.
ΔE302/3-torsinA recruits WT-torsinA to the NE. (A) Distribution of GFP immunoreactivity in the GFP-WT-torsinA cell line transfected with WT-, ΔE302/3-, or E171Q-torsinA. Arrows indicate transfected cells (identified by β-galactosidase expression). (B) Quantification of the percentage of transfected cells with clearly enhanced NE GFP immunolabeling (compared to untransfected GFP-WT-torsinA cells). Quantification was performed by a blinded observer assessing the NE concentration of GFP labeling in 100 β-galactosidase-positive cells. Each cDNA transfection was performed three times; error bars represent SEM. **, Significant (P < 0.01) difference from WT transfected GFP-WT-torsinA cells.
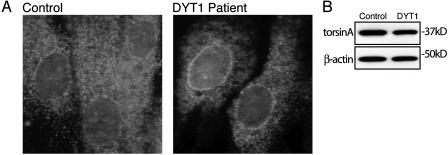
We find that ΔE302/3-torsinA is specifically located in the NE upon in vitro transfection and in transgenic mouse neurons. We subsequently assessed whether this NE mislocalization occurs in tissue from patients with DYT1 dystonia to explore the potential relevance of this finding to the human disease. We examined the subcellular distribution of endogenous torsinA in primary fibroblasts derived from DYT1 patients (heterozygous for the ΔE302/3 mutation) and controls. Control fibroblasts exhibit diffuse torsinA labeling, with the exception of an additional thin band of immunoreactivity around the nucleus (Fig. 6A). In contrast, torsinA immunoreactivity in patient fibroblasts was strikingly concentrated around the nucleus, and there was noticeably reduced ER labeling compared with control cells. Immunoblot of cell lysates confirmed that control and DYT1 patient cultures express similar amounts of torsinA (Fig. 6B).
Fig. 6.
TorsinA abnormally localizes to the NE in tissue from DYT1 patients. (A) Immunofluorescent torsinA labeling of control and DYT1 patient fibroblasts. (B) SDS/PAGE of Triton X-100-extracted protein probed with anti-torsinA and anti-β-actin antibodies.
Discussion
Our observations that (i) a greater amount of WT-torsinA is located in the NE than is typical for an ER protein and (ii) E171Q-torsinA concentrates specifically in the NE suggest that torsinA normally interacts with NE substrate(s). Null mutations in the Caenorhabditis elegans torsinA homologue ooc-5 lead to early defects in nuclear rotation (30, 31), consistent with a role for torsinA at the NE. Also consistent with our observations is the demonstration that torsinA is present in purified NE membranes isolated from rodent liver (16). Thus, the behavior of torsinA appears to be unique in two respects: it is, to our knowledge, the first example of a non-transmembrane protein that concentrates in the NE, and significant amounts of the protein reside simultaneously both in the bulk ER and the NE.
The finding that ΔE302/3-torsinA abnormally concentrates in the NE of patient-derived fibroblasts, neurons, and BHK21 cells suggests that DYT1 dystonia may be caused by the abnormal interaction of ΔE302/3-torsinA with a normal torsinA NE substrate. Furthermore, ΔE302/3-torsinA appears to recruit WT protein to the NE, which has the potential to explain the dominant nature of the disease. Our data do not allow us to conclude with certainty whether the disease might result from an accumulation of torsinA at the NE, or loss of torsinA from the ER. However, the possibility that torsinA may normally interact with a substrate in the NE suggests that this is the more likely site of disease pathogenesis. Future studies using brain tissue from DYT1 patients will be necessary to further evaluate this hypothesis.
There are several diseases caused by mutations of genes encoding inner nuclear membrane proteins, including Hutchinson–Gilford Progeria, Emery–Dreifuss muscular dystrophy, Charcot–Marie–Tooth type 2 neuropathy, and Dunnigan-type familial partial lipodystrophy (19, 32). Similar to DYT1 dystonia, most of these diseases manifest predominantly as tissue-specific disorders (of muscle, peripheral nerve, or adipocytes), despite affecting widely expressed proteins, and may display decreased penetrance and variable expressivity (33, 34). The torsinA ΔE302/3 mutation also displays reduced penetrance and widely variable expressivity, even among family members. Thus, our data and the clinical features of the disease are consistent with the possibility that DYT1 dystonia may be a previously uncharacterized NE disease and the first, to our knowledge, to selectively affect CNS function.
Acknowledgments
We thank Helen Shio for her valuable help in performing ImmunoGold electron microscopy; R. Ally, L. Zhang, and F. Bianchi for excellent technical help; R. Hen, H. Worman, J. Graff, L. Greene, E. Schon, L. Santarelli, and J. Roseman for helpful comments and suggestions; and X. Breakefield and B. Lauring for the generous gift of anti-torsinA antibodies. W.T.D. also thanks J. Graff and S. Fahn for their professional example and supportive mentorship. This work was supported by grants from the Dystonia Medical Research Foundation and the Parkinson's Disease Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; NE, nuclear envelope.
References
- 1.Fahn, S., Marsden, C. D. & Calne, D. B. (1987) in Movement Disorders 2, eds. Marsden, C. D. & Fahn, S. (Butterworths, London), pp. 332–58.
- 2.Berardelli, A., Rothwell, J. C., Hallett, M., Thompson, P. D., Manfredi, M. & Marsden, C. D. (1998) Brain 121, 1195–1212. [DOI] [PubMed] [Google Scholar]
- 3.Ozelius, L. J., Hewett, J. W., Page, C. E., Bressman, S. B., Kramer, P. L., Shalish, C., de Leon, D., Brin, M. F., Raymond, D., Corey, D. P., et al. (1997) Nat. Genet. 17, 40–48. [DOI] [PubMed] [Google Scholar]
- 4.Neuwald, A. F., Aravind, L., Spouge, J. L. & Koonin, E. V. (1999) Genome Res. 9, 27–43. [PubMed] [Google Scholar]
- 5.Ogura, T. & Wilkinson, A. J. (2001) Genes Cells 6, 575–597. [DOI] [PubMed] [Google Scholar]
- 6.Patel, S. & Latterich, M. (1998) Trends Cell Biol. 8, 65–71. [PubMed] [Google Scholar]
- 7.Vale, R. D. (2000) J. Cell Biol. 150, F13–F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dron, M., Meritet, J. F., Dandoy-Dron, F., Meyniel, J. P., Maury, C. & Tovey, M. G. (2002) Genomics 79, 315–325. [DOI] [PubMed] [Google Scholar]
- 9.Hewett, J., Gonzalez-Agosti, C., Slater, D., Ziefer, P., Li, S., Bergeron, D., Jacoby, D. J., Ozelius, L. J., Ramesh, V. & Breakefield, X. O. (2000) Hum. Mol. Genet. 9, 1403–1413. [DOI] [PubMed] [Google Scholar]
- 10.Kustedjo, K., Bracey, M. H. & Cravatt, B. F. (2000) J. Biol. Chem. 275, 27933–27939. [DOI] [PubMed] [Google Scholar]
- 11.Hewett, J., Ziefer, P., Bergeron, D., Naismith, T., Boston, H., Slater, D., Wilbur, J., Schuback, D., Kamm, C., Smith, N., et al. (2003) J. Neurosci. Res. 72, 158–168. [DOI] [PubMed] [Google Scholar]
- 12.Baron, U., Freundlieb, S., Gossen, M. & Bujard, H. (1995) Nucleic Acids Res. 23, 3605–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tremblay, P., Meiner, Z., Galou, M., Heinrich, C., Petromilli, C., Lisse, T., Cayetano, J., Torchia, M., Mobley, W., Bujard, H., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Farrell, C., Hernandez, D. G., Evey, C., Singleton, A. B. & Cookson, M. R. (2002) Neurosci. Lett. 327, 75–78. [DOI] [PubMed] [Google Scholar]
- 15.Kessel, R. G. (1992) Int. Rev. Cytol. 133, 43–120. [DOI] [PubMed] [Google Scholar]
- 16.Schirmer, E. C., Florens, L., Guan, T., Yates, J. R., III, & Gerace, L. (2003) Science 301, 1380–1382. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, Q., Skepper, J. N., Yang, F., Davies, J. D., Hegyi, L., Roberts, R. G., Weissberg, P. L., Ellis, J. A. & Shanahan, C. M. (2001) J. Cell Sci. 114, 4485–4498. [DOI] [PubMed] [Google Scholar]
- 18.Rhodin, J. A. G. (1975) Histology: A Text and Atlas (Oxford Univ. Press, Oxford).
- 19.Burke, B. & Stewart, C. L. (2002) Nat. Rev. Mol. Cell Biol. 3, 575–585. [DOI] [PubMed] [Google Scholar]
- 20.Holmer, L. & Worman, H. J. (2001) Cell Mol. Life Sci. 58, 1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worman, H. J. & Courvalin, J. C. (2000) J. Membr. Biol. 177, 1–11. [DOI] [PubMed] [Google Scholar]
- 22.Klein, C., Brin, M. F., de Leon, D., Limborska, S. A., Ivanova-Smolenskaya, I. A., Bressman, S. B., Friedman, A., Markova, E. D., Risch, N. J., Breakefield, X. O. & Ozelius, L. J. (1998) Hum. Mol. Genet. 7, 1133–1136. [DOI] [PubMed] [Google Scholar]
- 23.Whiteheart, S. W., Rossnagel, K., Buhrow, S. A., Brunner, M., Jaenicke, R. & Rothman, J. E. (1994) J. Cell Biol. 126, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babst, M., Wendland, B., Estepa, E. J. & Emr, S. D. (1998) EMBO J. 17, 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi, T., Tanaka, K., Inoue, K. & Kakizuka, A. (2002) J. Biol. Chem. 277, 47358–47365. [DOI] [PubMed] [Google Scholar]
- 26.Hartman, J. J. & Vale, R. D. (1999) Science 286, 782–785. [DOI] [PubMed] [Google Scholar]
- 27.Pak, M., Hoskins, J. R., Singh, S. K., Maurizi, M. R. & Wickner, S. (1999) J. Biol. Chem. 274, 19316–19322. [DOI] [PubMed] [Google Scholar]
- 28.Singh, S. K., Guo, F. & Maurizi, M. R. (1999) Biochemistry 38, 14906–14915. [DOI] [PubMed] [Google Scholar]
- 29.Arlt, H., Tauer, R., Feldmann, H., Neupert, W. & Langer, T. (1996) Cell 85, 875–885. [DOI] [PubMed] [Google Scholar]
- 30.Basham, S. E. & Rose, L. S. (1999) Dev. Biol. 215, 253–263. [DOI] [PubMed] [Google Scholar]
- 31.Basham, S. E. & Rose, L. S. (2001) Development (Cambridge, U.K.) 128, 4645–4656. [DOI] [PubMed] [Google Scholar]
- 32.De Sandre-Giovannoli, A., Bernard, R., Cau, P., Navarro, C., Amiel, J., Boccaccio, I., Lyonnet, S., Stewart, C. L., Munnich, A., Le Merrer, M. & Levy, N. (2003) Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- 33.Bonne, G., Mercuri, E., Muchir, A., Urtizberea, A., Becane, H. M., Recan, D., Merlini, L., Wehnert, M., Boor, R., Reuner, U., et al. (2000) Ann. Neurol. 48, 170–180. [PubMed] [Google Scholar]
- 34.Vytopil, M., Ricci, E., Dello Russo, A., Hanisch, F., Neudecker, S., Zierz, S., Ricotti, R., Demay, L., Richard, P., Wehnert, M., et al. (2002) Neuromuscul. Disord. 12, 958–963. [DOI] [PubMed] [Google Scholar]