Abstract
The Mediator is an evolutionarily conserved, multiprotein complex that is a key regulator of protein-coding genes. In metazoan cells, multiple pathways that are responsible for homeostasis, cell growth and differentiation converge on the Mediator through transcriptional activators and repressors that target one or more of the almost 30 subunits of this complex. Besides interacting directly with RNA polymerase II, Mediator has multiple functions and can interact with and coordinate the action of numerous other co-activators and co-repressors, including those acting at the level of chromatin. These interactions ultimately allow the Mediator to deliver outputs that range from maximal activation of genes to modulation of basal transcription to long-term epigenetic silencing.
The unique transcription programmes of protein-coding genes are dependent on regulatory sequences that are bound by a wide range of gene- and tissue-specific transcriptional factors, which respond to developmental and environmental signals. However, the programme is interpreted largely through the precisely orchestrated recruitment of co-activators by these specific transcription factors1. To take effect, the regulatory signals must be properly relayed to the general RNA polymerase II (Pol II) machinery that is responsible for transcribing protein-coding genes. This machinery is involved in the formation and function of a pre-initiation complex (PIC), which — in addition to Pol II — consists of the general transcription factors (GTFs) TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH. Consistent with a broad two-step mechanism for transcription (FIG. 1), studies over the past two decades have identified numerous co-activators that facilitate the initial penetration of the chromatin barrier to allow the transcriptional machinery access to the regulatory sequences or directly target PIC components. Of the limited number of co-activators that are known to directly target Pol II, the 30-subunit Mediator complex has emerged as potentially the most crucial by virtue of its widespread and multifaceted role across the transcriptome (BOX 1).
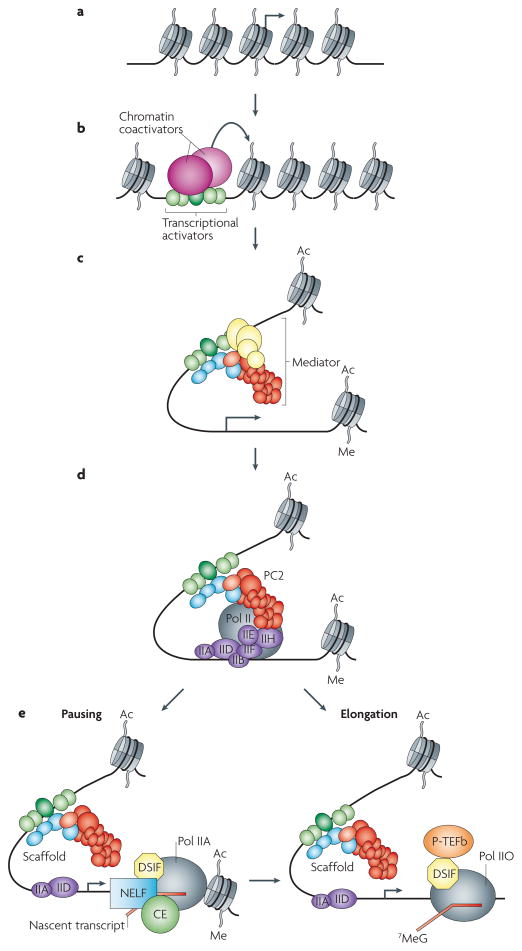
Figure 1. Involvement of Mediator in multiple steps of transcription.
Current models of transcriptional activation involve multiple steps. a | An idealized pathway is shown that begins with chromatin, in which nucleosomes exist in a characteristic beads-on-a-string array. This contrasts to transcriptionally inert chromatin, a highly compacted structure in which DNA is tightly packaged not just through wrapping around nucleosomes but through additional higher-order structures that entail linker histones and other proteins such as heterochromatin protein 1 (not shown). b | The activation pathway is initiated by one or more transcriptional activators that bind to their cognate sites in the regulatory region of the gene. These factors recruit a series of chromatin co-activators that can covalently modify nucleosomes at specific histone residues (not identified) and mobilize nucleosomes through ATP-requiring reactions. c | The resulting intermediate contains chromatin that is characterized by distinct covalent modifications, such as acetylation (Ac) and methylation (Me), and by a relative dearth of nucleosomes. The activators then recruit Mediator. In some cases, the intact Mediator that consists of the core and the kinase module might be recruited at this stage. d | Pre-initiation complex (PIC) assembly, entailing the various general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH) and RNA polymerase II (Pol II), and transcription initiation then ensue with concomitant restructuring of the Mediator that results from loss of the kinase module. e | After Pol II clears the promoter, there are two possible outcomes. As shown on the right, the process can proceed directly to the elongation phase at which it is associated with elongation factors that include DSIF and P-TEFb, whose entry into the transcription elongation complex (TEC) may coincide with capping (7-methyl-guanosine; 7 MeG) of the nascent RNA. The RPB1 carboxy-terminal domain (CTD) also undergoes substantial phosphorylation at Ser2 and Ser5 (Pol IIO) through the sequential actions of TFIIH and P-TEFb. Importantly, a scaffold containing a vestigial subset of general transcription factors and Mediator remains at the promoter to potentially facilitate subsequent rounds of transcription. Alternatively, as shown on the left, at many loci, Pol II may be subject to promoter-proximal pausing (at approximately nucleotide +50). This Pol II would be associated with DSIF but phosphorylated only at Ser 5 (Pol IIA). Under appropriate conditions, the paused Pol II complex can also mature into an elongation complex. Although depicted linearly, the intermediates are not likely to be as clear-cut as shown. Indeed, as discussed in the text, Mediator couples many of the steps. Moreover, some aspects of the reaction are reversible, again this occurs under the control of Mediator.
Box 1. Generality of Mediator action.
A long-standing issue concerns whether Mediator is required for expression of all genes in a genome. The earliest data from genome-wide analyses in yeast indicated that inactivation of the core MED17 subunit affects transcription of the majority of genes in yeast cells; indeed, the phenotype of these mutants was found to be essentially indistinguishable from that of cells bearing mutations in the RNA polymerase II (Pol II) RPB1 subunit114. This led to the working hypothesis that Mediator is required for the transcriptional regulation of all genes. However, subsequent studies yielded conflicting results115–118. On balance, the evidence favours the idea that Mediator is recruited to the promoters of essentially all genes in yeast; however, the relative steady-state distribution of Mediator versus Pol II might not be the same for all genes. The variable Mediator/Pol II ratio on genes might also reflect a further role for Mediator that does not necessarily entail close coupling between Mediator and Pol II activity. For example, a relatively low Mediator/Pol II ratio at a gene might indicate a Mediator requirement in the first and not subsequent rounds of transcription. Thus, as Mediator acts at all (or nearly all) genes in yeast and by extrapolation (with some qualifications) also in metazoan cells, it might even be viewed as a general transcription factor (GTF). Such a concept of Mediator as a GTF is directly supported by in vitro studies, in yeast and mammalian systems, which revealed a Mediator requirement for basal (activator-independent) transcription29,119–121. Indeed, as discussed in the main text, a major part of Mediator function is through its direct effects on the general transcription machinery. Even so, recent studies in metazoan systems point to the possibility of Mediator-independent activation mechanisms and, hence, an apparently conditional requirement for Mediator. These include the identification of Gdown1, which was initially isolated as a polypeptide that strongly associates with the 12-subunit Pol II122 and also of the transcription elongation factor DSIF94 as factors contributing to an increased degree of dependence on Mediator for activated transcription. Although the precise mechanisms by which these factors elicit Mediator requirements are presently unclear, the results could reflect checkpoint-type mechanisms that restrict gene transcription to situations in which the relevant activators responding to their particular signals deliver Mediator to the promoters of the target genes.
It also is emerging that some components of the general transcription machinery might not be invariant, at least in higher metazoans. For example, in a cultured cell model for skeletal muscle differentiation in which myotubes can be distinguished from myoblasts, but evidently not in bulk skeletal tissue from mice123, TFIID is seemingly lost in the myotubes and replaced by an alternative and much pared down version of TFIID that contains the TBP-related factor, TRF3 as the TATA-recognizing component and just one additional TAFII, TAF3 (REF. 124). Concomitantly, expression of some Mediator subunits in myotubes is much reduced relative to levels in precursor cells. The possibility has been raised that in at least some highly specialized cases, such as in terminally differentiated, non-dividing cells, the transcription of some genes in these cells might be Mediator-independent. Thus, overall, although there is evidence for widespread requirement for Mediator — indeed even as a quasi-GTF — there may be occasions when the transcription machinery operates independently of the Mediator. Nonetheless, this might not necessarily imply that genes subject to such control are entirely Mediator-independent throughout their transcriptional history.
Following its biochemical isolation from yeast and metazoan cells2,3, characterization of the Mediator has mostly focused on understanding how it relays signals from site-specific factors to the PIC. Specific interactions occur both between individual Mediator subunits and transcriptional activators and between the Mediator and Pol II. These findings led to the general model that regulatory signals are processed through the ability of the Mediator to interface between the two machineries and thereby promote the stable formation of the PIC. However, consistent with the structural complexity of the Mediator, it is becoming increasingly apparent that its role in transcription is not simply restricted to such a passive function. Thus, the simple adaptor function of the Mediator in transcription needs to be reappraised in favour of a model in which Mediator can sense and assimilate a multitude of signals and then deliver a properly calibrated output to the transcription machinery. In this updated view, Mediator is more of an integrative hub than a device for just maximizing transcription levels.
Focusing primarily on the metazoan complex, and also drawing on studies in yeast as warranted, this Review discusses the many activities that have recently been ascribed to the Mediator. We first highlight the means by which these Mediator activities, which reside in distinct modules, are brought into play as a result of interactions with diverse factors. We then discuss well-studied examples of Mediator interactions, outlining some of the emerging general principles that might underpin Mediator action. Besides facilitating activator function, Mediator can also stimulate activator- independent basal transcription, act at steps subsequent to PIC assembly and repress transcription in specific contexts that include effects at the chromatin level. We argue that these characteristics enable Mediator to function as a signal-processing centre for numerous genetic programmes.
Structure of Mediator
The initial discovery and description of the Mediator as a crucial eukaryotic co-activator represented a remarkable convergence of diverse lines of investigation in the transcription field. Thus, numerous studies in yeast and metazoan systems that aimed to identify the cofactors which enable the functions of diverse transcriptional activators identified Mediator as a Pol II- and activator-interacting multiprotein complex4–6. It has since become apparent through bioinformatics analyses that most of the subunits that comprise Mediator complexes contain discrete domains that have been conserved throughout eukaryotic evolution7,8. Illustrating the idea that genes found to encode putative Mediator subunits in diverse eukaryotic organisms give rise to corresponding functional complexes, a homology-based approach led to the isolation of a bona fide Mediator complex from Arabidopsis thaliana, a plant species9. Consistent with a major role in transcriptional regulation, the newly isolated plant complex was also found to contain factors that had previously been implicated in the expression of several key developmental programmes by genetic studies.
Primarily through studies of the yeast Mediator, combined genetic, biochemical and structural approaches have indicated that individual subunits reside in one of four distinct modules designated ‘head’, ‘middle’, ‘tail and ‘kinase’ (REFS 7,10–12)(FIG. 2). Thus, despite its structural complexity, Mediator possesses an underlying modular architecture that reflects a division of labour at the functional level. Whereas the head, middle and tail modules form a relatively stable ‘core’ structure, the kinase module — which consists of cyclin-dependent kinase 8 (CDK8), cyclin C and the MED12 and MED13 subunits — associates reversibly with the Mediator complex. Indeed, the two main variants of the human Mediator, exemplified by the PC2 form (which was isolated solely on the basis of its ability to mediate activator-dependent transcription in vitro) and by the larger thyroid receptor- associated protein (TRAP) form (which was isolated following its ligand-dependent association with the thyroid hormone receptor13), differed mainly with respect to the absence of the kinase module in the PC2 complex14,15. It has been suggested that the biochemical isolation of these variant forms might reflect a natural interconversion process that takes place at target genes as part of the transition from an inactive (or repressive) form of the Mediator to a more active form3,15. Broadly, and consistent with the core (PC2) complex being the active form of the Mediator, the head and middle modules are believed to be involved in interactions with the core Pol II machinery, whereas the tail subunits interact with various activators. The kinase module may bind, in part, the same general surface of the core Mediator complex as does Pol II, and this module might therefore occlude Pol II from the complex to exert a repressive function16,17.
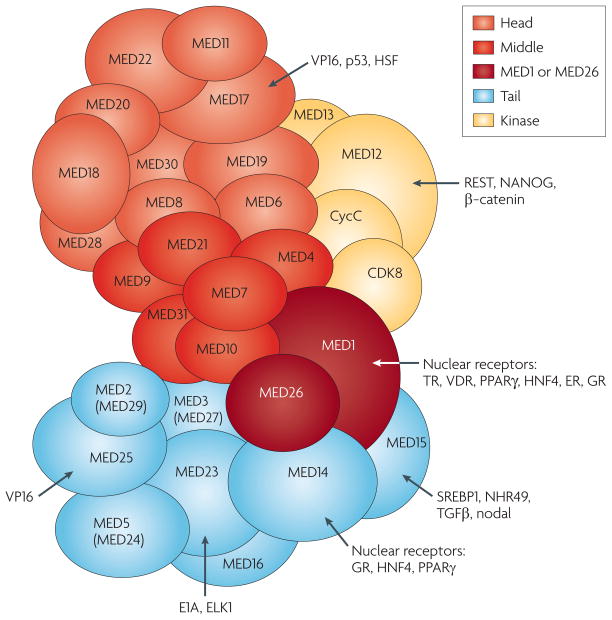
Figure 2. Modular structure of Mediator and interactions with diverse factors.
A composite depiction of the subunit structure of the human Mediator complex is shown. Note that although the relative placement of the subunits in the subcomplexes is based on published binary interaction and partial structural data10,52,125,126, it is primarily for illustration; the precise locations have yet to be mapped. The MED1 and MED26 subunits, which are not present in all isolates, are likely to be located at the junction of the middle and tail modules. MED28 and MED30 have been provisionally placed in the middle module. Transcription factors selected for discussion in the main text and their target subunits in Mediator are also indicated. The unified nomenclature proposed by Bourbon et al.127 has been used here. However, very recent bioinformatic analyses strongly suggest that MED27 and MED29, which were earlier considered to be metazoan-specific, are related to yeast MED3 and MED2, respectively. Here they are referred to as MED3 (also known as MED27) and MED2 (also known as MED29)7. Similarly, MED5 and MED24 may be related and referred to as MED5 (also known as MED24). ER, oestrogen receptor; GR, glucocorticoid receptor; HNF4, hepatocyte nuclear factor; NHR49, nuclear hormone receptor 49; PPARγ, peroxisome proliferator-activated receptor-γ; SREBP1, sterol regulatory element-binding protein 1; TGFβ, transforming growth factor-β; VDR, vitamin D3 receptor.
In addition to variability that arises directly from an intrinsic modularity of the complex, other sources of heterogeneity have been identified, especially in mammalian systems. These include preferential incorporation into the core complex of one of two paralogues of the individual subunits (for example, MED13 versus MED13L or CDK8 versus CDK11)8,18. Additional variation arises from substoichiometric or tissue-specific expression of a given subunit (for example, MED1) relative to other Mediator subunits19,20.
Despite evidence of strong evolutionary pressure on the complex — which has led not only to preservation across eukaryotes of genes encoding many of the subunits (as mentioned above) but also of the gross architecture of the complex11 — individual subunits of the mammalian complex have diverged considerably from their yeast orthologues. In fact, there is only patchy homology in the subunits7. A potentially important observation is that these regions of homology are interspersed in intrinsically disordered regions that can acquire structural features on interactions with other proteins. This is consistent with developing ideas on the malleability of the transcription machinery21. As the metazoan Mediator subunits display a much higher prevalence of such homologous regions, this argues in favour of the greater range of potential interactions for these Mediator subunits, in accord with the higher degree of complexity associated with metazoans. In addition, there are a few bona fide Mediator subunits that seem to be exclusive either to yeast or to metazoans. Most notably, MED26, which is greatly enriched in the PC2 and CRSP forms of the mammalian Mediator3,22, as well as MED23 and MED25, which are targeted by some transcription factors, are not found in yeast (FIG. 2).
An end point of diverse signalling pathways
The functional versatility of the metazoan Mediator has become increasingly evident through its implication in a wide range of physiological processes and through demonstrations of physical interactions between numerous site-specific DNA-binding activators and discrete targets in the Mediator23. The relatively well-characterized examples below have been selected to illustrate some major molecular strategies that the Mediator seems to use to absorb the physiological signals borne by these regulators.
Targeting of MED1 by ligand-inducible nuclear receptors
The initial isolation of the metazoan Mediator complex as a TRAP complex established the paradigm that Mediator function entails interfacing between a given activator and the transcriptional machinery13. Nuclear receptors have since continued to provide important mechanistic insights into Mediator function. These transcription factors constitute a special class of activators whose individual members are important regulators of diverse aspects of animal physiology. They are typically regulated by small-molecule ligands that, on association with the cognate receptor, induce discrete conformational changes that in turn unleash a cascade of events, culminating in the activation of the target gene promoter24,25. In general, the activation function 2 (AF2) domain of receptors is predominantly responsible for interactions with diverse co-activators that contain NR boxes, which are characterized by the signature amino-acid sequence motif LXXLL. Beginning with the identification of the Mediator MED1 subunit (which contains two such NR boxes) as a target of the ligand-bound thyroid hormone receptor, many diverse receptors that include (but are not limited to) the vitamin D receptor26, peroxisome proliferator-activated receptor-γ (PPARγ)27,28, hepatocyte nuclear factor 4α (HNF4α)29, the glucocorticoid receptor30 and the oestrogen receptor31 have been shown to directly interact with this subunit. Furthermore, the AF2–NR box interaction was found to be necessary and sufficient for anchoring the entire Mediator complex to the receptor32. The physiological implications of the high degree of specificity underlying this interaction became clear through genetic ablation of Med1 in mice33. Fibroblasts isolated from Med1−/− embryos (which survive until 11.5 days postcoitum and display many normal activation pathways) showed a selective loss of some nuclear receptor-mediated pathways, consistent with the demonstration that, except for the MED1 subunit, the residual complex is mostly intact. The affected pathways included PPARγ-dependent differentiation of fibroblasts to adipocytes27.
Alternative or redundant mechanisms might also exist for receptor-dependent Mediator recruitment in a cellular context28. These could involve (weaker) interactions of other Mediator subunits such as MED14, which has been found to interact with nuclear receptors29,30,34. Alternatively, concurrent or conditional interactions of Mediator with other receptor-bound co-factors might contribute to its recruitment. MED1 can also serve as a target for non-receptor type factors such as GATA1 (also known as ERYF1)35.
Channelling of MAPK signalling through MED23
MED23, which was originally identified as a target of the viral oncoprotein E1A36, is an example of a Mediator subunit that responds to a highly specialized signalling pathway. It is required for Mediator recruitment to a small subset of genes that are mostly under the control of the MAPK-regulated factor ELK1 (REF. 37). In mouse embryonic stem (ES) cells and fibroblasts, these ELK1-regulated genes include early growth response 2 (Egr2, also known as Krox20), which encodes one of the first transcription factors to be induced in an insulin- triggered adipogenesis cascade38. Consequently, like Med1−/− fibroblasts, Med23−/− fibroblasts also fail to undergo adipogenesis in culture after the appropriate treatment. Thus, the MED23 subunit is also one of the end points of the insulin-signalling pathway, which induces RAS–MAPK-dependent activation of ELK1.
MED15 as a regulatory node in lipid metabolism
MED15 provides an interesting paradigm for Mediator subunits that may have evolved to process transcriptional signals for regulating distinct cellular processes. MED15 was originally identified as the Mediator subunit that is targeted by mammalian sterol regulatory element-binding protein 1α (SREBP1α)39, which is a sensor of cellular cholesterol and plays a part in lipid homeostasis by activating crucial genes that regulate the process. More recent evidence suggests that MED15 may be thought of as a master regulator of lipid homeostasis. Like the chromatin-modifying co-activators p300 and CBP, MED15 contains a KIX domain fold that mediates its interaction with SREBP1α. This domain is conserved in the Caenorhabditis elegans orthologue (MDT-15) of MED15 and is similarly used by the worm counterpart of SREBP1 to target the Mediator39. Further suggesting that MED15 is a node for diverse pathways relevant to lipid metabolism in C. elegans, the orphan nuclear hormone receptor 49 (NHR49) — an activator that is unrelated to SREBP1 but nonetheless regulates other genes involved in lipid metabolism — was also found to interact with MED15 (REF. 40). Remarkably, even the yeast orthologue of MED15, which was identified genetically in screens for selective sugar use and originally called GAL11, serves as the target for a zinc finger-containing activator that senses fatty acid levels in this simple organism41. At the same time, it should be noted that MED15 has also been implicated as the target subunit for certain developmental pathways that involve signalling by transforming growth factor-β (TGFβ), activin and nodal42. Together, these observations point to the potential of the Mediator for integration of these cellular processes just as the demonstration of direct roles for MED1, MED15 and MED23 subunits in some aspect of lipid metabolism control is also consistent with the potential function of Mediator as a processing centre.
The tail module as a major locus for signal transduction
Although the precise location in the Mediator of many of the metazoan-specific subunits has yet to be determined, MED16, MED23 and MED24 form a tight-knit submodule in the Mediator complex43,44. Based on further yeast studies7,10, MED15 and the MED16–MED23–MED24 triad might constitute part of the tail in metazoans (FIG. 2). Even the nuclear receptor target MED1, which at first seems to be an exception by virtue of its location in the middle region of Mediator, might be localized relatively close to the junction with the tail subunits10. Thus, many of the above-described pathways terminate in the Mediator tail. This concentration of activator targets to a relatively restricted locus on the metazoan Mediator (FIG. 2) is highly reminiscent of the yeast Mediator, whose tail subunits were initially implicated through genetic analyses as co-factors for gene activation pathways involved in cellular processes as diverse as mating type switching and selective sugar use45. The evolutionary selection for a general architecture of a signal processor, in which even the location of the primary ‘receiving end’ has been maintained, suggests that the underlying mechanisms whereby appropriate transduction of the signals from the activator to the ‘business end’ takes place have also likely been conserved.
Nonetheless, it should also be noted that other activators seem to directly target subunits in the head, middle and kinase modules. For the head and middle module interactions, examples include the tumour suppressor p53 (REF. 46), the viral activator VP16 (REF. 46) and the Drosophila melanogaster heat shock factor (HSF)47, each of which interacts with MED17 in the head module and at least one additional target. Thus, p53 potentially also interacts with MED1 (REF. 48), VP16 with MED25 (REFS 49,50) and HSF with MED23 (REF. 51). Indeed, the targeting by VP16 of MED25 — which is one of a few metazoan subunits with no clear orthologues in the yeast complex — illustrates an alternative model for Mediator interaction, in which association of an otherwise labile Mediator subunit with the bulk complex might potentially take place only in the presence of the interacting activator. Further, interactions of VP16 with both MED25 and MED17 seem to increase the fractional content of MED25 in the bulk Mediator complex, which constitutively occupies the target gene promoter that was investigated49. Most simply, this could reflect direct activator-facilitated recruitment of only the MED25 polypeptide to the PIC. However, an alternative possibility is that the activator facilitates a complete replacement of a MED25-free Mediator complex with one that contains this polypeptide. In fact, nuclear receptor-interacting MED1 is also present at substoichiometric levels in the bulk complex and its content is relatively enriched at Mediator complexes in PICs that are formed following receptor activation and binding to the target genes19. Perhaps this might occur partly as a result of concomitant MAPK-dependent phosphorylation of MED1 (REF. 52).
Involvement of the kinase module in developmental pathways
Many developmental pathways, especially in C. elegans, D. melanogaster and zebrafish models, have been shown to be under the control of the several subunits (CDK8, cyclin C, MED12 and MED13) that make up the kinase module. Intriguingly, a screen for C. elegans genes that might be involved in diverse signalling pathways revealed that the gene encoding MED12 is one of six genes (all encoding subunits of factors implicated in transcription control) that are pivotal for multiple pathways and were thus postulated to constitute major regulatory hubs53. Consistent with this hypothesis, screens for eye development in D. melanogaster had earlier revealed MED12 and MED13 as crucial end points of the Wnt signalling pathway in which interactions are finally established with the promoter-bound transcription factors β-catenin and pygopus54. Direct interactions of Mediator with mammalian β-catenin, leading to activation of target genes, have also been demonstrated55. Further, MED12 is required for neuronal development in zebrafish56 and, relevant to the regulation of neuronal gene expression in mammals, was also shown to be targeted by the negatively acting transcription factor REST57. Overall, and from the standpoint of the Mediator as a signal-sorting centre, it is interesting that so many signalling pathways that are crucial for development and functioning of neural systems have targets in the kinase module. It should also be noted that in the kinase module, MED12 and MED13 have distinct overall effects from CDK8 and cyclin C on D. melanogaster eye, leg and wing development58, suggesting further functional bifurcation in the kinase module. Just as other subunits that respond predominantly to signals from related pathways can also respond to heterologous signals, pathways unrelated to development might potentially also be controlled by the Mediator kinase module59,60.
Effector functions of Mediator
The near ubiquity of Mediator involvement across the transcriptome and its targeting by a wide range of transcription factors with highly specific outcomes raise the issue of how Mediator tailors input information into the right output for a given gene. As described below, Mediator possesses multiple functions that transcription factors can selectively exploit to bring about the desired effects.
Mediator interactions with Pol II and the general transcription machinery
At the simplest level, diversity in outputs may be achieved through the induction of activator-specific conformational states of the Mediator that, in turn, might have different effects on Pol II function61. Electron microscopy analyses of the yeast complex have shown that Mediator makes extensive contacts with Pol II11. Among the many prominent patches of these interactions are contacts with the RPB3 and RPB11 subunits, which — intriguingly — are the eukaryotic orthologues of the bacterial RNA polymerase α-subunit, the main target of transcriptional activators62. Furthermore, overall similarities are evident between the thyroid hormone receptor-bound human Mediator and Pol II- bound yeast Mediator11,63. As each of these complexes presumably reflects an activated form of the Mediator, the structures indicate an evolutionarily conserved ability of the complex to acquire discrete conformational states in response to specific signals.
The ability of Mediator to recruit Pol II is likely mechanistically linked to an ability to directly regulate assembly and function of the PIC as a whole (BOX 1). Potentially revealing in this regard is the demonstration that excess amounts of TFIIB, a GTF that is closely associated with the Pol II active site where it is well positioned to directly affect the initiation process, can partially bypass a requirement for Mediator in in vitro systems64. Further, TFIIH, the GTF that plays an important part in promoter melting and promoter clearance, is also regulated by Mediator at the level of its recruitment to the PIC65,66 and through modulation of its kinase activity67,68. Note also that the kinase module has been associated primarily with negative (repressive) functions of the Mediator. Various mechanisms ranging from physical occlusion of Pol II binding to core Mediator16,17 to CDK8-mediated phosphorylation of the Pol II RPB1 carboxy-terminal domain (CTD)69 and of the cyclin H subunit of TFIIH68 have been proposed previously to account for this activity. However, consistent with substantial evidence from unrelated systems60,70,71, it is also likely that the CDK8 module might even have a positive role through as yet unknown mechanisms.
Although recruitment of Pol II to the PIC is undoubtedly a key Mediator function, theoretical considerations alone dictate that, to achieve the level of customization that exists for the expression programme of each gene, other mechanisms must also contribute. This is most dramatically evident from the divergent outputs elicited, for example, when Mediator is targeted by a nuclear receptor or by REST, with one factor leading to gene activation and the other leading to silencing. Recent demonstrations of Mediator interplay with other diverse co-activators, especially those that work at the level of chromatin and in steps that transpire after Pol II has been recruited (so-called post-recruitment effects), have been particularly informative in explaining how Mediator can deliver such a broad repertoire of outputs.
Mediator and regulation through chromatin
Recent evidence strongly suggests that despite the earlier demonstrated ability of Mediator to efficiently function on naked DNA templates in in vitro assays, which is consistent with its known effects on PIC formation and function, Mediator can also deal with constraints arising from the chromatin environment inside the cell. Broadly, the chromatin state is regulated by nucleosome mobility through the action of ATP-requiring remodelling factors and through alterations in the distribution of characteristic covalent marks on distinct residues (for example, acetylation and methylation on lysines) in the amino-terminal tails of histones that make up nucleosomes72. Among these modifications, acetylation by the histone acetyltransferases (HATs) p300, CBP and GCN5 (a subunit of the SAGA complex) has been strongly correlated with gene activation in various pathways that include ligand-induced nuclear receptor pathways.
In line with models in which chromatin remodelling and PIC formation proceed sequentially1, evidence is emerging that Mediator might directly coordinate transitions between the chromatin machinery and the nascent PIC (FIG. 3). Initial studies showed that activator- recruited Mediator and p300 functionally synergize in activating chromatin templates29,73. More recent studies have further elaborated a detailed mechanism in which Mediator can direct the orderly exchange of chromatin co-activators and PIC components owing to its ability to sequentially interact with both p300 and TFIID74. In this model, the activator, p300 and Mediator initially form a ternary complex on the chromatin template. Subsequent histone acetylation and p300 autoacetylation lead to p300 ejection and formation of a new TFIID-containing complex that in turn leads to PIC formation. However, in other contexts, Mediator and the chromatin factor may be competing for the same site on the activator, as would be expected when LXXLL-containing chromatin co-activators (for example, p160 family members that recruit p300 and other histone-modifying factors) and Mediator (through MED1) compete for AF2 domains of promoter-bound nuclear receptors. In these cases, sequential co-activator exchange is more likely to occur75,76.
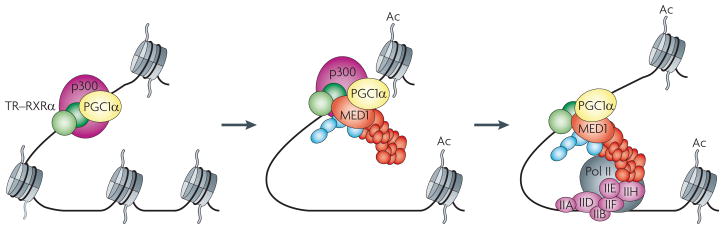
Figure 3. Modulation of Mediator function by ancillary factors: coordination of chromatin remodelling and PIC formation.
Although it plays a crucial part in assembling the pre-initiation complex (PIC), which is formed on nucleosome-free templates, Mediator also contributes to chromatin remodelling through functional interactions with chromatin co-factors. A potential activation pathway is shown that follows the binding of ligand-bound thyroid hormone receptor (TR) in association with its heterodimerization partner retinoic acid receptor-α (RXRα) to their cognate site embedded in chromatin. In the first step, the histone acetyltransferase p300, is recruited. On some genes, this interaction is stimulated by peroxisome proliferator-activated receptor-γ co-activator 1α (PGC1α); other contributors (for example, p160 co-activators) are not shown. As both chromatin co-activators and Mediator bind to the activation function 2 (AF2) domains of nuclear receptors, it was previously suggested that their binding would be mutually exclusive and that Mediator-driven PIC formation would provide the driving force for co-factor exchange on the receptor. More recent evidence suggests that Mediator and p300 can co-exist in a transient ternary intermediate (shown in the second step in the figure) that could facilitate transitions resulting in functional PICs74 (shown in the third step in the figure). Given further that PGC1α dynamically interacts with the Mediator through the Mediator complex subunit 1 (MED1) (structural rearrangements are depicted as an altered orientation of PGC1α), it might further modulate this coordination role of the Mediator77,78. Any potential role of the kinase module in this particular scenario remains unaddressed and has been omitted for clarity.
Importantly, this role of Mediator in coupling chromatin remodelling and PIC formation might be further facilitated, or perhaps fine-tuned, by other gene- and tissue-specific co-activators. Most notably, these include PPAR-γ co-activator 1α (PGC1α) — a co-activator for several nuclear receptors — which displays multipartite interactions with nuclear receptors, p300 and Mediator, and stimulates activation from both chromatin and DNA templates77. Consistent with the fact that PGC1α, like Mediator, also binds nuclear receptors through its LXXLL domain, it is further proposed that the receptor–Mediator–PGC1α interactions are dynamic and that the corresponding ternary complex undergoes structural rearrangements as it sequentially adjusts from a chromatin-remodelling mode to a PIC-supporting mode78. Thus, by acting like a chaperone, PGC1α seems to facilitate transitions between one Mediator-anchored transcriptional intermediate to another (FIG. 3).
In a slight variation of the mechanism described above, Mediator function also has been found to be closely associated with that of the evolutionarily conserved SAGA co-activator complex, whose subunits include various TAFIIs and components that have HAT, kinase and deubiquitylase activities79. Studies in yeast have already provided strong genetic evidence for the convergence of Mediator and SAGA functions at some genes80. In metazoan cells, cooperative physical interactions between Mediator, SAGA and the MYC oncoprotein have been reported to lead to the activation of a target gene (telomerase reverse transcriptase)81. In this case, precisely how Mediator and SAGA work together to bring about activation is not clear, as the ternary complex is recruited to a pre-acetylated chromatin environment and TFIID and Pol II are already present at the promoter. However, it is likely that the cooperative interactions reflect close coupling of SAGA and Mediator functions at the chromatin and PIC levels along the lines discussed above. Other activators might also co-recruit SAGA and Mediator, as indicated by the fact that GCN5 is also detectable in mammalian Mediator preparations isolated following affinity purification on VP16 (REF. 82). Furthermore, a variant GCN5-containing complex (ATAC) is also found in tight association with the Mediator through a bridging factor that might be especially (but not exclusively) enriched in mouse ES cells83.
Unlike the acetylation of histones effected by p300 and GCN5, some chromatin modifications are more closely associated with longer-term and potentially heritable epigenetic states that have important consequences for developmental and tissue-restricted regulation of genes84. In the broadest terms, transcriptionally active genes that constitute euchromatin are primarily characterized by the presence of domains containing nucleosomes enriched with histone H3 methylated at lysine 4 (H3K4). At the other extreme, transcriptionally silent loci such as those in heterochromatin primarily consist of domains bearing methylated H3K9 and H3K27.
Recent studies have revealed that at least one epigenetic pathway is directly under Mediator control57 (FIG. 4). The repressed state of neuronal genes in extraneuronal cells is achieved through formation of a MED12-anchored ternary complex between the silencing factor REST85, Mediator and the methyltransferase G9a, which methylates H3K9 at target genes57. Dimethylated H3K9 can in turn serve as a platform for repressive, heterochromatin protein 1, which recruits DNA methyltransferase 1. These events are accompanied by REST-mediated recruitment of histone deacetylase 1 and lysine-specific demethylase 1, an H3K4 demethylase, which together lead to selective long-term epigenetic silencing of neuronal genes. Thus, consistent with early yeast genetic studies showing that CDK8 can also recruit a histone deacetylase through interactions with TUP1 (REF. 86), the kinase module of the Mediator may have an additional general repressive role at the epigenetic level.
Figure 4. Modulation of Mediator function by ancillary factors: epigenetic silencing.
In concert with additional negatively acting factors, Mediator can contribute to long-term silencing of certain developmentally regulated loci. Starting with an active pre-initiation complex (PIC; which would likely contain the PC2 form of the Mediator), REST binding to the cognate site commits the gene to a heterochromatin fate as development proceeds. A ternary complex containing REST, the histone methyltransferase G9a and the intact Mediator is first assembled. The multipartite interactions are anchored through the MED12 subunit of the kinase module. Following G9a action and dimethylation of histone H3 lysine 9 (H3K9) to H3K9me2, heterochromatin protein 1 (HP1) and DNA methyltransferase 1 (DNMT1) are recruited to the site. Ultimately, the gene is embedded in transcriptionally inert heterochromatin marked by H3K9me2 and methylated DNA (CH3). Other factors (for example, histone deacetylase 1 and lysine-specific demethylase 1) that contribute to silencing are not shown. The figure is based on the findings described in REF. 57.
Epigenetic regulation is also believed to underlie the remarkable ability of ES cells to both self-renew and maintain pluripotency. Many loci in these cells are characterized by bivalent chromatin domains that contain both methylated H3K4 and H3K27 marks, thereby contributing to the poised status of these cells87. As ES cells are induced to differentiate, these domains are appropriately resolved depending on whether a given gene is epigenetically activated or silenced. Intriguingly, MED12 is also targeted by NANOG88. NANOG is one of the key transcription factors required for maintenance of the ES state (others include OCT4 and SOX2) and controls the expression of its own gene89. Global expression profiling following Nanog- or Med12-knockdown further revealed common sets of genes that are activated or repressed by these factors, indicative of substantial functional overlap88. Thus, it is tempting to suggest that MED12 (and hence the Mediator as a whole) might be directly involved in maintaining the poised state of ES cells through manipulation of the epigenetic landscape. By analogy with how REST effects H3K9 methylation, a mechanism entailing the G9a methyltransferase may also be expected to operate in this context. However, other as yet unelaborated Mediator mechanisms could also contribute, especially in cases in which the target gene is activated rather than repressed.
Post-recruitment roles of Mediator
Beyond its effects on chromatin and the coupling of chromatin remodelling and modification to PIC formation, Mediator exerts direct effects on the PIC. Indeed, in certain contexts, PICs may actually be assembled on nucleosome-free promoter regions90 so that Mediator effects at the PIC level would dominate. In principle, these effects could be both at the level of promoting efficient formation of the PIC, especially through the well-characterized ability of Mediator to interact with Pol II, and at the level of modulating the function of a preformed PIC. Whereas cumulative evidence to date suggests that both mechanisms operate, the additional ability to function at a post-recruitment level would expand the potential of Mediator to function as a signal transducer that fine-tunes transcription levels rather than as a binary switch that merely turns the system on and off.
That Mediator might also modulate the activity of the PIC even after Pol II incorporation became evident from observations in in vitro and cell-based experimental systems15,29,37. More generally, numerous kinetic chromatin immunoprecipitation (ChIP) experiments in a range of metazoan systems indicate the existence of complete PICs without concurrent expression on many genes, and together are consistent with the possibility that in these cases Mediator is likely to be acting on a preformed PIC in response to a specific signal (for example, serum-stimulated phosphorylation of ELK1 and subsequent contact with Mediator through MED23 (REF. 37)). Mechanistically, these effects could take place at the level of initiation (defined as the synthesis of the first phosphodiester bond) or, as phosphodiester bond formation is not generally rate limiting, more likely at the level of transition of the initiating Pol II complex to one that is fully competent for transcription elongation91. The latter level may not be restricted to promoter escape but might perhaps also encompass more downstream events that take place as the Pol II encounters additional checkpoints (for example, capping (FIG. 1)).
Transcriptional regulation at steps subsequent to PIC assembly is a major regulatory feature, at least in metazoan systems, and has come to attention most dramatically through the discovery of the so-called paused Pol II molecules that can be detected at the majority of transcription units regardless of whether or not a full-length RNA is transcribed from them92,93. Based on the original heat-shock model91, these are likely to reflect Pol II molecules that have initiated but are stalled at a promoter-proximal location (approximately +50 relative to the transcription start site (TSS)) and are awaiting a stimulatory signal to allow them to transition to a productive transcription elongation complex (TEC). Although definitive evidence is currently lacking, it would not be surprising if Mediator proves to be a major conduit for the transmission of such a signal. Several known functions of Mediator already predispose it for such a role. A functional link between Mediator and elongation factor DSIF94 (BOX 1), whose association with Pol II as it escapes from the promoter marks it as an elongating Pol II91, likely reflects an extension of Mediator function well beyond PIC formation and into steps occurring at or close to its maturation to a TEC. Indeed, physical associations (possibly indirect) between Mediator and the positive elongation factor P-TEFb, which is recruited to the TEC and promotes elongation, have also been observed59,95. This property also correlates with the ability of certain serum-inducible genes to be fully transcribed only when the CTD of the Pol II subunit RPB1 is properly phosphorylated in a CDK8-dependent fashion59. Furthermore, genetic analyses in yeast provide compelling evidence for common roles of the MED31 subunit of Mediator and transcription elongation factor SII96,97, which in turn functions in close association with DSIF98. Synthetic gene array analyses in yeast have revealed further links between Med31 and the Set2 H3K36 methyl transferase, which also functions as an elongation factor99.
However, it is unlikely that Mediator travels with the Pol II as it transcribes the gene body. On the contrary, there is evidence from in vitro studies in yeast systems that following Pol II escape from the promoter, Mediator, likely anchored by activators, is actually retained as part of a scaffold complex (FIG. 1) from which it can facilitate subsequent rounds of transcription100. Thus, in addition to helping initially to establish a functional PIC, Mediator could contribute by ‘holding the place’ for recycling Pol II molecules that enter the PIC later on. It previously has been suggested that the subunit composition of the scaffold resembles that of PC2, which might reflect an in situ-generated form of the Mediator that has lost the kinase module as part of the remodelling process that accompanies the initiation to elongation transition3,15. Correspondingly, it has been suggested that re-incorporation of the module into the scaffold-bound Mediator may serve as a device to turn off ongoing transcription after an appropriate number of rounds of transcription17.
It also is possible that post-recruitment alterations in Mediator composition are subject to further control by gene-specific factors. For example, at the retinoic acid receptor-β gene, ligand-bound retinoic acid receptor functions to recruit another regulatory molecule, poly(ADP- ribose) polymerase 1, which promotes displacement of the kinase module from a pre-assembled, but inactive, Mediator- and Pol II-containing partial PIC. Entry of TFIIH completes the PIC and is correlated with gene activity65. This is in accord with the known functions of TFIIH in promoter escape and the initiation to elongation transition, as well as indications that recruitment of TFIIH to the PIC and regulation of its enzymatic activities are under Mediator control.
Mediator and transcriptional enhancers
Although Mediator recruitment is often viewed in the context of the promoter region — defined as DNA sequences that lie in relative close proximity to the TSS — transcription activators, which are responsible for recruiting Mediator, may bind to far distal sites located in enhancer elements. This thus raises the issue not just of the precise location of the Mediator in the context of the target gene101 but also of the temporal sequence whereby Mediator-related events unfold, especially in relation to the post-recruitment scenarios discussed above. Whereas no generalities have emerged as to which of the two (Mediator or Pol II) is recruited first at these distant locations, close spatial juxtaposition of these factors at some point is obviously a prerequisite for transcription, at least for activated transcription. Indeed, thyroid hormone-dependent induction of the cellular retinoic acid binding protein 1 gene is accompanied by gross changes in the chromatin environment that ensue when the distal thyroid hormone receptor-binding element located in the enhancer is brought into close proximity with the promoter region in a MED1-dependent manner102. More recent evidence further indicates that on many active genes, especially in mouse ES cells, Mediator plays a direct part in the formation of a DNA loop that links enhancers and promoters. This is effected through direct interactions between Mediator and components of the cohesin complex and the related loading factor NIPBl, previously implicated in chromosome dynamics103. It is therefore apparent that Mediator might itself contribute to enhancer–promoter communication, which is fast emerging as an important aspect of gene regulation104,105. There also seems to be close mechanistic coupling between enhancers and core promoters106. Thus, potential Mediator-nucleated higher-order structures formed as a result of these interactions might be particularly favourable with regard to receiving and integrating inputs from diverse transcription factors that typically bind to enhancers and transmitting them to the corresponding core promoter-specific PIC.
Conclusions and future directions
In more than a decade since the discovery of Mediator and its emergence as a fundamental component of the machinery that regulates the transcription of protein-coding genes, extensive work has reinforced how its modular architecture and broad dynamic range of functions allow it to fulfil this central role. Underscoring how various gene networks are tightly controlled and the role of the Mediator therein, reports linking various Mediator subunits to human disease (ranging from congenital malformations107 to mental retardation108 to cancer109–111) have also begun to appear.
Yet, in some ways, we are still in the early stages of understanding all the interactions and activities of which the Mediator is potentially capable. As highlighted here, many regulatory pathways in metazoan systems have now been shown to be channelled through the Mediator. In each case, the Mediator can function to translate the received input into a specific transcriptional output, which characterizes the particular pathway. But the complete transduction mechanism for any given signal remains to be elucidated and other important aspects remain to be fully developed. As an example, one of the earliest known facts about the Mediator was its functional interaction with the CTD of the RPB1 subunit of Pol II112. However, subsequent conflicting data about how relevant this domain is for physical interactions with the Mediator or its function have not yet been resolved113. With the realization that the CTD is mainly a device for coupling transcription to post-transcriptional processes and that many genes might be regulated at the level of the paused Pol II, a resolution of this issue would provide important insights.
It is almost certain that many more Mediator interactions with transcriptional activators and repressors responsible for various cellular pathways will continue to be described in the coming years. But for a deeper understanding of how Mediator processes the signals that these factors relay and translates the physical interaction into a transcriptional response, future directions must also include systematic analyses at least at three levels. First, high-resolution structural analyses of the various modules (which are already underway for the yeast complex but not discussed here owing to the focus of this Review), and ultimately of the entire Mediator, would significantly illuminate the function of this complex as such studies have done for other multiprotein assemblies such as Pol II and the ribosome. Second, genome-wide analyses should be supplemented with detailed snapshots of selected model gene systems using modern cell-based approaches (for example, high-resolution ChIP coupled with RNAi-mediated knockdown) with a view to describing the full range of changes in the composition and interactions of the Mediator as the transcriptional programme of a gene of interest is executed. Finally, as a crucial test for any hypotheses that emerge from these analyses, it will be necessary to recapitulate and dissect these pathways in cell-free systems reconstituted from pure factors.
Acknowledgments
We apologize to colleagues whose work could not be cited directly owing to limitations of scope and space. We thank members of our laboratory for their many contributions to our understanding of the Mediator. Our work on the Mediator is supported in part by NIH grant 1RC1GM09029 to S.M. and R.G.R.
Glossary
- Pre-initiation complex
The multiprotein assembly containing Pol II and the general transcription factors, TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH, that forms at core promoter elements before the onset of transcription. It is formally analogous to the ‘closed complex’ described for the prokaryotic RNA polymerase
- Chromatin
The nucleoprotein structure that packages DNA in the nucleus of eukaryotic cells. The basic unit of chromatin is the nucleosome, a protein core made up of two molecules each of histones H2A, H2B, H3 and H4, around which 146 bp of DNA is wrapped. Different chromatin states are defined by a range of post-translational modifications of core histones and by the incorporation of various histone isoforms
- Basal transcription
Low levels of transcription that can occur in the absence of an activator, especially in in vitro systems
- Initiation
The step in the transcription cycle in which Pol II in the PIC synthesizes the first phophodiester bond. It takes place subsequent to promoter melting or open complex formation
- Chromatin remodelling
Typically an ATP-dependent enzymatic process that alters histone–DNA interactions or regulates the position of nucleosomes
- Chromatin modification
Includes covalent modification (acetylation, phosphorylation, methylation and ubiquitylation) of histones on discrete residues
- Elongation
Refers to the phase in the transcription cycle in which the polymerase that has escaped the promoter extends oligomeric RNA chains into full-length products
- Promoter escape
The events in the transcription cycle that lead to relinquishing of multiple interactions holding the PIC together and entry of Pol II into the elongation phase of transcription
- Capping
The process by which eukaryotic mRNA is modified by the addition of an m7G(5′)ppp(5′)N structure at the 5′ terminus. Capping is essential for several important steps of gene expression, including mRNA stabilization, splicing, mRNA export from the nucleus and initiation of translation
- Enhancer
A regulatory DNA element that usually binds several transcription factors and can activate transcription from a promoter at relatively large distances and in an orientation-independent manner
- Core promoter
The regulatory region of a gene that specifies the transcription start site and on to which Pol II and the general transcription factors assemble to initiate transcription. Depending on the promoter, core promoter sequence elements vary and may extend from approximately 40 bp upstream (for example, the TATA box) to more than 40 bp downstream of the transcription start site
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Malik S, Roeder RG. Dynamic regulation of Pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 5.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 6.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. References 5 and 6 review seminal studies in yeast that identified the Mediator complex in this organism. [DOI] [PubMed] [Google Scholar]
- 7.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato S, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. This paper helped to resolve the problem of compositional heterogeneity that accompanied early studies reporting Mediator isolation. It also helped establish the near-final subunit composition of the mammalian Mediator complex. [DOI] [PubMed] [Google Scholar]
- 9.Backstrom S, Elfving N, Nilsson R, Wingsle G, Bjorklund S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Guglielmi B, et al. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai G, Imasaki T, Takagi Y, Asturias FJ. Mediator structural conservation and implications for the regulation mechanism. Structure. 2009;17:559–567. doi: 10.1016/j.str.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagi Y, et al. Head module control of Mediator interactions. Mol Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. This paper first identified the human Mediator complex in association with ligand-bound thyroid hormone receptor and helped establish the paradigm of the Mediator as an interface between transcriptional activators and the Pol II machinery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik S, Gu W, Wu W, Qin J, Roeder RG. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- 15.Malik S, Baek HJ, Wu W, Roeder RG. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elmlund H, et al. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutsui T, et al. Human mediator kinase subunit CDK11 plays a negative role in viral activator VP16-dependent transcriptional regulation. Genes Cells. 2008;13:817–826. doi: 10.1111/j.1365-2443.2008.01208.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, et al. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell. 2005;19:89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Jiang P, et al. Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proc Natl Acad Sci USA. 2010;107:6765–6770. doi: 10.1073/pnas.1001814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toth-Petroczy A, et al. Malleable machines in transcription regulation: the Mediator complex. PLoS Comput Biol. 2008;4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 23.Blazek E, Mittler G, Meisterernst M. The Mediator of RNA polymerase II. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- 24.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 26.Rachez C, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 27.Ge K, et al. Transcription coactivator TRAP220 is required for PPARγ 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. This physiological study showed how distinct subunits in the Mediator complex can control specific developmental and signalling pathways. [DOI] [PubMed] [Google Scholar]
- 28.Ge K, et al. Alternative mechanisms by which Mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor γ-stimulated adipogenesis and target gene expression. Mol Cell Biol. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik S, Wallberg AE, Kang YK, Roeder RG. TRAP/SMCC/Mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol. 2002;22:5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang YK, Guermah M, Yuan CX, Roeder RG. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors α and β through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc Natl Acad Sci USA. 2002;99:2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik S, et al. Structural and functional organization of TRAP220, the TRAP/Mediator subunit that is targeted by nuclear receptors. Mol Cell Biol. 2004;24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. This mouse knockout study showed how distinct subunits in the Mediator complex can control specific developmental and signalling pathways. [DOI] [PubMed] [Google Scholar]
- 34.Grontved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers Mediator to the N-terminal domain of peroxisome proliferator-activated receptor γ and is required for full transcriptional activity and adipogenesis. Mol Cell Biol. 2010;30:2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stumpf M, et al. The Mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci USA. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 37.Wang G, et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, et al. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev Cell. 2009;16:764–771. doi: 10.1016/j.devcel.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 40.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thakur JK, et al. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem. 2009;284:4422–4428. doi: 10.1074/jbc.M808263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato Y, Habas R, Katsuyama Y, Naar AM, He X. A component of the ARC/Mediator complex required for TGF β/Nodal signalling. Nature. 2002;418:641–646. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- 43.Ito M, Okano HJ, Darnell RB, Roeder RG. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002;21:3464–3475. doi: 10.1093/emboj/cdf348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens JL, Cantin GT, Wang G, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 Mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. This knockout study showed how distinct subunits in the Mediator complex can control specific developmental and signalling pathways. [DOI] [PubMed] [Google Scholar]
- 45.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 46.Ito M, et al. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 47.Park JM, Werner J, Kim JM, Lis JT, Kim YJ. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- 48.Meyer KD, Lin SC, Bernecky C, Gao Y, Taatjes D. J p53 activates transcription by directing structural shifts in Mediator. Nature Struct Mol Biol. 2010;17:753–760. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mittler G, et al. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim TW, et al. MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc Natl Acad Sci USA. 2004;101:12153–12158. doi: 10.1073/pnas.0401985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belakavadi M, Pandey PK, Vijayvargia R, Fondell JD. MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Mol Cell Biol. 2008;28:3932–3942. doi: 10.1128/MCB.02191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nature Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 54.Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the Mediator complex subunits Med12 and Med13. Proc Natl Acad Sci USA. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/β-catenin signaling. J Biol Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, Yang N, Uno E, Roeder RG, Guo S. A subunit of the mediator complex regulates vertebrate neuronal development. Proc Natl Acad Sci USA. 2006;103:17284–17289. doi: 10.1073/pnas.0605414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. This study extended the repressive functions of the kinase module of the Mediator to a role in establishing an epigenetically silenced state in differentiated neurons. It also provided insights into how defects in Mediator function can lead to human disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loncle N, et al. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 2007;26:1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belakavadi M, Fondell JD. Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol. 2010;30:2437–2448. doi: 10.1128/MCB.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 62.Benoff B, et al. Structural basis of transcription activation: the CAP-α CTD–DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 63.Taatjes DJ, Schneider-Poetsch T, Tjian R. Distinct conformational states of nuclear receptor-bound CRSP–Med complexes. Nature Struct Mol Biol. 2004;11:664–671. doi: 10.1038/nsmb789. [DOI] [PubMed] [Google Scholar]
- 64.Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 65.Pavri R, et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of Mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 66.Esnault C, et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 67.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 68.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing Mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 69.Hengartner CJ, et al. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 70.Furumoto T, et al. A kinase subunit of the human Mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells. 2007;12:119–132. doi: 10.1111/j.1365-2443.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- 71.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 73.Acevedo ML, Kraus WL. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor α-dependent transcription with chromatin templates. Mol Cell Biol. 2003;23:335–348. doi: 10.1128/MCB.23.1.335-348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. This paper exemplifies biochemical studies that have revealed how Mediator can work with chromatin co-activators. [DOI] [PubMed] [Google Scholar]
- 75.Fondell JD, Guermah M, Malik S, Roeder RG. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci USA. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci USA. 2002;99:7934–7939. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/Mediator function through coactivator PGC-1α. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 78.Chen W, Yang Q, Roeder RG. Dynamic interactions and cooperative functions of PGC-1α and MED1 in TRα-mediated activation of the brown-fat-specific UCP-1 gene. Mol Cell. 2009;35:755–768. doi: 10.1016/j.molcel.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qiu H, et al. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol Cell Biol. 2005;25:3461–3474. doi: 10.1128/MCB.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu X, Vorontchikhina M, Wang YL, Faiola F, Martinez E. STAGA recruits Mediator to the MYC oncoprotein to stimulate transcription and cell proliferation. Mol Cell Biol. 2008;28:108–121. doi: 10.1128/MCB.01402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meyer KD, et al. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krebs AR, et al. ATAC and Mediator coactivators form a stable complex and regulate a set of non-coding RNA genes. EMBO Rep. 2010;11:541–547. doi: 10.1038/embor.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 85.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nature Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 86.Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 88.Tutter AV, et al. Role for Med12 in regulation of Nanog and Nanog target genes. J Biol Chem. 2009;284:3709–3718. doi: 10.1074/jbc.M805677200. [DOI] [PubMed] [Google Scholar]
- 89.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nature Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 92.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 94.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc Natl Acad Sci USA. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 96.Malagon F, Tong AH, Shafer BK, Strathern JN. Genetic interactions of DST1 in Saccharomyces cerevisiae suggest a role of TFIIS in the initiation–elongation transition. Genetics. 2004;166:1215–1227. doi: 10.1534/genetics.166.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guglielmi B, Soutourina J, Esnault C, Werner M. TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA. 2007;104:16062–16067. doi: 10.1073/pnas.0704534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palangat M, Renner DB, Price DH, Landick R. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc Natl Acad Sci USA. 2005;102:15036–15041. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Krogan NJ, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 101.Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park SW, et al. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol Cell. 2005;19:643–653. doi: 10.1016/j.molcel.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 103.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010 Aug 18; doi: 10.1038/nature09380. This recent study extends the role of the Mediator to enhancer–promoter communication, which is emerging as an important aspect of transcriptional control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hatzis P, Talianidis I. Dynamics of enhancer–promoter communication during differentiation-induced gene activation. Mol Cell. 2002;10:1467–1477. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 105.Szutorisz H, Dillon N, Tora L. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci. 2005;30:593–599. doi: 10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 106.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 107.Muncke N, et al. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries) Circulation. 2003;108:2843–2850. doi: 10.1161/01.CIR.0000103684.77636.CD. [DOI] [PubMed] [Google Scholar]
- 108.Philibert RA, Madan A. Role of MED12 in transcription and human behavior. Pharmacogenomics. 2007;8:909–916. doi: 10.2217/14622416.8.8.909. [DOI] [PubMed] [Google Scholar]
- 109.Firestein R, et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morris EJ, et al. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu Y, et al. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc Natl Acad Sci USA. 1999;96:10848–10853. doi: 10.1073/pnas.96.19.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gu W, et al. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 114.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 115.Andrau JC, et al. Genome-wide location of the coactivator Mediator: binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 116.Zhu X, et al. Genome-wide occupancy profile of Mediator and the Srb8–11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 117.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nature Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 118.Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci USA. 2009;106:16734–16739. doi: 10.1073/pnas.0905103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mittler G, Kremmer E, Timmers HT, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 122.Hu X, et al. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci USA. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gazdag E, et al. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev. 2009;23:2210–2223. doi: 10.1101/gad.535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deato MD, et al. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baumli S, Hoeppner S, Cramer P. A conserved Mediator hinge revealed in the structure of the MED7. MED21 (Med7.Srb7) heterodimer. J Biol Chem. 2005;280:18171–18178. doi: 10.1074/jbc.M413466200. [DOI] [PubMed] [Google Scholar]
- 126.Lariviere L, et al. Structure and TBP binding of the Mediator head subcomplex Med8–Med18–Med20. Nature Struct Mol Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 127.Bourbon HM, et al. A unified nomenclature for protein subunits of ediator complexes linking transcriptional regulators to RNA polymerase II. Mol Cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]