Abstract
During postembryonic development of higher plants, the shoot apical meristem produces lateral organs in a regular spacing (phyllotaxy) and a regular timing (plastochron). Molecular analysis of mutants associated with phyllotaxy and plastochron would greatly increase understanding of the developmental mechanism of plant architecture because phyllotaxy and plastochron are fundamental regulators of plant architecture. pla1 of rice is not only a plastochron mutant showing rapid leaf initiation without affecting phyllotaxy, but also a heterochronic mutant showing ectopic shoot formation in the reproductive phase. Thus, pla1 provides a tool for analyzing the molecular basis of temporal regulation in leaf development. In this work, we isolated the PLA1 gene by map-based cloning. The identified PLA1 gene encodes a cytochrome P450, CYP78A11, which potentially catalyzes substances controlling plant development. PLA1 is expressed in developing leaf primordia, bracts of the panicle, and elongating internodes, but not in the shoot apical meristem. The expression pattern and mutant phenotype suggest that the PLA1 gene acting in developing leaf primordia affects the timing of successive leaf initiation and the termination of vegetative growth.
In higher plants, all above-ground organs originate from the shoot apical meristem (SAM) (1–3). During postembryonic development, SAM produces lateral organs in a regular spacing (phyllotaxy) and a regular timing (plastochron); this fact has fascinated many plant biologists (4–9). The pattern of lateral organ initiation determines the plant morphology directly. Therefore, the diversity of shoot architecture among plant species would be attributable mainly to the regulatory modification of lateral organ initiation patterns.
Many studies have attempted to elucidate the mechanism that regulates leaf initiation pattern (2, 10). An inhibitory field model was proposed through surgical experiments (6, 7). This model predicts that a diffusible substance emanating from the center of the SAM and preexisting leaf primordia inhibits the initiation of a new leaf primordium. Alternatively, a biophysical model suggests that the phyllotactic pattern is generated spontaneously as a result of physical force in the SAM (8, 11, 12). This model is supported by studies in which phyllotaxy was modified by local application of expansin, which promoted cell wall extensibility (13–15). Both models have been widely accepted, but their molecular basis remains elusive.
Genetic and molecular analyses have identified several genes associated with lateral organ initiation. In maize, the terminal ear 1 (te1) mutant shows rapid leaf production, aberrant phyllotaxy, and abnormal phytomers (16). The TE1 gene contains several RNA binding protein motifs and appears to inhibit phytomer/leaf initiation in the SAM. Another mutant, Arabidopsis altered meristem program 1 (amp1), was reported to have altered plastochron and phyllotaxy. The AMP1 gene encodes putative glutamate carboxypeptidases. Potential substrates of AMP1 are small acidic peptides or folate polyglutamate, suggesting that the AMP1 gene product modulates the level of a small signaling molecule involved in leaf initiation (17, 18). These mutations and genes would be useful, but pleiotropic defects in these mutants in which both plastochron and phyllotaxy are simultaneously affected make it difficult to interpret the gene function explicitly. Mutations that specifically affect plastochron or phyllotaxy would be more useful for comprehensive understanding of the regulatory mechanism.
To date, only a few mutants have been identified that affect the plastochron specifically, but not the phyllotaxy. The rice plastochron1 (pla1) mutant exhibits the rapid initiation of vegetative leaves without affecting phyllotaxy and ectopic shoot production in the reproductive phase because of the conversion of primary rachis meristems to vegetative shoot meristems (19). This unique phenotype suggests that the PLA1 gene regulates the rate of leaf initiation and the vegetative–reproductive phase change.
Here, we describe molecular cloning of the rice PLASTOCHTRON1 gene and the expression pattern of its mRNA. PLA1 encodes a member of cytochrome P450 proteins, CYP78A11, and is expressed exclusively in developing young leaf primordia and elongating internodes, but not in SAM. Based on the expression pattern and its mutant phenotype, we suggest a possible role of PLA1 as a timekeeper of plant development.
Materials and Methods
Plant Materials. We used four allelic mutants of pla1: pla1-1, pla1-2, pla1-3, and pla1-4. pla1-1 was derived from γ-ray irradiation; the other three were derived from chemical mutagenesis with N-methyl-N-nitrosourea. Background strains for pla1-1 and pla1-2 mutants were Fukei71 and Kinmaze, respectively; pla1-3 and pla1-4 were from Taichung65. These strains were grown in pots under natural conditions. Transgenic plants were grown in a biohazard greenhouse at 30°C during the day and 25°C at night.
Sequence Analysis of Candidate Genomic Region. The genomic region covering the P450 gene was amplified through 30 PCR cycles (10 s at 98°C, 60 s at 58°C, and 120 s at 72°C) with the 5′ primer (5′-TCCCACCCCACTTCATCACA-3′) and the 3′ primer (5′-CCACGTCTTACCTACGCTAA-3′) that were designed from the genomic sequence of Nipponbare. Amplified products from pla1 mutant alleles and corresponding wild-type plants were purified by SUPREC-02 (Takara Bio, Kyoto), and used as templates for sequence reaction. Nucleotide sequences of PCR products were determined by using primers designed from the Nipponbare sequence data of the candidate region of the P450 gene. Sequencing reaction was performed with a BigDye terminator cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions.
Complementation Test. A 6-kb SpeI restriction fragment containing only the P450 ORF with a 3.1-kb upstream sequence was obtained from the bacterial artificial chromosome clone B44A10, provided by the Clemson University Genome Institute (Clemson, SC). The 6-kb SpeI fragment was ligated to an Agrobacterium binary vector, pBGH1, a derivative of the pIG121HM (20) with the hygromycin phosphotransferase (htp) gene. The resultant plasmid, designated pBGH/P450, was introduced into Agrobacterium strain EHA101 and then infected into calli of the pla1-2 mutant (21). As a control, the pBGH1 vector without P450 sequence was also introduced into pla1-2. Southern blot analysis with the htp gene probe confirmed that all transgenic plants regenerated from hygromycin-resistant calli that carried the introduced gene.
Screening of cDNA Libraries. A 1.8-kb genomic DNA fragment with the P450 putative ORF was labeled to screen two rice cDNA libraries. Approximately 106 plaques each were screened from a 2-week-old seedling library and an immature panicle library. Phage clones that hybridized to the probe were isolated, and then plasmids with inserts were excised by an in vivo protocol recommended by the manufacturer (Stratagene).
RNA Extraction and cDNA Synthesis. Total RNA was extracted from seedlings 2 weeks after germination by a single-step method (22) with minor modifications. Poly(A)+ RNA was recovered from total RNA by using Oligotex-dT30 (Takara Bio). A first-strand cDNA was synthesized from poly(A)+ RNA by using Superscript II reverse transcriptase (GIBCO/BRL). Reverse transcription was performed at 30°C for 10 min, 50°C for 1 h, and 80°C for 2 min. The cDNA fragment containing the entire P450 ORF was amplified through 30 PCR cycles (10 s at 98°C, 60 s at 58°C, and 120 s at 72°C) with primers used for amplification of P450 genomic fragment in the Sequence Analysis of Candidate Genomic Region (see above). Amplified products were purified by SUPREC-02 (Takara Bio) and used as templates for sequencing reaction.
In Situ Hybridization. Paraffin-sectioned shoot apices were subjected to in situ hybridization as described (19, 23), except that hybridization was carried out at 55–60°C to compensate for the high GC content of the PLA1 gene. Templates for RNA probes were generated from 1.8-kb genomic fragments of CYP78A11. Digoxigenin-labeled UTP (Boehringer Mannheim) probes of antisense and sense were used.
Results
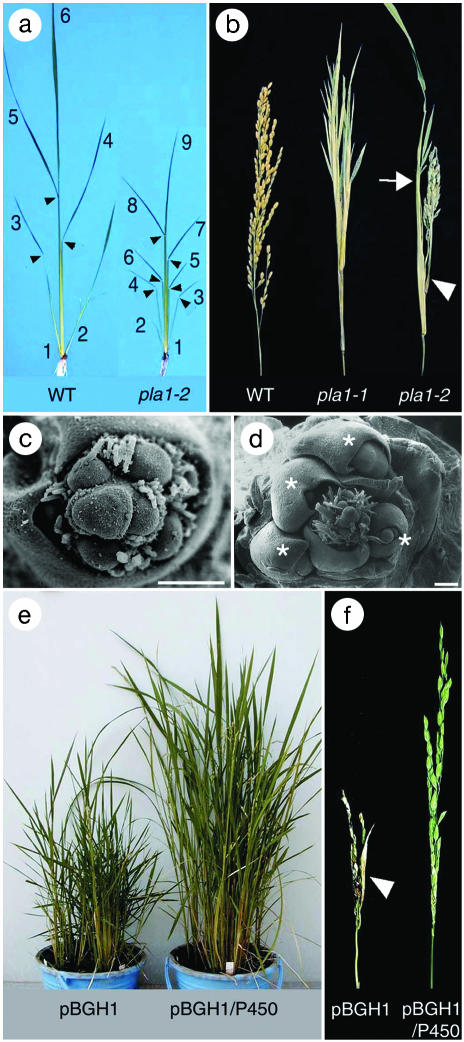
Phenotype of pla1 Mutant. The plastochron1 mutants (19) were identified by their rapid emergence of vegetative leaves (Fig. 1a). During vegetative growth, pla1 plants doubled the number of leaves compared with wild-type siblings. The large number of leaves in pla1 plants was a consequence of rapid leaf initiation because the duration of vegetative phase was comparable among pla1 and wild-type plants (19) (Fig. 1a and Table 1). The SAM of pla1 plants was enlarged via activated cell divisions (19) (Table 1), but leaf size and plant height were reduced to about half that of the wild type (Fig. 1). Many pla1 leaves showed strong bending at the lamina joint (Fig. 1a). Despite the size change in SAM and leaves, the spatial pattern of leaf initiation of pla1 plants was completely normal (19). Therefore, pla1 is specifically involved in temporal patterning but not in spatial patterning. This finding contrasts with other mutations, which simultaneously affect both plastochron and phyllotaxy (16, 17, 24).
Fig. 1.
Phenotypes of wild-type, pla1 mutant, and transgenic pla1-2 plants. (a) Seedlings of wild-type (WT) and pla1-2 plants 17 days after germination, showing that many more leaves are formed in pla1-2 than in wild type. Arrowheads indicate lamina joint. (b) Panicles of wild type, pla1-1 with vegetative shoots instead of primary branches, and pla1-2 with truncated panicle, one shoot (arrowhead), and enlarged bract (arrow). (c) Scanning electron microscopy of a wild-type young panicle. (d) Scanning electron microscopy of a pla1-1 young panicle. Asterisks indicate ectopic vegetative shoots with an enlarged bract. (e) pla1-2-like transgenic plants carrying pBGH1 alone. Normal transgenic plants carrying pBGH1/P450 are shown to the right. (f) Panicles of transgenic plants: left, a pla1-2-like panicle with enlarged bract (arrowhead) in transgenic plant carrying pBGH1; right, normal panicle in transgenic plant carrying pBGH1/P450.
Table 1. Phenotype of plastochron1 alleles.
| Line | Plastochron | No. of ectopic shoots | SAM width, μm | SAM height, μm |
|---|---|---|---|---|
| Fukel71* | 4.8 ± 0.1 | 0 | 61.7 ± 1.1 | 34.8 ± 1.3 |
| Kinmaze* | 4.8 ± 0.0 | 0 | 63.2 ± 1.4 | 35.0 ± 1.0 |
| Taichung65† | 5.0 ± 0.0 | 0 | 62.9 ± 1.6 | 34.1 ± 0.8 |
| pla1-1* | 2.5 ± 0.1 | 3.2 ± 0.3 | 69.3 ± 1.5 | 37.4 ± 1.6 |
| pla1-2* | 2.8 ± 0.0 | 1.3 ± 0.2 | 69.6 ± 1.0 | 37.3 ± 0.9 |
| pla1-3* | 3.1 ± 0.1 | 1.1 ± 0.2 | 69.5 ± 1.0 | 36.9 ± 1.0 |
| pla1-4† | 2.3 ± 0.1 | 3.2 ± 0.2 | 73.5 ± 0.8 | 40.6 ± 0.8 |
Average data of five plants of each genotype.
pla1-1 and pla1-2 were derived from Fukei71 and Kinmaze, respectively.
pla1-3 and pla1-4 were derived from Taichung65.
The pla1 mutation also affects the panicle development in the reproductive phase. Many primordia of primary rachis branches (inflorescence branches) were converted into vegetative shoots in the strong allele pla1-1 (Fig. 1b). Ectopic shoots eventually produced small panicles. In the weak allele pla1-2, only a few proximal primary branch primordia developed into vegetative shoots with abnormally elongated bracts; and the remaining primordia formed a small panicle in which the internodes of rachis and branches were extremely truncated (Fig. 1b). The elongated bracts subtending ectopic vegetative shoots are foliage leaf-like because they have a leaf blade and a sheath interrupted by a ligule (data not shown). Because the number of primary branches converted into vegetative shoots was correlated with the leaf initiation rate in the vegetative phase, it is considered that vegetative growth activity (leaf initiation rate) is reflected in the extent of vegetative program expression in the reproductive phase (Table 1).
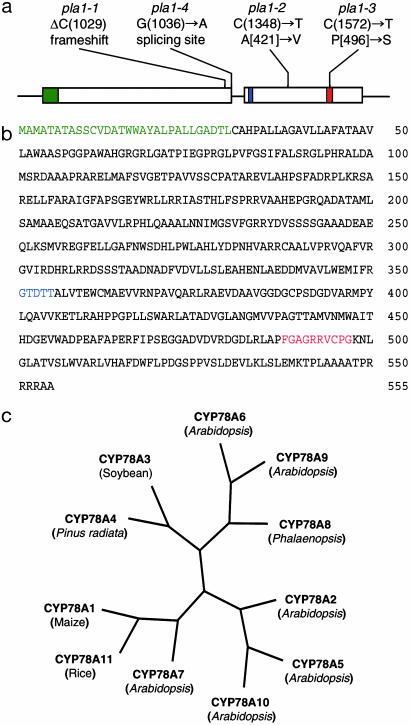
Molecular Cloning of PLA1. In the previous work (25), we mapped the PLA1 locus on the long arm of chromosome 10 between the two RFLP markers. By using high-density markers and 578 F2 homozygous plants for pla1-2, the PLA1 gene was revealed to locate on a bacterial artificial chromosome clone of B44A10. Multiple markers designated on the bacterial artificial chromosome enabled tracking of the gene within the interval of 74-kb between markers B44A10–142 and B44A10–16 (25). Gene prediction analysis by genscan (http://genes.mit.edu/GENSCAN) and hmm (http://rgp.dna.affrc.go.jp/RiceHMM/index.html) programs identified three candidate genes within the 74-kb sequence. Nucleotide sequences of the candidate genes were compared among the wild-type and all four mutant alleles. Among these genes, a gene carrying cytochrome P450 sequence showed four independent mutations in each of the four pla1 alleles (Fig. 2a). The pla1-1 allele has one base deletion in the first exon, resulting in frameshift of ORF. Single base changes causing amino acid substitution from alanine to valine and proline to serine were detected in pla1-2 and pla1-3, respectively. pla1-4 has one base change from G to A at the splicing donor site. Among the four alleles, pla1-1 and pla1-4, which showed severe phenotypes, are considered to be null alleles because they seem to produce no functional proteins, whereas pla1-2 and pla1-3, which showed mild phenotypes, had the amino acid substitution that may cause partial loss of protein function. Thus, PLA1 is likely to encode a cytochrome P450 protein.
Fig. 2.
Structure of the PLA1 gene. (a) Exon/intron structure of PLA1. Two open boxes indicate protein coding regions composed of two exons split by a small intron. Mutations in four alleles are indicated as follows: (i) deletion of C at the position of 1,029 bp in pla1-1 resulting in frameshift of the ORF, (ii) nucleotide transition of C to T at position 1348 in pla1-2 resulting in the substitution of A to V at the position of 421 aa, (iii) transition of C to T at position 1687 in pla1-3 1-2 resulting in the substitution of P to S at position 496, and (iv) transition of G to A at position 1151 in pla1-4 resulting in the disruption of the splicing donor site. (b) Deduced amino acid sequence of PLA1 protein. In a and b, hydrophobic regions, oxygen binding motifs, and heme binding motifs are indicated by green, blue, and red, respectively. (c) Phylogenetic relationship among CYP78A proteins. The phylogenetic tree was generated based on the entire amino acid sequences by using the clustal w program (www.ddbj.nig.ac.jp/e-mail/clustalw-e.html).
We conducted a complementation experiment to confirm whether the P450 gene rescued the pla1 phenotype. A 6-kb SpeI restriction fragment containing only the P450 ORF with a 3.1-kb upstream sequence was cloned into pBGH1. The resultant plasmid pBGH/P450 was introduced into the pla1-2 mutant. As a control, the pBGH1 vector without the P450 sequence also was introduced into pla1-2. Ten plants transformed with the pBGH1/P450 and 14 plants transformed with pBGH1 developed to maturity. Southern blot analysis detected one to five bands in the transgenic plants, indicating that all transformants carried at least one copy of the introduced gene (date not shown). These transgenic lines were subjected to phenotypic observation.
In vegetative phase, all of the control transgenic plants carrying only the vector were indistinguishable from the pla1-2 homozygous plant (Fig. 1e). In contrast, 10 transgenic plants with pBGH1/P450 developed normally in both vegetative and reproductive phases (Fig. 1 e and f). The phenotypic differences in vegetative and reproductive growth were not apparent among transformants with different copy numbers of P450. We concluded that the PLA1 gene encodes the cytochrome P450 protein because abnormal characters of pla1-2 mutant were rescued only when P450 was introduced.
Structure of the PLA1 Gene. To determine the coding region and gene structure of PLA1, two cDNA libraries made from young seedlings 2 weeks after germination and from developing panicles were screened by a 1.8-kb genomic fragment containing the P450 ORF sequence. However, no cDNA clones corresponding to PLA1 were obtained, probably because of low abundance of PLA1 mRNA. Then, the PLA1 cDNA sequence was examined by RT-PCR method. The PLA1 transcript was amplified from first strand cDNAs of shoot apices by using primers set to cover the P450 ORF sequence. The nucleotide sequence of RT-PCR product was determined and compared with the genomic sequence. PLA1 cDNA has two exons split by a small intron (Fig. 2a). The predicted PLA1 protein consists of 555 aa with a predicted molecular mass of 59 kDa (Fig. 2b). The deduced amino acid sequence of this protein contains a hydrophobic region in the N terminus and putative oxygen and heme-binding domains that are characteristic of microsomal cytochrome P450 (26) (Fig. 2b). According to the recommendation of the Cytochrome P450 Gene Nomenclature Committee (27), the PLA1 protein was designated as CYP78A11 (GenBank accession no. AB096259). The CYP78A class comprises 11 members including rice CYP78A11. Rice CYP78A11 forms a cluster with maize CYP78A1 (GenBank accession no. L23209) and Arabidopsis CYP78A7 (GenBank accession no. AC016893) (Fig. 2c). CYP78A11 exhibits the highest similarity to the CYP78A1 of maize (71% amino acid identity) (28), and strong similarity to Arabidopsis CYP78A7 (61% identity) and Pinus radiata CYP78A4 (59% identity; GenBank accession no. AF049067). Genes in the CYP78A class belong to the group A cytochrome P450 in plants (http://drnelson.utmem.edu/CytochromeP450.html). They are phylogenetically more closely related to each other than to non-group A cytochrome P450s and seem to be involved in plant-specific reactions (29).
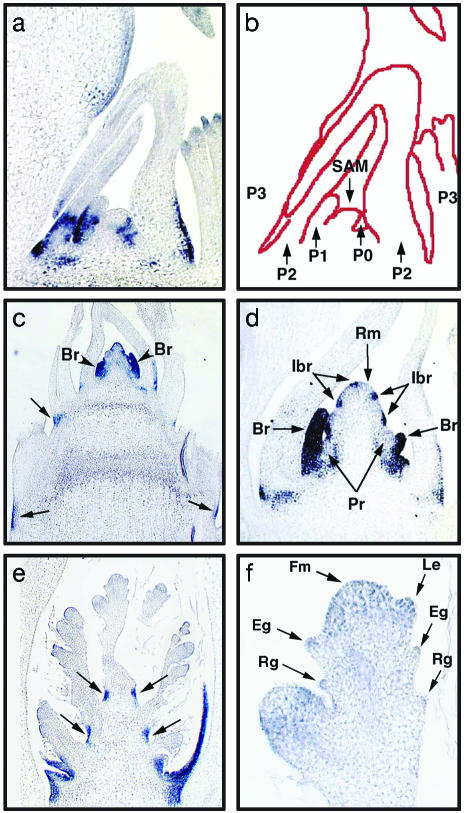
In Situ Expression Pattern of PLA1. For further investigation of PLA1 function, PLA1 expression was examined with RT-PCR. PLA1 mRNA was detected in seedlings, shoot apices, and young panicles but not in mature leaves, calli, and roots (data not shown), suggesting that PLA1 was expressed in shoot apices that contained developing leaf primordia and SAM. Temporal and spatial expression patterns of PLA1 were examined in detail by in situ hybridization. For convenience, developmental stages of leaves are designated as follows. The youngest leaf primordium is termed plastochron1 (P1); the second and third youngest primordia are termed P2 and P3, respectively. The preprimordial founder cell population in the meristem is termed P0 (30). The first PLA1 expression was observed in P0 leaf founder cells (Fig. 3 a and b). In the P1 leaf, the expression was detected at the leaf margin and the abaxial side of the proximal region that later differentiated into the sheath (Fig. 3 a and b). In the P2 leaf, PLA1 mRNA was localized at the leaf margin and the abaxial side of the basal region (Fig. 3 a and b). In the P3 leaf, PLA1 expression was restricted to the lamina joint just above the blade-sheath boundary. The PLA1 expression was not observed beyond the P3 stage. These results show that PLA1 is expressed in developing young leaves but not in the SAM.
Fig. 3.
In situ expression of PLA1 in vegetative and reproductive apex of wild-type plant. Dark blue stains represent PLA1 gene expression. (a) Median longitudinal section of shoot apex 1 month after germination. (b) Schematic representation of a.(c) Longitudinal section of shoot apex just after transition to reproductive phase. Two bracts of primary branches are formed. Arrows indicate PLA1 expression in the internodes of an elongating stem. (d) Longitudinal section of a young panicle at a slightly later stage of c.(e) Longitudinal section of a developing panicle in which spikelets are being formed. Arrows indicate PLA1 expression in the rachis internodes. (f) Longitudinal section of young spikelet. P0, plastochron0 leaf founder cells; P1, plastochron1 leaf; P2, plastochron2 leaf; P3, plastochron3 leaf; Br, bract; Ibr, incipient bract; Rm, rachis meristem; Pr, Primary branch primordium; Fm, floral meristem; Le, lemma primordium; Eg, empty glume primordium; Rg, rudimentary glume primordium.
PLA1 expression was also observed in early reproductive phase. After transition to reproductive growth, a young rachis (inflorescence) meristem produces several bracts of primary rachis branches in a helical pattern. In young inflorescence apices, PLA1 expression was observed strongly in developing bract leaves and their incipient primordia before their emergence but not in the rachis and primary rachis branch meristems (Fig. 3 c and d). PLA1 expression disappeared in developing spikelets or was down-regulated to a very low level (Fig. 3 e and f). No specific expression was observed in the leaf-like organs such as glumes and lemmas. In addition, PLA1 expression was observed in the peripheral region of elongating internodes in stem and rachis (Fig. 3 c and e).
Discussion
In this study, we have cloned the PLA1 gene, which regulates the rate of leaf initiation and the duration of vegetative phase. Apart from several other genes such as TE1 (16) and SHO (24), which show abnormal patterns of leaf initiation in both phyllotaxy and plastochron and other abnormal traits, PLA1 is the first cloned gene exclusively associated with temporal regulation of leaf initiation. Plastochron affects the number of leaves and then the number of tillers (branches), which determines the number of panicles. Thus, the PLA1 gene is agronomically important when considering the innovation of plant forms and seed production.
Molecular cloning revealed that the PLA1 gene encodes a cytochrome P450 protein, CYP78A11. The CYP78A subclass (http://drnelson.utmem.edu/CytochromeP450.html) comprises 11 members, including rice CYP78A11. Several of them show flower- or meristem-specific expression (28, 32, 33). Overexpression of Arabidopsis CYP78A5 (33) and CYP78A9 (34) cause abnormal flower development, suggesting the roles of these genes in reproductive organ formation. However, because of the lack of loss-of-function mutants, biological functions of CYP78A genes are almost unknown. The identification of CYP78A11 from PLA1 provides a new tool for further study of CYP78A genes.
Some members of the plant P450 family are known to be involved in important biochemical pathways, such as biosynthesis of phenylpropanoids, alkaloids, terpenoids, lipids, cyanogenic glycosides, and glucosinolates, and plant growth regulators such as gibberellins, jasmonic acid, and brassinosteroids (reviewed in refs. 35–37). CYP78A11 showed highest homology to maize CYP78A1 (71% identity) that is preferentially expressed in developing inflorescences (28). Biochemical analysis showed that CYP78A1 catalyze the 12-monooxygenation of a fatty acid (38). It has also been shown that plants synthesize many fatty acid derivatives, several of which act as signaling molecules such as jasmonic acid (39). Like maize CYP78A1, CYP78A11 might be involved in a metabolic pathway of fatty acid synthesis required for leaf development.
In situ experiments showed that the PLA1 gene is expressed in the abaxial side of young leaf primordia, bracts of primary rachis branches, and peripheral cells of internodes in stem and rachis, but not in the SAM. This expression pattern is consistent with mutant phenotypes such as short leaves, overgrowth of bracts, and dwarfism of the stem and rachis. These phenotypes suggest that the PLA1 gene promotes the growth of leaves and internodes. Biosynthesis or distribution of hormone(s) such as auxins or brassinosteroids is likely to be affected in pla1 mutant because the growth of leaves and internodes is known to be regulated by such plant hormones (40–42). In addition, the angle at lamina joint is enlarged in pla1. It is known that auxins and brassinosteroids control bending at the lamina joint (43–46). Although it is yet unclear that CYP78A members are related to plant hormones, the mutant phenotypes strongly suggest that PLA1 regulate leaf and internode growth via the regulation of plant hormone(s).
It is unexpected that the PLA1 gene is not expressed in the SAM, despite the rapid initiation of leaf primordia, enlargement of the SAM, and activated cell divisions that are observed in pla1. As for plastochron, PLA1 seems to negatively regulate the initiation of leaf primordia. It has been argued that the preexisting leaf primordia inhibit the leaf primordia initiation in their vicinity (6, 7). This inhibitor model has been discussed mainly in relation to phyllotaxy, and not to plastochron. However, it is possible that preexisting primordia also suppress the precocious initiation of new leaves. The PLA1 expression in the basal region of leaf primordia does not contradict this assumption. On the basis of this model, however, it is difficult to explain the enlargement of SAM and activated cell divisions. In addition, PLA1 is expressed in the abaxial side of leaf base, and not in the adaxial side, making it difficult to assume the direct inhibitory effect of PLA1 on leaf initiation. It is well known that enlarged SAM and/or activated cell divisions promote the production of lateral organs. For example, clv mutations in Arabidopsis and abphyl mutation in maize cause SAM enlargement together with the increase of lateral organs (47, 48). Thus, the rapid leaf initiation would result from the enlarged SAM mediated by activated cell divisions. Because the phyllotaxy of pla1 is normal, positional regulation of leaf primordia is not affected, suggesting that in pla1, enlarged SAM is differently organized from that of abphyl. It was reported that exogenous application of a gibberellic acid resulted in the enlargement of SAM via activated cell divisions, and that rapid leaf initiation and phyllotactic change in Xanthium pennsylvanicum (49–51). Also, the inhibition of auxin transport affects the phyllotaxy and leaf emergence rate in Arabidopsis and tomato (52, 53). In several species, developing young leaves, but not SAM, are estimated as one source of these phytohormones (53, 54). Together with other phenotypes (short leaves, dwarfism, and leaf bending), hormonal regulation appears to be modified in pla1 to promote cell divisions in the SAM, most probably through its non-cell-autonomous function.
Another interesting feature of pla1 is the heterochrony attributable to ectopic expression of vegetative program in the reproductive phase. Although many heterochronic mutants thus far reported are associated with juvenile–adult phase change (31, 55–58), pla1 is specifically associated with vegetative–reproductive phase change without affecting flowering time. The earliest sign of heterochrony in pla1 is the overgrowth of bracts. The mutant bracts are foliage leaf-like in that they are differentiated into blades and sheathes interrupted by ligules. PLA1 expression in bract leaves was much stronger and broader than in foliage leaves, suggesting the differential roles of PLA1 between bract and foliage leaves. In fact, pla1 mutation caused size reduction of foliage leaves, but overgrowth and conversion to foliage leaf in bracts. The negative regulation of bract growth by PLA1 may play a crucial role in the suppression of the vegetative program in the reproductive phase.
The primary function of PLA1 would reside in leaf and stem development; however, the lack of PLA1 function eventually affects the temporal regulation of leaf initiation and the termination of vegetative program. Further analysis of the PLA1 gene and the mutants would provide valuable insights on the relationship between leaf development and SAM organization, and on the mechanism driving vegetative–reproductive phase transition.
Acknowledgments
We thank Dr. Wing of the Clemson University Genome Institute for kindly providing a bacterial artificial chromosome library. We also thank A. Ishii, T. Makino, and S. Sakai for technical assistance.
Abbreviation: SAM, shoot apical meristem.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB096259).
References
- 1.Walbot, V. (1985) Trends Genet. 1, 165–169. [Google Scholar]
- 2.Steeves, T. A. & Sussex, I. M. (1989) in Patterns in Plant Development (Cambridge Univ. Press, Cambridge, U.K.), pp. 46–310.
- 3.Sussex, I. M. (1989) Cell 56, 225–229. [DOI] [PubMed] [Google Scholar]
- 4.Smith, L. G. & Hake, S. (1992) Plant Cell 4, 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyndon, R. F. (1998) in The Shoot Apical Meristem: Its Growth and Development (Cambridge Univ. Press, Cambridge, U.K.), pp. 142–193.
- 6.Snow, M. & Snow, R. (1931) Philos. Trans. R. Soc. London B 221, 1–31. [Google Scholar]
- 7.Wardlaw, C. W. (1949) Growth 13, Suppl., 93–131. [Google Scholar]
- 8.Green, P. B., Steele, C. S. & Rennich, S. C. (1996) Ann. Bot. 77, 515–527. [Google Scholar]
- 9.Lintilhac, P. M. (1984) in Positional Controls in Plant Development, eds. Barlow, P. W. & Carr, D. J. (Cambridge Univ. Press, Cambridge, U.K.), pp. 83–106.
- 10.Callos, J. D. & Medford, J. I. (1994) Plant J. 6, 1–7. [Google Scholar]
- 11.Selker, J. L., Steucek, G. L. & Green, P. B. (1992) Dev. Biol. 153, 29–43. [DOI] [PubMed] [Google Scholar]
- 12.Green, P. B. (1994) J. Exp. Bot. 45, 1775–1788. [Google Scholar]
- 13.Fleming, A. J., McQueen, M. S., Mandel, T. & Kuhlemeier, C. (1997) Science 276, 1415–1418. [Google Scholar]
- 14.Fleming, A. J., Caderas, D., Wehrli, E., McQueen, M. S. & Kuhlemeier, C. (1999) Planta 208, 166–174. [Google Scholar]
- 15.Pien, S., Wyrzykowska, J., McQueen, M. S., Smart, C. & Fleming, A. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11812–11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veit, B., Briggs, S. P., Schmidt, R. J., Yanofsky, M. F. & Hake, S. (1998) Nature 393, 166–168. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhury, A. M., Letham, S., Craig, S. & Dennis, E. S. (1993) Plant J. 4, 907–916. [Google Scholar]
- 18.Helliwell, C. A., Chin-Atkins, A. N., Wilson, I. W., Chapple R., Dennis, E. S. & Chaudhury, A. (2001) Plant Cell 13, 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh, J.-I., Hasegawa, A., Kitano, H. & Nagato, Y. (1998) Plant Cell 10, 1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohta, S., Mita, S., Hattori, T. & Nakamura, K. (1990) Plant Cell Physiol. 31, 805–813. [Google Scholar]
- 21.Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. (1994) Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- 22.Chomcznski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 23.Kouchi, H. & Hata, S. (1993) Mol. Gen. Genet. 238, 109–119. [DOI] [PubMed] [Google Scholar]
- 24.Itoh, J.-I., Kitano, H., Matsuoka, M. & Nagato, Y. (2000) Plant Cell 12, 2161–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn, B. O., Miyoshi, K., Itoh, J.-I., Nagato, Y. & Kurata, N. (2002) Theor. Appl. Genet. 105, 654–659. [DOI] [PubMed] [Google Scholar]
- 26.Nebert, D. W. & Gonzalez, F. J. (1987) Annu. Rev. Biochem. 56, 945–993. [DOI] [PubMed] [Google Scholar]
- 27.Nelson, D. R., Koymans, L., Kamataki, T., Stegeman, J. J., Feyereisen, R., Waxman, D. J., Waterman, M. R., Gotoh, O., Coon, M. J., Estabrook, R. W., et al. (1996) Pharmacogenetics 6, 1–42. [DOI] [PubMed] [Google Scholar]
- 28.Larkin, J. (1994) Plant Mol. Biol. 25, 343–353. [DOI] [PubMed] [Google Scholar]
- 29.Durst, R. & Nelson, D. R. (1995) Drug Metab. Drug Interact. 12, 189–206. [DOI] [PubMed] [Google Scholar]
- 30.Sharman, B. C. (1942) Ann. Bot. (London) 6, 245–284. [Google Scholar]
- 31.Asai, K., Satoh, N., Sasaki, H., Satoh, H. & Nagato, Y. (2002) Development (Cambridge, U.K.) 129, 265–273. [DOI] [PubMed] [Google Scholar]
- 32.Nadeau, J. A., Zhang, X. S., Li, J. & O'Neill, S. D. (1996) Plant Cell 8, 213–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zondlo, S. C. & Irish, V. F. (1999) Plant J. 19, 259–268. [DOI] [PubMed] [Google Scholar]
- 34.Ito, T. & Meyerowitz, E. M. (2000) Plant Cell 12, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolwell, G. P., Bozak, K. & Zimmerlin, A. (1994) Phytochemistry 37, 1491–1506. [DOI] [PubMed] [Google Scholar]
- 36.Schuler, M. A. (1996) Crit. Rev. Plant Sci. 15, 235–284. [Google Scholar]
- 37.Chapple, C. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 311–343. [DOI] [PubMed] [Google Scholar]
- 38.Imaishi, H., Matsuo, S., Swai, E. & Ohkawa, H. (2000) Biosci. Biotechnol. Biochem. 64, 1696–1701. [DOI] [PubMed] [Google Scholar]
- 39.Weber, H. (2002) Trends Plant Sci. 7, 217–224. [DOI] [PubMed] [Google Scholar]
- 40.Reid, J. B. (1993) J. Plant Growth Regul. 12, 207–226. [Google Scholar]
- 41.Hooley, R. (1994) Plant Mol. Biol. 26, 1529–1555. [DOI] [PubMed] [Google Scholar]
- 42.Ross, J. J., Murfet, I. C. & Reid, J. B. (1997) Physiol. Plant. 100, 550–560. [Google Scholar]
- 43.Maeda, E. (1965) Physiol. Plant. 18, 813–827. [Google Scholar]
- 44.Takeno, K. & Pharis, R. P. (1982) Plant Cell Physiol. 23, 1275–1281. [Google Scholar]
- 45.Wada, K., Kondo, H. & Marumo, S. (1985) Agric. Biol. Chem. 49, 2249–2251. [Google Scholar]
- 46.Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S. Ashikari, M., Kitano, H. & Matsuoka, M. (2000) Plant Cell 12, 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark, S. E., Running, M. P. & Meyerowitz, E. M. (1993) Development (Cambridge, U.K.) 119, 397–418. [DOI] [PubMed] [Google Scholar]
- 48.Jackson, D. & Hake, S. (1999) Development (Cambridge, U.K.) 126, 315–329. [DOI] [PubMed] [Google Scholar]
- 49.Maksymowych, R. & Maksymowych, A. B. (1973) Am. J. Bot. 60, 901–906. [Google Scholar]
- 50.Maksymowych, R., Codero, R. & Erickson, R. O. (1976) Am. J. Bot. 63, 1047–1053. [Google Scholar]
- 51.Maksymowych, R. & Erickson, R. O. (1977) Science 196, 1201–1203. [DOI] [PubMed] [Google Scholar]
- 52.Reinhardt, D., Mandel, T. & Kuhlemeier, C. (2000) Plant Cell 12, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avsian-Kretchmer, O., Cheng, J.-C., Chen, L., Moctezuma, E. & Sung, Z. R. (2002) Plant Physiol. 130, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi, Y.-H., Yoshizawa, K., Kobayashi, M. & Sakurai, A. (1995) Plant Cell Physiol. 36, 997–1001. [Google Scholar]
- 55.Dudley, M. & Poethig, R. S. (1993) Genetics 133, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans, M. M., Passas, H. J. & Poethig, R. S. (1994) Development (Cambridge, U.K.) 120, 1971–1981. [DOI] [PubMed] [Google Scholar]
- 57.Moose, S. P. & Sisco, P. H. (1994) Plant Cell 6, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poethig, R. S. (1988) Genetics 119, 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]