Abstract
The Vf gene from the wild species Malus floribunda 821 is the most studied apple scab resistance gene. Several molecular markers mapping around this gene were the starting point for a positional cloning project. The analysis of the bacterial artificial chromosome clones spanning the Vf region led to the identification of a cluster of genes homologous to the Cladosporium fulvum resistance gene family of tomato. One of these genes, HcrVf2 (homologue of the C. fulvum resistance genes of the Vf region), was used to transform the susceptible apple cultivar Gala. Four independent transformed lines resistant to apple scab were produced, proving that HcrVf2 is sufficient to confer scab resistance to a susceptible cultivar. The results show that direct gene transfer between cross-compatible species can be viable when, as in apple, the use of backcrosses to introduce resistance genes from wild species cannot exactly reconstitute the heterozygous genotype of clonally propagated cultivars.
Apple scab, which is caused by the ascomycete Venturia inaequalis (Cke.) Wint., is the most important disease in all apple-growing districts with high spring and summer rainfall. Its control in commercial orchards can require up to 15 fungicide treatments per year. An alternative approach is the use of resistant cultivars (cvs.), and sources of scab resistance have been found in small-fruited wild species (1). However, the transfer of these genes by classical breeding to cultivated apples is difficult because of the long juvenile phase, self-incompatibility, and the impossibility of exactly reproducing the heterozygous state of cultivated varieties. Starting from the wild species Malus floribunda 821 carrying the Vf gene, breeders have developed several scab-resistant apple cvs. (2), but not one has met with commercial success. Indeed, when compared with such commercially popular cvs. as Golden Delicious and Gala, the main horticultural and fruit-quality traits of these scab-resistant cvs. are notably different and undoubtedly less acceptable. The direct transfer of the Vf gene to apple cvs. thus is seen as a potential strategy for disease control. Moreover, the fact that two Vf-virulent races of V. inaequalis have been identified makes Vf resistance a good system in which to study gene-for-gene interaction between apple and scab (3–5).
The mode of Vf inheritance corresponds to a single dominant gene or to the combined action of a cluster of tightly linked genes (1). However, the Vf gene is not viewed strictly as dominant, because homozygous Vf plants are usually more resistant than heterozygous ones (6, 7). When inoculated with scab, Vf-carrying apple plants show varying resistance reactions, ranging from the absence of symptoms to chlorotic/necrotic, clearly sporulating lesions. Plants without the Vf gene do not develop a defense reaction, and heavily sporulating, unrestricted lesions are visible. It has been proposed (8, 9) that the variation in resistance reactions of Vf-carrying plants may be due to the presence and action of modifier genes.
To elucidate the Vf gene's complex mode of action, positional cloning was performed, and the steps leading to the identification of a cluster of resistance genes encoding receptor-like proteins have been reported (10–13). The amino acid sequence of these genes [homologues of Cladosporium fulvum resistance genes of the Vf region (HcrVf) (11)] contains an extracellular leucine-rich repeat domain and a putative transmembrane domain similar to those of the Cf resistance genes of tomato (14). The cluster is composed of at least four HcrVf genes, all transcribed but one producing only truncated mRNA (15). Fully transcribed genes HcrVf1, -2, and -4 cosegregated with apple scab resistance in >2,000 assayed plants and thus are viewed as candidate Vf resistance genes. This article reports the complete evidence that the HcrVf2 gene derived from a wild Malus species can induce apple scab resistance in transformed cv. Gala plants and thus reports the cloning of a resistance gene from a woody plant.
Materials and Methods
Construction of the Plasmid Binary Vector. The entire ORF of HcrVf2 (GenBank accession no. AJ297740) and the constitutive cauliflower mosaic virus 35S promoter were cloned into the binar y vector pCambia-2301 (GenBank accession no. AF234316). Clones were verified by sequencing, and the clone pCORF2 was chosen because, except for one base pair change in the signal peptide, it revealed complete identity with HcrVf2 (16). This clone was subsequently transferred into the Agrobacterium tumefaciens EHA105 strain containing the helper plasmid pCH32 (17) by following the methods of Höfgen and Willmitzer (18).
Plant Material and Apple Transformation. The HcrVf2 gene was introduced into the scab-susceptible apple cv. Gala by A. tumefaciens-mediated transformation using the engineered EHA105 strain. Plantlets grown in vitro were preconditioned by vacuum infiltration in liquid regeneration media TN505 (16) or TN22 [Murashige and Skoog (MS) medium supplemented with 2 mg·liter–1 napthalene acetic acid and 2 mg·liter–1 thidiazuron; P. Negri, personal communication]. Infection and cocultivation of leaves (19) and internodes (20) were followed by decontamination (200 mg·liter–1 cefotaxime) and selection (50 mg·liter–1 kanamycin) on the two media. Regenerating shoots were proliferated on A17 (21) and rooted on half-strength MS medium supplemented with 2 mg·liter–1 indolebutyric acid (P. Negri, personal communication). Transformed lines were named Ga2, for Gala transformed with HcrVf2, and numbered consecutively as they regenerated. As controls for greenhouse apple scab inoculation, in vitro-grown shoot tip cultures of Gala, Florina, and Enterprise were rooted, and the plants were acclimated to the greenhouse.
Evaluation of Apple Scab Resistance. Apple scab resistance of the five transgenic lines Ga2-2, Ga2-5, Ga2-7, Ga2-8, and Ga2-21 was tested by greenhouse inoculation. Young leaves of actively growing plants were sprayed with a suspension of V. inaequalis conidia (at least 200,000 conidia per ml) derived from cvs. Gala, Red Chief, and Golden Delicious growing in an untreated orchard. Conidia vitality was verified after 24 h by determining the fraction of germinated conidia (≈80%). Inoculum ability to infect Gala was tested by including, in all experiments, cv. Gala and/or a regenerated Gala line lacking the HcrVf2 gene (Ga2-7); the varieties Florina and Enterprise served as scab-resistant controls. A completely randomized scheme was adopted for plant location in the greenhouse. The environmental conditions were set as follows: 19 ± 5°C and 100% humidity for 48 h postinoculation and 70–80% thereafter. Three independent infection cycles were carried out in an attempt to test all transformed plants and controls twice.
Macroscopic symptoms and infection severity were evaluated on each leaf 10, 15, and 21 days postinoculation according to Chevalier (22) and Croxall (3, 23) scales. The maximum level of symptoms recorded 21 days postinoculation and their median and confidence limits were computed to describe the overall response of each line.
The presence of germinated V. inaequalis conidia on the leaf surface of inoculated plants was assessed at the end of the second inoculation round by observing by optical microscopy one inoculated leaf per plant. At 21 days postinoculation, leaves were prepared for microscopy by following the methods of Silfverberg-Dilworth et al. (24).
DNA Extraction and Detection of T-DNA Integration. T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] integration was confirmed by Southern blot analysis. HindIII-digested genomic DNA from greenhouse-acclimated plants was transferred to nylon membranes (Hybond-N+, Amersham Pharmacia Biosciences) and hybridized with probes produced by radiolabeling PCR-generated fragments of the entire HcrVf2 ORF and a section of the neomycin phosphotransferase II (nptII) gene (Fig. 1).
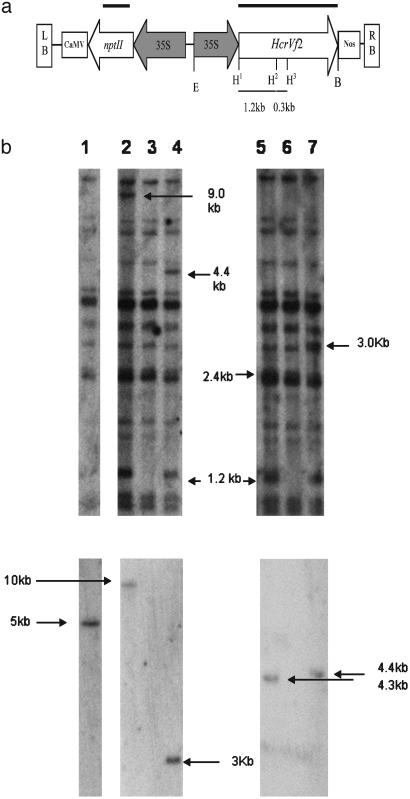
Fig. 1.
(a) Schematic representation of T-DNA from pCORF2 used in the transformation experiment. nptII and the putative scab resistance gene HcrVf2 are regulated by the cauliflower mosaic virus 35S promoters (filled arrows). E, EcoRI; B, BstEII; H, HindIII; LB, left border; RB, right border; CaMV, cauliflower mosaic virus 35S poly(A) signal; Nos, Nos poly(A) signal. H1, H2, and H3 represent restriction sites 1, 2, and 3, respectively, and thin bars represent the distance between HindIII restriction sites. Thick bars represent Southern blot probes. (b) T-DNA integration and copy number. Southern blot hybridization of HindIII-digested genomic DNA. Lanes: 1, Ga2-7; 2, Ga2-2; 3, Gala; 4, Ga2-5; 5, Ga2-21; 6, Gala; 7, Ga2-8. (Upper) T-DNA was hybridized with HcrVf2 probe, showing, in addition to the 1.2-kb specific fragment of the HcrVf2 gene, a second fragment for the estimation of the copy number. [The 300-bp fragment produced by HindIII sites 2 and 3 (H2 and H3 in a) was too small and was not transferred to the membrane.] Ga2-2, Ga2-5, Ga2-8, and Ga2-21 each contain a single copy of the pCORF2 T-DNA. Ga2-7 contains no HcrVf2 gene. T-DNA integration is shown by fragments of 9.0, 4.4, 3.0, and 2.4 kb hybridizing to this probe for the lines Ga2-2, Ga2-5, Ga2-8, and Ga2-21, respectively. (Lower) nptII probe. All transgenic lines show one single hybridization band of specific size demonstrating the presence of and integration to the nptII gene.
RNA Extraction and RT-PCR. For transgenic and control plants, the presence or absence of the HcrVf2 and nptII mRNAs was determined 21 days postinoculation. Total RNA was extracted from 30 mg of young leaf tissue and quantified. cDNAs were synthesized from 0.2 μg of total RNA by using the ImProm-II Reverse Transcription system (Promega) with an oligo(dT) primer. PCR was performed by using HcrVf2-specific primers (RT1 forward and RT2 reverse) (11) and nptII-specific primers (25). PCR using total RNA as template was performed to detect any DNA contamination.
Results
Transfer of HcrVf2 Gene to the Scab-Susceptible Apple cv. Gala. Agrobacteria-mediated transformation of cv. Gala with HcrVf2 was successful and yielded several independent transformed lines. Of these, five proliferating well in culture in vitro were acclimated to the greenhouse and tested for scab resistance. Integration of the T-DNA (Fig. 1a) and uniformity among regenerated plants of the same line were confirmed by Southern hybridization of DNA extracted from single plants of the lines Ga2-2, Ga2-5, Ga2-7, Ga2-8, and Ga2-21. Two probes, one from HcrVf2 and the other from the selectable marker nptII, which confers resistance to kanamycin, were used. Twelve fragments hybridizing to the HcrVf2 probe were present in untransformed Gala DNA. Additional hybridization signals supported transgene integration in transformed regenerated plants (Fig. 1b). Four transgenic lines (not Ga2-7) showed two additional bands. One gene fragment (1.2 kb) was produced by HindIII sites within the T-DNA and another, of at least 1.5 kb, was produced by a HindIII site in the T-DNA and an additional site within the apple genome. This latter fragment, the same size for all individual plants of each line, was 9.0, 4.4, 3.0, and 2.4 kb for the lines Ga2-2, Ga2-5, Ga2-8, and Ga2-21, respectively. Southern analysis based on the nptII probe showed a single band with a specific size for each transgenic line, including Ga2-7 (Fig. 1b).
The hybridization results support the presence of one complete copy of the T-DNA in the transgenic lines Ga2-2, Ga2-5, Ga2-8, and Ga2-21. Plants of the line Ga2-7 most likely contain a partial integration of the T-DNA, because only fragments also hybridizing with Gala DNA hybridized to the HcrVf2 probe, whereas one clear hybridization signal was evident for the nptII probe. Moreover, PCR amplification of 35S::HcrVf2 from this line was negative and that from the nptII positive, thus supporting the conclusion of a partial integration of the transformation construct in line Ga2-7 (data not shown).
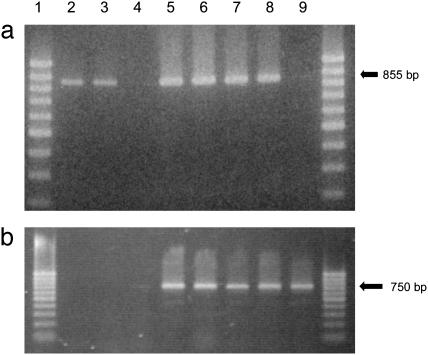
Transcription of HcrVf2 is demonstrated by RT-PCR, because the expected fragment (855 bp) was amplified from the Vf-resistant cvs. Enterprise and Florina, as well as all plants of lines Ga2-2, Ga2-5, Ga2-8, and Ga2-21. No amplification was noted from Gala or Ga2-7 plants (Fig. 2a). The nptII gene transcription was confirmed in all transgenic lines but not in control cvs. (Fig. 2b). PCR carried out directly with RNA showed that no DNA contamination was present in any of the samples (data not shown).
Fig. 2.
Transcription evaluated by RT-PCR. Lanes: 1,100-bp ladder; 2, Enterprise; 3, Florina; 4, Gala; 5, Ga2-2; 6, Ga2-5; 7, Ga2-8; 8, Ga2-21; 9, Ga2-7. (a) HcrVf2 primers show that the gene is expressed in Vf cvs. and in all transformed lines except Ga2-7. (b) nptII primers show that the gene is expressed only in transformed lines.
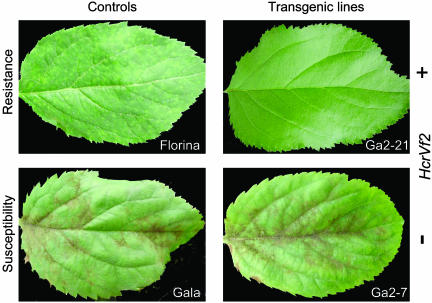
Scab Resistance Evaluation. To assess scab resistance in transgenic lines, plants were evaluated 21 days postinoculation with V. inaequalis conidia. The untransformed cv. Gala and the line Ga2-7 showed clear symptoms of susceptibility, whereas untransformed Vf-resistant varieties and HcrVf2 transgenic plants proved resistant (Table 1). Resistant plants were inoculated twice, and the results were consistently similar, as indicated by the median value and confidence limits (Table 1). Gala showed high scab susceptibility to the inoculum, with sporulation observed as early as 10 days postinoculation. Symptom severity peaked after 21 days, with heavy sporulation observed on all inoculated young leaves; in addition, Gala showed no resistance reaction (Fig. 3). Similar symptoms were observed in plants from the line Ga2-7, which contains a functional copy of the nptII, but not the HcrVf2, gene. Additional inoculations of Ga2-7 and Gala plants proved impossible because of the extreme damage suffered after the first inoculation round.
Table 1. Pooled results of the three greenhouse scab inoculation rounds 21 days postinoculation.
| Symptom*
|
Severity†
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Confidence limit
|
Confidence limit
|
|||||||||
| Line or cv. | HcrVf2 gene presence | No. of inoculated plants | Med. | L | U | Max. | Med. | L | U | Max. |
| Ga2-2 | + | 6 | 0 | 0 | 2 | 3b | 0 | 0 | 2 | 3 |
| Ga2-5 | + | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 |
| Ga2-8 | + | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 |
| Ga2-21 | + | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 |
| Ga2-7 | - | 4 | 4 | 4 | 4 | 4 | 6 | 4 | 7 | 7 |
| Gala | - | 7 | 4 | 4 | 4 | 4 | 6 | 4 | 7 | 7 |
| Enterprise | + | 2 | 2 | 0 | 3b | 3b | 3 | 0 | 4 | 4 |
Median values of symptoms and their confidence limits were calculated by using the data from the three youngest leaves present at the time of each plant's inoculation. Med., median; Max., maximum; L, lower; U, upper.
Symptoms were assessed after Chevalier et al. (22) with 0 = no macroscopically visible symptom, 1 = hypersentive pinpoints, 2 = chlorotic areas with no sporulation, 3a = chlorotic and necrotic areas with no sporulation, 3b = chlorotic and necrotic areas with sparse sporulation, and 4 = sporulation covering entire leaf. These values represent degrees of resistance, with 4 representing susceptibility. Confidence limit α = 0.01.
Infection severity values were assessed on a modified Croxall scale (3, 23): 1 indicates a percentage of leaf area with sporulating symptoms from 0% to 1%; 2, from 1% to 5%; 3, from 5% to 10%; 4, from 10% to 25%; 5, from 25% to 50%; 6, from 50% to 75%; and 7, from 75% to 100%. Confidence limit α = 0.01.
Fig. 3.
Leaf symptoms 21 days after inoculation with V. inaequalis conidia. Transgenic line Ga2-21 (Upper Right), containing HcrVf2, represents typical symptoms observed on all HcrVf2 transformed lines. Transgenic line Ga2-7 (Lower Right) contains only the selective nptII gene and is thus a transgenic control. The susceptible cv. Gala (Lower Left) and resistant cv. Florina (Upper Left) are included for the comparison of scab symptoms.
On leaves of the Vf-carrying cv. Enterprise, small chlorotic lesions appeared after 10 days; ≈25% of the leaf surface of one to two leaves per plant showed chlorotic lesions after 21 days. During the second inoculation round, necrosis and a very limited level of sporulation were observed on two Enterprise leaves. Leaves of the other Vf-carrying cv., Florina, were inoculated only once and showed chlorotic lesions after 21 days.
The highest level of resistance was observed in transgenic lines Ga2-5, Ga2-8, and Ga2-21, which had either no symptoms or rare, pinpoint pits as resistance reactions (Fig. 3). The transgenic line Ga2-2 showed no symptoms on approximately half of the young inoculated leaves and small chlorotic lesions after 10 days on the other leaves; in a few cases, after 21 days the chlorosis became more severe, and small necrotic lesions with restricted sporulation were observed on one leaf of one plant. Although Ga2-2 occasionally showed some restricted fungal development, the line still can be considered more resistant than the Vf-carrying cv. Enterprise, as indicated by the median (Table 1).
The presence of germinated V. inaequalis conidia was confirmed on the leaves of plants showing either a resistance reaction or no symptoms from lines Ga2-2, Ga2-5, Ga2-8, and Ga2-21 (Table 2). In most cases (97% of conidia observed), no fungal development beyond that of appressoria and penetration peg formation was observed (class γ). In a few rare cases, subcuticular primary stroma formed, but expansion did not exceed 40 μm (class β). On leaves from two plants of the line Ga2-2 that had small chlorotic lesions, small isolated areas of stroma with sporulating conidiophores were found (class α). However, an equivalent fungal development was seen on a leaf of the Vf-resistant cv. Enterprise.
Table 2. Presence of germinated V. inaequalis conidia on leaf surface of resistant plants.
| No. of leaf samples*
|
No. of conidia
|
Fungal development†
|
||||
|---|---|---|---|---|---|---|
| Line or cv. | Average per leaf sample | Total | α | β | γ | |
| Ga2-2 | 6 | 97 | 582 | 55 | 8 | 519 |
| Ga2-5 | 4 | 137 | 548 | 0 | 1 | 547 |
| Ga2-8 | 4 | 100 | 401 | 0 | 0 | 401 |
| Ga2-21 | 3 | 80 | 240 | 0 | 0 | 240 |
| Enterprise | 1 | 54 | 54 | 10 | 33 | 11 |
This table does not contain the data from susceptible plants because on Gala and transgenic Ga2-7 leaf samples the new stroma and conidiophores were too dense, making conidia counting impossible.
For each plant, one leaf section (≈1.5 × 1 cm) was examined.
Fungal development for each conidium was defined according to the following classes: α, isolated areas of stroma with sporulating conidiophores; β, primary stroma expanding no more than ≈40 μm; and γ, no fungal development beyond that of appressoria and penetration peg.
Discussion
The reported data show that the HcrVf2 gene can confer resistance to apple scab when transformed in the scabsusceptible cv. Gala. Of the five transgenic lines, the Ga2-5, Ga2-8, and Ga2-21 showed a very high level of resistance. Line Ga2-2 showed a level of resistance at least comparable with that of Vf cvs., although the peak symptoms observed in these plants were more severe than those seen in plants of the other transgenic lines. Because all of the transgenic resistant lines contain only a single copy of the transgene, the differences between Ga2-2 and the other scab resistance transformants are most likely attributable to the integration site of T-DNA, which may modify transgene expression (26, 27) and, consequently, influence the level of resistance. The slight differences observed between inoculations were probably due to environmental variation, because they were observed in both transformed and control plants. The fact that transgenic line Ga2-7, containing the nptII, but not the HcrVf2, gene, is scab-susceptible proves that neither the method used to regenerate plants nor the nptII gene affects plant resistance to V. inaequalis.
The interaction of V. inaequalis with Malus genotypes follows the gene-for-gene rule (28, 29). Our data, in fact, clearly show that, when transformed with the HcrVf2 gene, the scabsusceptible cv. Gala can develop an incompatible interaction with a mixed-strain inoculum of V. inaequalis.
HcrVf2 is one of at least three resistance genes mapping at the Vf locus (11). The influence of the HcrVf1 and HcrVf4 genes on scab resistance must be investigated to determine whether HcrVf2 is the only gene able to trigger a resistance response. Of the >30 plant resistance genes characterized so far, at least half are members of gene clusters (30). In tomato, the Cf genes confer resistance to C. fulvum, and different members of the Cf gene family recognize different avr genes (31, 32). The two resistance genes Cf-4 and Cf-9 each are found within a cluster of five ORFs, and within each cluster two ORFs confer resistance to C. fulvum (Cf-4 and Hcr9-4E, Cf-9 and Hcr9-9A or Hcr9-9B, respectively). Interestingly, Hcr9-9A and Hcr9-9B are active only in adult plants, whereas Cf-9 and Cf-4 are active in seedlings, too, showing a differentiated expression pattern between the members of a cluster (33, 34). The resistance gene Xa21 in rice, which confers resistance to Xanthomonas oryzae pv. oryzae, encodes a receptor-like kinase with an extracellular leucine-rich repeat and a serine/threonine kinase in the putative intracellular domain. Of seven ORFs studied within the Xa21 gene family, only two were found to contain the complete sequence encoding for receptor kinase-like proteins (35), and only Xa21 has been shown to confer resistance (36). The Pto resistance gene in tomato mediates resistance to strains of Pseudomonas syringae pv. tomato expressing avrPto. Pto is the only gene of a cluster of seven found to interact with the AvrPto protein and, hence, to confer resistance, although several genes of the cluster have been shown to be transcribed. Note, however, that only one avr ligand has been identified from pv. tomato and that the other ORF found at the Pto locus may interact with other strains of the pathogen (37). The examples cited show that, although HcrVf2 clearly can confer resistance to apple scab, it will be important to clarify the function of all genes within the Vf cluster.
The cloning of an apple scab resistance gene represents the basis for further investigation of the resistance mechanism. It also represents a step toward a gene therapy (restoring resistance where lost) of the scab-susceptible cvs. that currently dominate the apple industry. This strategy will allow the transfer of resistance from a wild apple species to any commercial apple genotype while maintaining the horticultural and fruit-quality traits growers and consumers prize most. It may also be possible to achieve greater resistance durability by the simultaneous transfer of several resistance genes from wild apple species. Going one step further, it may be possible to use apple promoters and novel techniques that, by eliminating selective marker genes (38, 39), generate transgenic varieties without any foreign genes and, hence, may make genetically modified plants more acceptable to growers and consumers alike.
Acknowledgments
We thank F. Salamini (Max Planck Institute, Koln, Germany) for critical reading of the manuscript; E. Chevreau (Institut National de la Recherche Agronomique, Angers, France) and P. Negri (University of Bologna) for advice on apple transformation methodology; F. Gennari and R. Paris (University of Bologna) for help during inoculation; E. Muzzi (University of Bologna) for suggestions on statistical analysis; and Johannes Fütterer (Swiss Federal Institute of Technology) for advice on preparation of the binary vector. This work was supported by Swiss National Foundation Grant 31-58914.99, Strategic Project L. 449/97 of the Italian Ministry of Instruction, Universities, and Research (MIUR), and the Italian Ministry of Agriculture and Forestry (MiPAF).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: cv., cultivar.
References
- 1.Williams, E. B. & Kuc, J. (1969) Annu. Rev. Phytopathol. 7, 223–246. [Google Scholar]
- 2.Janick, J., Cummins, J. N., Brown, S. K. & Hemmat, M. (1996) Fruit Breeding: Tree and Tropical Fruits (Wiley, New York), pp. 1–77.
- 3.Parisi, L., Lespinasse, Y., Guillaumes, J. & Kruger, J. (1993) Phytopathology 83, 533–537. [Google Scholar]
- 4.Bénaouf, G. & Parisi, L. (2000) Phytopathology 90, 236–242. [DOI] [PubMed] [Google Scholar]
- 5.MacHardy, W. E., Gadoury, D. M. & Gessler, C. (2001) Plant Dis. 85, 1036–1051. [DOI] [PubMed] [Google Scholar]
- 6.Gessler, C., Patocchi, A., Kellerhals, M. & Gianfranceschi, L. (1997) in IOBC/WPRS Bulletin, eds. Berrie, A. M., Xu, X.-M., Harris, D. C., Roberts, A. L., Evans, K., Barbara, D. J. & Gessler, C., (Internatl. Org. Biol. Integrated Contr. Noxious Animals Plants, W. Palaearctic Region. Sect., Croydon, England), Vol. 20, pp. 105–109. [Google Scholar]
- 7.Tartarini, S., Sansavini, S., Vinatzer, B. A., Gennari, F. & Domizi, C. (2000) in Eucarpia Symposium on Fruit Breeding and Genetics, Acta Horticulturae 538, eds. Geibel, M., Fischer, M. & Fischer, C. (Internatl. Soc. Horticultur. Sci., Dresden, Germany), Vol. II, pp. 549–552. [Google Scholar]
- 8.Rousselle, G. L., William, E. B. & Hough, L. F. (1974) in XIX International Horticultural Congress, eds. Antoszewski R., Harrison, L. & Zych, C. C. (Internatl. Soc. Horticultur. Sci., Warsaw), Vol. 3, pp. 19–26. [Google Scholar]
- 9.Gessler, C. (1989) in IOBC/WPRS Bulletin, eds. Gessler, C., Butt, D. & Koller, B. (Internatl. Org. Biol. Integrated Contr. Noxious Animals Plants, W. Palaearctic Region. Sect., Brissago, Switzerland), Vol. 12, pp. 168–190. [Google Scholar]
- 10.Vinatzer, B. A., Zhang, H. B. & Sansavini, S. (1998) Theor. Appl. Genet. 97, 1183–1190. [Google Scholar]
- 11.Vinatzer, B. A., Patocchi, A., Gianfranceschi, L., Tartarini, S., Zhang, H. B., Gessler, C. & Sansavini, S. (2001) Mol. Plant–Microbe Interact. 14, 508–515. [DOI] [PubMed] [Google Scholar]
- 12.Patocchi, A., Vinatzer, B. A., Gianfranceschi, L., Tartarini, S., Zhang, H. B., Sansavini, S. & Gessler, C. (1999) Mol. Gen. Genet. 262, 884–891. [DOI] [PubMed] [Google Scholar]
- 13.Patocchi, A., Gianfranceschi, L. & Gessler, C. (1999) Theor. Appl. Genet. 99, 1012–1017. [DOI] [PubMed] [Google Scholar]
- 14.Jones, D. A., Thomas, C. M., Hammond-Kosack, K. E., Balint-Kurti, P. J. & Jones, J. D. G. (1994) Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- 15.Xu, M. L. & Korban, S. S. (2002) Genetics 162, 1995–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbieri, M., Belfanti, E., Tartarini, S., Vinatzer, B., Sansavini, S., Dilworth, E., Gianfranceschi, L., Hermann, D., Patocchi, A. & Gessler, C. (2003) Hortic. Sci. 38, 329–331. [Google Scholar]
- 17.Hamilton, C. M. (1997) Gene 200, 107–116. [DOI] [PubMed] [Google Scholar]
- 18.Hofgen, R. & Willmitzer, L. (1988) Nucleic Acids Res. 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, D. J., Uratsu, S., Cheng, J., Negri, P., Viss, P. & Dandekar, A. M. (1993) Plant Cell Rep. 12, 559–563. [DOI] [PubMed] [Google Scholar]
- 20.Sriskandarajah, S. & Goodwin, P. (1998) Plant Cell Tissue Organ Cult. 53, 1–11. [Google Scholar]
- 21.James, D. J. & Dandekar, A. M. (1991) in Plant Tissue Culture Manual, ed. Lindsey, K. (Kluwer Academic, Dordrecht, The Netherlands), Book 8, pp. 1–18.
- 22.Chevalier, M., Lespinasse, Y. & Renaudin, S. (1991) Plant Pathol. 40, 249–256. [Google Scholar]
- 23.Croxall, H. E., Gwynne, D. C. & Jenkins, J. E. (1952) Plant Pathol. 2, 39–41. [Google Scholar]
- 24.Silfverberg-Dilworth, E., Patocchi, A. & Gessler, C. (2003) in IOBC/WPRS Bulletin, eds. Gessler, C. & Triloff, P. (Internatl. Org. Biol. Integrated Contr. Noxious Animals Plants, W. Palaearctic Region. Sect., Lindau, Germany), in press.
- 25.Hamilton, C. M., Frary, A., Lewis, C. & Tanksley, S. D. (1996) Proc. Natl. Acad. Sci. USA 93, 9975–9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iglesias, V. A., Moscone, E. A., Papp, I., Neuhuber, F., Michalowski, S., Phelan, T., Spiker, S., Matzke, M. & Matzke, A. J. M. (1997) Plant Cell 9, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar, S. & Fladung, M. (2001) Planta 213, 731–740. [DOI] [PubMed] [Google Scholar]
- 28.MacHardy, W. E. (1996) Apple Scab (Am. Phytol. Soc., St. Paul).
- 29.Flor, H. H. (1971) Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- 30.Hulbert, S. H., Webb, C. A., Smith, S. M. & Sun, Q. (2001) Annu. Rev. Phytopathol. 39, 285–312. [DOI] [PubMed] [Google Scholar]
- 31.Hammond-Kosack, K. E., Tang, S. J., Harrison, K. & Jones, J. D. G. (1998) Plant Cell 10, 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, C. M., Jones, D. A., Parniske, M., Harrison, K., Balint-Kurti, P. J., Hatzixanthis, K. & Jones, J. D. G. (1997) Plant Cell 9, 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parniske, M., Hammond-Kosack, K. E., Golstein, C., Thomas, C. M., Jones, D. A., Harrison, K., Wulff, B. B. & Jones, J. D. (1997) Cell 91, 821–832. [DOI] [PubMed] [Google Scholar]
- 34.Takken, F. L. W., Thomas, C. M., Joosten, M., Golstein, C., Westerink, N., Hille, J., Nijkamp, H. J. J., De Wit, P. & Jones, J. D. G. (1999) Plant J. 20, 279–288. [DOI] [PubMed] [Google Scholar]
- 35.Song, W. Y., Pi, L. Y., Wang, G. L., Gardner, J., Holsten, T. & Ronald, P. C. (1997) Plant Cell 9, 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., Gardner, J., Wang, B., Zhai, W. X., Zhu, L. H., et al. (1995) Science 270, 1804–1805. [DOI] [PubMed] [Google Scholar]
- 37.Chang, J. H., Tai, Y. S., Bernal, A. J., Lavelle, D. T., Staskawicz, B. J. & Michelmore, R. W. (2002) Mol. Plant–Microbe Interact. 15, 281–291. [DOI] [PubMed] [Google Scholar]
- 38.Bar, M., Leshem, B., Giloa, L. & Gidoni, D. (1996) Theor. Appl. Genet. 93, 407–413. [DOI] [PubMed] [Google Scholar]
- 39.Joersbo, M., Donaldson, I., Kreiberg, J., Petersen, S. G., Brunstedt, J. & Okkels, F. T. (1998) Mol. Breed. 4, 111–117. [Google Scholar]