Abstract
The development of transgenic mosquitoes that are refractory to the transmission of human diseases such as malaria, dengue, and yellow fever has received much interest due to the ability to transform a number of vector mosquito species with transposable elements. Transgenic strains of mosquitoes have been generated with molecular techniques that exhibit a reduced capacity to transmit pathogens. These advancements have led to questions regarding the fitness of transgenic mosquitoes and the ability of transformed mosquitoes to compete and effectively spread beneficial genes through nontransformed field populations, the core requirement of a genetically based control strategy aimed at reducing the spread of mosquito-borne human disease. Here we examine the impact of transgenesis on the fitness of Aedes aegypti, a mosquito that transmits yellow fever. Mosquitoes were altered with two types of transgene, the enhanced GFP gene and two transposase genes from the Hermes and MOS1 transposable elements. We examined the effects of these elements on the survivorship, longevity, fecundity, sex ratio, and sterility of transformed mosquitoes and compared results to the nontransformed laboratory strain. We show that demographic parameters are significantly diminished in transgenic mosquitoes relative to the untransformed laboratory strain. Reduced fitness in transgenic mosquitoes has important implications for the development and utilization of this technology for control programs based on manipulative molecular modification.
Advances in insect biotechnology, in particular the development of genetic transformation methods, have led to renewed interest in genetic-based insect control strategies (1). Altering the pest status of insects using recombinant DNA technologies is being considered as a possible solution to certain medical and agricultural insect problems that have proven difficult to solve using more conventional chemical, cultural, and biological control practices aimed at population suppression or eradication. Anopheles gambiae, the major vector of human malaria in Africa, is one high-profile target of this new form of genetic insect control (2). With this project, it is envisioned that the mosquito's susceptibility to malaria parasites would be genetically altered; insects possessing this new genotype would be introduced into native susceptible mosquito populations in such a way as to lead to the replacement of endogenous susceptible mosquito populations, thereby reducing malaria transmission. Other vector-borne diseases may also be targets of this form of genetic control. To implement these ideas, effective methods are needed to introduce novel genes into insect genomes, and transgenes must be identified that can lead to the elimination of pathogen transmission. In addition, techniques for successfully introducing laboratory-generated genotypes into native populations are required. Although significant progress is being made in developing transgenic insect technologies and effector genes, very little work has been done on introducing laboratory-generated genotypes into native populations to determine establishment, population growth, and persistence. Genetic transmission-enhancing (drive) systems are envisioned as playing an important role in these population replacement strategies, but the nature and characteristics of such systems have not been well defined. The strength of the drive system required would depend, in part, on how fast a genotype must be disseminated within a natural population as well as the fitness (i.e., survivorship and reproductive rates) of transgenic insects. Transgenes conferring fitness advantages could act to promote the spread of particular genotypes, whereas transgenes resulting in fitness costs could counteract any transmission advantage associated with a drive system. Therefore, creating insects with appropriate fitness will be critical to the success of these transgenic-based genetic control strategies.
Previous examples using genetically manipulated mosquitoes offer a guide to the role that fitness can play in mating competitiveness in the field. The genetic control of Culex tarsalis using sex-linked translocations of single autosomal recessive mutations has been previously tested under laboratory and field conditions (3, 4). Laboratory strains containing a novel genetic makeup were not competitive with wild-type C. tarsalis, and the genetic alteration that was developed in the laboratory for introduction into field populations failed to establish. This early work demonstrated the importance of determining fitness of genetically manipulated mosquitoes designed to spread genes through field populations.
It has generally been assumed that transgenic organisms will have lower fitness than nontransgenic conspecifics in the absence of selection (5), although this is a hotly debated topic and lies at the center of discussions concerning the risks of releasing transgenic organisms. Our current knowledge of transgenesis on fitness is based on studies in Drosophila. These studies have indicated that gene vector integration and transgene expression can be sources of positive and negative fitness impacts (6, 7). The impact of current nondrosophilid transformation systems, including vectors and genetic marker systems, on host competitiveness has only recently been studied (8). The ability of transgenic strains of Anopheles stephensi to compete with wild-type An. stephensi in small population cages was examined, and none of the four transgenic lines was found to be competitive; indeed, in all populations, the frequency of the transgene dramatically declined (8). The fitness of transgenic An. stephensi was not examined (8). Fitness studies are important because an understanding of how current insect transformation technology affects fitness will permit the rational design of competitive transgenic insects for use in genetic control programs. Here we examine in detail the reproductive and developmental fitness of three lines of transgenic Aedes aegypti relative to the nontransformed conspecific. To assess the fitness of transgenic Ae. aegypti, we compared laboratory-generated demographic data across transgenic and the nontransformed line to determine the competitiveness of transgenic mosquitoes. Analyses presented here may indicate how successfully transgenic mosquitoes will compete with their wild-type counterparts in the field.
Materials and Methods
Mosquito Strains. The three transgenically modified Ae. aegypti lines were all generated from the Orlando laboratory strain maintained at the University of California, Riverside. The Orlando strain has been maintained in the laboratory since at least 1961 and has not been invigorated with field-collected material since that time. For the purposes of this study, the transgenic lines were denoted enhanced GFP (EGFP), autoHermes, and pBacMOS. Strain EGFP contains the Hermes transposable element into which the EGFP gene has been inserted and placed under the control of the actin5C promoter of Drosophila melanogaster. The EGFP strain has been maintained in the laboratory for >3 years and was previously described (9). Strain autoHermes contains the same construct as the EGFP strain with the addition of the Hermes transposase gene placed under the control of the D. melanogaster hsp70 promoter (10). This strain contains an autonomous Hermes element and has been maintained in the laboratory for >2 years. The strain pBacMOS contains the piggyBac transposable element into which has been inserted the EGFP gene under the control of the eye and anal papillae-specific 3xP3 promoter (11). In addition, it contains the MOS1 transposase gene placed under the control of the D. melanogaster hsp70 transposase gene (10). pBacMOS does not contain introduced mariner elements and has been maintained in the laboratory for >2 years.
Because we were interested in determining the effects of transgenesis on the fitness of Ae. aegypti, we chose to examine the effects of EGFP and transposase expression, because these two genes might constitute the genotype of a mosquito genetically engineered to be refractory to pathogen transmission and to drive genes through populations. EGFP and related fluorescent proteins are popular choices for marking transgenics and could be used to follow the spread of introduced transgenes through populations. Transposable elements are potential genetic drive mechanisms for spreading beneficial transgenes through mosquito populations, and active transposase must be synthesized to mobilize target elements. The strategy used in this study was to examine EGFP and transposase expression by using existing transgenic strains that had been successfully reared in the laboratory for some time. These strains, based on the sole criterion of being able to be maintained in laboratory culture, might be judged to be “robust” or “fit.”
Establishment of Experimental Cohorts. Thirty pupae of each experimental line (Orlando strain and transgenics) were placed individually in plastic cups (30-ml Condiment Cups, Tyco Plastics, Eagan, MN) with 20 ml of 1-day-old tap water and 0.005 g of diet [two crushed dog-bone biscuits (Original Milkbone, Kraft, East Hanover, NJ) and one wheast (Redstar, Milwaukee). Each cup with pupae was placed in a larger paper cup (477-ml food containers, Solo, Urbana, IL) and covered with mesh netting (120 nylon strands per cm2). Adult mosquitoes that emerged from these pupae were designated as F0, and male–female pairs from each experimental line that emerged were set up in 477-ml paper mating cups with a plastic 30-ml oviposition cup lined with dry filter paper (3.5 × 10 cm; Midwest Scientific, Valley Park, MO).
Seven to nine days after emergence of F0 females and males and subsequent pairing for mating, female mosquitoes were given access to a mouse anesthetized with 0.2 ml of Ketased solution [1.5 ml of purified water/0.1 ml of xylazine (Phoenix Scientific, St. Joseph, MO)/0.4 ml of Ketased (Abbott)] for 10 min and allowed to blood feed through the mesh covering the oviposition cup. Two days after blood feeding, oviposition cups were moistened with 1-day-old tap water, and every 24 h, oviposition cups were removed and replaced with new cups holding moistened filter paper. Cups containing mosquito eggs were labeled with a unique female identifying number that indicated mosquito line, replicate, and date of oviposition. Collected eggs were allowed to embryonate for 7 days at 75% humidity, 26°C, 12:12-hr (light/dark) photoperiod. After embryonation, individual egg papers were submerged in 150 ml of 1-day-old tap water with 0.1 g of larval mosquito diet in a 600-ml glass jar (Kern Home Canning Jars, Alltrista, Muncie, IN). After 24 h, 150 first-instar larvae (designated here as the F1 generation) from each experimental line were placed individually into plastic 30-ml larval rearing cups with 20 ml of 1-day-old tap water and 0.005 g of larval food. For Orlando strain mosquitoes, 150 larvae (15 larvae from each of 10 females) hatched after 24 h and were used for the F1 cohort. For two transgenic lines (EGFP and pBacMOS), egg hatching was delayed, and the F1 larval cohort from 10 F0 females was removed as larvae emerged over a 7-day period. For the autoHermes line, only four F0 females produced viable eggs, and 150 larvae were used from these four females to initiate the experimental F1 cohort.
Preimaginal Development Times. The mean number of days for eggs laid by F0 females to hatch was determined and compared across mosquito lines with ANOVA in sas (sas 1980, SAS Institute, Cary, NC). Tukey's studentized range test (P = 0.05) was used to determine whether hatching times of eggs varied significantly across experimental lines. Individual F1 larvae that emerged from eggs were placed in plastic 30-ml larval rearing cups and monitored every 24 h. The number of days spent in each preimaginal life stage (i.e., 1st, 2nd, 3rd, and 4th instars, pupae) and numbers surviving in each stage were recorded daily. The mean number of days in each preimaginal stage was determined and compared across mosquito lines with ANOVA in sas (sas 1980). Tukey's studentized range test (P = 0.05) was used to determine whether developmental times by stage varied significantly across experimental lines. The first 40 adult female and male mosquitoes to emerge for each line were paired and placed into individual 477-ml oviposition cups. To reduce possible inbreeding effects, adults within experimental lines were paired only if they originated from different F0 females.
Adult Longevity, Female Fecundity, Partial Life Table Construction, and Offspring Sex Ratio. F1 adult survivorship rates were determined every 24 h. Female mosquitoes were blood fed with an anesthetized mouse 12–14 days after emergence. Daily fecundity and percentage of egg hatch after 48 h and 1 week were recorded for each F1 female that produced eggs from each blood feeding for four consecutive gonotrophic cycles. One week after each oviposition cycle, blood meals (i.e., an anesthetized mouse) were offered to surviving females in each experimental line, and fecundity rates were recorded. The percentage of females either failing to lay eggs or laying egg clutches in which all eggs failed to hatch in each gonotrophic cycle was considered sterile; sterility rates were calculated and compared across experimental lines by using χ2 (P = 0.05) in sas (sas 1980). Mean lifetime fecundity (i.e., viable eggs across all gonotrophic cycles) for each female in each transgenic line was calculated and compared across mosquito lines with ANOVA in sas (sas 1980). Tukey's studentized range test (P = 0.05) was used to determine whether female fecundity varied significantly across experimental lines. Preimaginal survivorship rates and sex ratio of offspring were recorded for each of the four consecutive gonotrophic cycles for each reproductive female in each experimental line. Preimaginal survivorship rates for each experimental line were used to construct partial life tables (12). The sex of the resulting adults from each gonotrophic cycle for each experimental line was recorded.
Demographic Growth Parameters. Larval to adult survivorship data, daily fecundity of individual females, and sex ratio of progeny reared from females at each experimental temperature were used to construct lxmx life tables from which demographic growth parameters were calculated. Daily development and survivorship data and daily progeny production for female mosquitoes from each experimental line were used to produce a birth cohort of females. The proportion of larvae produced by F1 females reared that were female was used to adjust daily progeny production in the mx column to estimate the number of daughters produced daily by surviving females. The following demographic parameters were calculated from lxmx life tables:
Net reproductive rates [Ro = Σlxmx (where lxmx is the net female maternity, lx is the fraction of females alive at age x, and mx is the number of daughters born to surviving females at age x)] express the per-generation growth rate of the population as the number of daughters produced by females (Ro > 1.0 the population increases in size, Ro = 1.0 no increase in population size, Ro < 1.0 population growth is declining) (12).
Mean generation time (Tc = Σxlxmx/Ro) is the average interval separating births of one generation from the next (12).
The intrinsic rate of natural increase, rm [found as the solution to: 1 = Σlxmxexp(–rmx) (this equation was iterated for rm until a value of one was obtained)] is the maximum exponential rate of increase by a population growing within defined physical conditions (13).
Doubling time in days [Td = ln(2)/rm] is the time required by a population growing exponentially without limit to double in size when increasing at a given rm (12).
Mean demographic parameter estimates with SE were generated by jackknife analysis of lxmx life-table data. The jackknife method removes one observation at a time from the original data set and recalculates the statistic of interest from the truncated data set. These new estimates, or pseudovalues, form a set of numbers from which mean values and variances can be calculated and compared statistically (14–17). The jackknife method of resampling is well suited for estimating variance for population growth statistics (16). Mean jackknife estimates of demographic parameters were compared across temperatures by using ANOVA and Tukey's studentized range test (P = 0.05) to determine whether significant differences between population growth statistics for each experimental mosquito line existed.
Rearing Conditions. All experimental containers were maintained at 26°C with a 12:12-hr (light/dark) photoperiod, and 80% relative humidity in a secure insect-rearing room in the Insectary and Quarantine Facility at the University of California, Riverside. Temperature and relative humidity were recorded every 30 min with hobo dataloggers (Onset, Pocasset, MA).
Additional Nutritional Provisioning for Adult Mosquitoes. In addition to blood feeding, adult mosquitoes were supplied water via a moist cotton ball placed on the mesh top of each paper cup containing the pair, along with two raisins as a carbohydrate source. Cotton balls were moistened daily and covered with a plastic cup (22-ml Plastic Soufflé Cup, Iris, Smart & Final, Los Angeles) to reduce evaporation, and raisins were replaced every 3 days.
Results
Partial Life Tables and Preimaginal Development. For each gonotrophic cycle, percentage egg-to-adult survivorship was greatest for the Orlando strain when compared to the three transgenic mosquito lines (Table 1). The best-performing transgenic mosquito in terms of egg-to-adult survivorship across all gonotrophic cycles was the autoHermes strain. Across all gonotrophic cycles, Orlando strain females produced ≈43–75% more eggs than transgenic mosquitoes and 79–98% more adult offspring. For all mosquitoes across all gonotrophic cycles, mortality was greatest during embryogenesis. This was particularly pronounced for the EGFP strain during the second gonotrophic cycle, where zero eggs reached the pupal stage (Table 1, cycle 3). Percentage egg-to-adult survivorship was <10% for EGFP and pBacMOS across all gonotrophic cycles. For all experimental lines, egg-to-adult survivorship rates generally declined across successive gonotrophic cycles.
Table 1. Stage-specific survivorship of Orlando strain and transgenic Ae. aegypti.
| Number entering stage
|
Number dying in stage
|
Proportion dying in stage
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cycle | Life stage | Orlando | EGFP | autoHermes | pBacMOS | Orlando | EGFP | autoHermes | pBacMOS | Orlando | EGFP | autoHermes | pBacMOS |
| 1 | Egg | 3,698 | 912 | 1,796 | 1,665 | 806 | 774 | 1498 | 1581 | 0.22 | 0.85 | 0.83 | 0.95 |
| Larvae | 2,892 | 138 | 298 | 84 | 149 | 57 | 73 | 47 | 0.05 | 0.41 | 0.24 | 0.56 | |
| Pupae | 2,743 | 81 | 225 | 37 | 382 | 9 | 24 | 3 | 0.14 | 0.11 | 0.11 | 0.08 | |
| Adult | 2,361 | 72 | 201 | 34 | |||||||||
| % survivorship | 63.85 | 7.89 | 11.19 | 2.04 | |||||||||
| 2 | Egg | 2,082 | 618 | 1,230 | 1,015 | 721 | 610 | 793 | 992 | 0.35 | 0.99 | 0.64 | 0.98 |
| Larvae | 1,361 | 8 | 437 | 23 | 109 | 2 | 147 | 5 | 0.08 | 0.25 | 0.34 | 0.22 | |
| Pupae | 1,252 | 6 | 290 | 18 | 413 | 1 | 12 | 1 | 0.33 | 0.17 | 0.04 | 0.06 | |
| Adult | 839 | 5 | 278 | 17 | |||||||||
| % survivorship | 40.30 | 0.81 | 22.60 | 1.67 | |||||||||
| 3 | Egg | 2,341 | 577 | 1,393 | 1,405 | 1633 | 576 | 985 | 1,321 | 0.70 | 1.00 | 0.71 | 0.94 |
| Larvae | 708 | 1 | 408 | 84 | 84 | 1 | 91 | 18 | 0.12 | 1.00 | 0.22 | 0.21 | |
| Pupae | 624 | 0 | 317 | 66 | 54 | 0 | 55 | 2 | 0.09 | 0.00 | 0.17 | 0.03 | |
| Adult | 570 | 0 | 262 | 64 | |||||||||
| % survivorship | 24.35 | 0.00 | 18.81 | 4.56 | |||||||||
| 4 | Egg | 1,411 | 286 | 1,023 | 735 | 1106 | 278 | 921 | 725 | 0.78 | 0.97 | 0.90 | 0.99 |
| Larvae | 305 | 8 | 102 | 10 | 48 | 5 | 4 | 4 | 0.16 | 0.63 | 0.04 | 0.40 | |
| Pupae | 257 | 3 | 98 | 6 | 19 | 0 | 8 | 1 | 0.07 | 0.00 | 0.08 | 0.17 | |
| Adult | 238 | 3 | 90 | 5 | |||||||||
| % survivorship | 16.87 | 1.05 | 8.80 | 0.68 | |||||||||
Partial life tables for offspring produced by the Orlando strain and transgenic Ae. aegypti females resulting from consecutive gonotrophic cycles 1, 2, 3, and 4.
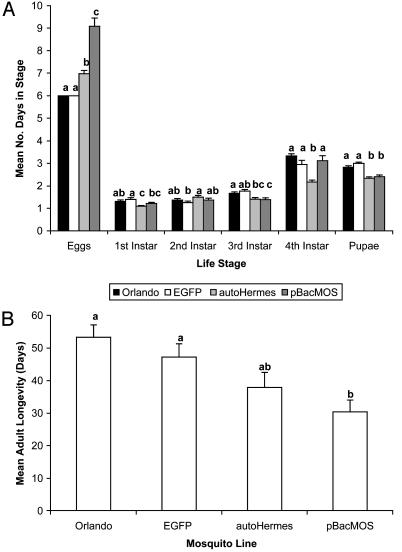
For F0 females, significant differences in mean numbers of days for eggs to hatch were observed across lines (F = 46.09, df = 3,164; P < 0.005), and autoHermes and pBacMOS eggs took significantly longer to hatch after oviposition (Fig. 1A). Significant differences were recorded in the developmental times of first (F = 7.51; df = 3, 191; P < 0.005); second (F = 2.67; df = 3. 191; P < 0.05); third (F = 6.32; df = 3, 191; P < 0.005); and fourth instar (F = 14.21, df = 3, 191; P < 0.005) larvae and pupae (F = 23.20; df = 3, 191; P < 0.005) (Fig. 1 A). There were no consistent trends across lines for preimaginal developmental times. Significant differences in adult longevity (F = 4.98; df = 3, 103; P < 0.005) were observed (Fig. 1B). The Orlando strain and EGFP mosquitoes lived significantly longer than pBacMOS adults.
Fig. 1.
(A) Mean developmental times in days for preimaginal stages of F0 female wild-type, EFGP, autoHermes, and pBacMOS Ae. aegypti. (B) Mean daily longevity of adult Orlando strain females, EFGP, autoHermes, and pBacMOS Ae. aegypti. Means followed by the same letters are not significantly different at the 0.05 level.
Preoviposition Periods, Lifetime Fecundity, Offspring Sex Ratio, and Sterility. For F1 females, significant differences across experimental lines were observed for preoviposition periods during the first (F = 49.23; df = 3,95; P < 0.005) (Table 2, cycle 1) and third (F = 6.16; df = 3,79; P < 0.005 (Table 2, cycle 3) gonotrophic cycles. Orlando strain eggs, on average, had longer preoviposition periods in comparison to transgenic mosquito eggs during gonotrophic cycles one and three. No significant differences were observed during gonotrophic cycles two (Table 2, cycle 2) and four (Table 2, cycle 4). Significant differences in mean fecundity estimates were observed during gonotrophic cycles one (F = 16.92; df = 3, 173; P < 0.005), two (F = 3.68; df = 3, 124; P < 0.05), three (F = 5.40; df = 3, 117; P < 0.05), and four (F = 3.41; df = 3, 102; P < 0.005) (Table 2). Orlando strain females laid significantly more eggs than transgenic mosquitoes and, of the transgenic mosquitoes, EGFP females consistently laid the fewest eggs, whereas autoHermes females exhibited the highest fecundity levels during each gonotrophic cycle.
Table 2. Reproductive statistics for Orlando strain and transgenic Ae. aegypti.
| Cycle | Parameter | Orlando | EGFP | autoHermes | pBacMOS |
|---|---|---|---|---|---|
| 1 | Preoviposition | 2.79 ± 0.07a | 1.95 ± 0.05b | 2.05 ± 0.05b | 1.36 ± 0.14c |
| Fecundity | 89.74 ± 7.57a | 22.86 ± 4.81b | 57.94 ± 8.26c | 46.53 ± 6.83bc | |
| Sex ratio | 39.05 | 50 | 41.79 | 61.76 | |
| % sterile females | 13.2 | 47.22 | 32.26 | 30.56 | |
| 2 | Preoviposition | 2.26 ± 0.11a | 2.06 ± 0.34a | 2.25 ± 0.23a | 2.44 ± 0.26a |
| Fecundity | 51.64 ± 7.66a | 21.02 ± 4.75b | 41.21 ± 7.61ab | 30.76 ± 7.04ab | |
| Sex ratio | 47.44 | 80 | 40.29 | 52.94 | |
| % sterile females | 33.33 | 46.67 | 31.03 | 45.55 | |
| 3 | Preoviposition | 2.23 ± 0.14ab | 2.67 ± 0.40a | 1.78 ± 0.15b | 2.92 ± 0.15a |
| Fecundity | 60.37 ± 8.05a | 19.44 ± 4.99b | 50.21 ± 8.68a | 41.87 ± 6.77ab | |
| Sex ratio | 38.25 | 0 | 43.13 | 51.56 | |
| % sterile females | 25.71 | 40.74 | 32.14 | 22.58 | |
| 4 | Preoviposition | 3.58 ± 0.75a | 1.73 ± 0.47a | 2.71 ± 0.50a | 2.47 ± 0.30a |
| Fecundity | 46.36 ± 8.80a | 13.57 ± 4.58b | 35.65 ± 7.84ab | 27.90 ± 6.50ab | |
| Sex ratio | 31.09 | 33.33 | 26.73 | 40 | |
| % sterile females | 28.57 | 52.17 | 46.15 | 48.28 |
Mean preoviposition period in days (±SE), mean lifetime fecundity (±SE), sex ratio of offspring (i.e., percentage female progeny produced), and percentage of F1 females that were sterile (i.e., either laid eggs that failed to hatch or did not lay eggs) for wild-type and transgenic Ae. aegypti for consecutive gonotrophic cycles 1, 2, 3, and 4. Means followed by the same letters within rows are not significantly different at the 0.05 level (ANOVA) of significance.
Sex ratio (i.e., percentage of females) of adult progeny produced during gonotrophic cycles was <50% for the Orlando strain and autoHermes, the two lines that produced the highest number of eggs across all egg laying events (Table 2). Sex ratio estimates for EGFP and pBacMOS were higher than autoHermes and the Orlando strain. However, these estimates are somewhat biased, because these strains produced substantially fewer progeny and most were females (e.g., from gonotrophic cycle two, EGFP females produced five adult offspring, of which four were female (Table 1, cycle 1), and this translated to 80% female offspring (Table 2, cycle 2).
Percentage sterility estimates varied across gonotrophic cycles (Table 2). A greater proportion of Orlando strain females produced viable eggs, especially in gonotrophic cycle one (Table 2, cycle 1). Sterility rates for Orlando strain females approximately doubled for gonotrophic cycles two through four. Of the transgenic mosquitoes, EGFP females exhibited the highest sterility rates across all four gonotrophic cycles (Table 2).
Demographic Growth Parameters. Significant differences were detected among the demographic parameter estimates generated from jackknifed lxmx data for each experimental mosquito line (Table 3). Mean net reproductive rate (Ro) (F = 42412; df = 3, 210; P < 0.005), and intrinsic rate of increase (rm) (F = 15616; df = 3, 210; P < 0.005) were highest for the Orlando strain and lowest for EGFP mosquitoes (Table 3). Mean generation times (Tc) were significantly shorter (F = 2840; df = 3, 210; P < 0.005) for EGFP and longest for Orlando strain mosquitoes. Mean estimates for population doubling times in days (Td) were significantly lower for the Orlando strain and highest for EGFP mosquito lines (F = 10912; df = 3, 210; P < 0.005) (Table 3).
Table 3. Demographic growth parameters for Orlando strain and transgenic Ae. aegypti.
| Parameter | Orlando | EGFP | autoHermes | pBacMOS |
|---|---|---|---|---|
| Ro | 87.23 ± 0.17a | 25.26 ± 0.09b | 47.72 ± 0.19c | 31.65 ± 0.07d |
| Tc | 41.26 ± 0.02a | 39.70 ± 0.02b | 40.23 ± 0.01c | 41.16 ± 0.008d |
| rm | 0.12 ± 0.00006a | 0.09 ± 0.0001b | 0.10 ± 0.0001c | 0.09 ± 0.0001d |
| Td | 6.01 ± 0.00a | 8.03 ± 0.009b | 6.73 ± 0.006c | 7.64 ± 0.008d |
Mean demographic growth parameters (±SE) generated from jackknifed lxmx data for cohorts of Orlando strain females and transgenic Ae. aegypti with fecundity data from four consecutive gonotrophic cycles. Means followed by the same letters within rows are significantly different at the 0.05 level (ANOVA). Ro, net reproductive rate; Tc, generation time in days; rm, intrinsic rate of increase per day; Td, doubling time in days.
Discussion
In this study, we were interested in determining the fitness costs of transgenesis on mosquitoes containing either the Hermes or MOS1 transposable elements. Although an earlier study measured competitiveness in mixed populations of wild-type insects in the laboratory (8), the present study examined and compared life-table characteristics of transgenic and nontransgenic mosquitoes. Three factors determining the spread of a genetic construct by a transposable element through a population have been proposed (18). These are: (i) the rate of transposition of the element used to spread the construct; (ii) the fitness cost to the host of being a transgenic organism; and (iii) the level of gene flow within the target population. The behavior of the transposable element, for example, whether it has a propensity to transpose close to its original site of insertion or to distant locations on the same chromosome or to other chromosomes, would also be a significant factor. Four Class II transposable elements have been used to generate transgenic mosquitoes, and little information exists as to their rates of transposition in mosquitoes. The transposition rate of an autonomous Hermes element in Drosophila has been estimated to be 2.4% (19), whereas the rate of MOS1-mediated germ-line excision of mariner elements in this species ranges from 0.05% to 14% (20). Transposition rates for autonomous piggyBac and Minos elements in both Drosophila and mosquitoes remain to be determined. The transposition rates of Hermes, MOS1, and piggyBac in the germ line of Ae. aegypti appear to be low based on experiments designed to measure remobilization rates of integrated gene vectors (21). The population structure of target species mosquitoes such as An. gambiae is complex. Restrictions to gene flow have led to the existence of several chromosomal forms with the An. gambiae complex. However, gene flow for An. gambiae within a chromosomal form can be high (22).
In this study, the fitness costs to all three transgenic lines were severe when compared to the Orlando strain for all measured demographic parameters. Across all gonotrophic cycles, each transgenic strain exhibited significantly reduced survivorship for all life stages, and mortality was greatest for the transition from egg to larval stage. Interestingly, even though postoviposition embryonic developmental times were longest for the autoHermes and pBacMOS strains, the preoviposition times were significantly shorter for both strains compared to the Orlando strain. The EGFP strain had shorter preoviposition times than all strains during gonotrophic cycles two and four, but embryonic developmental times were not significantly different from the Orlando strain. Adult longevity was also lowest for the autoHermes and pBacMOS strains.
Fecundity was significantly reduced in all transgenic strains relative to the Orlando strain but was most impaired for the EGFP strain. Thus even though this strain performed better than the autoHermes and pBacMOS strains with respect to embryonic development times and adult longevity, EGFP females were less fecund. There is not a uniformly negative effect on all fitness parameters across these three transgenic strains, and a strain that performs poorly in one measure of fitness may outperform other strains when other fitness characters are measured. Additionally, negative performance in one aspect can be correlated with another. For example, the EGFP strain exhibited a sharp reduction in fecundity that was matched with an increase in the percentage of sterile females relative to the other two transgenic strains and the Orlando strain.
The collective outcome of the impact of transgenesis on each of these fitness measurements is seen when demographic parameters of each of the strains are assessed. All three transgenic strains exhibited severe reductions in the their net reproductive rates, and both generation times and intrinsic rate of increase in population size are significantly different for all strains. Net reproductive rates (Ro) of the Orlando strain were 47–72% higher than the transgenic lines. Intrinsic rates of increase (rm) were 18–36% higher for the Orlando strain than the transgenic lines. Finally, the Orlando strain displayed a significantly shorter doubling time, 16–53%, than any of the three transgenic strains, with the autoHermes strain being the most underperforming strain.
These demographic data indicate that these transgenic strains of Ae. aegypti are not competitive with the nontransformed Orlando strain, and that there are serious fitness costs associated with being transgenic. Consequently, nontransformed mosquitoes would be predicted to rapidly outcompete all of the transgenic lines we have tested in this study. It will be of interest to determine whether the fitness costs associated with transgenesis reported here will be a general phenomenon seen in other transgenic strains of Ae. aegypti.
It must be emphasized that none of the three transgenic strains examined here were developed for use outside of the laboratory. Consequently, they do not contain all of the necessary components for enhanced field performance. For example, a proven gene-driving mechanism and an effector gene, thought to be necessary for a competitive field strain, were not incorporated into the transgenic lines examined here. The mosquitoes we experimented with were chosen because they exhibit some attributes of what a release strain might possess, and because these strains were amenable to laboratory rearing. It is highly likely that a released transgenic mosquito strain will be genetically marked so that it can be readily identified in the field. Fluorescent protein genes inserted into the mosquito genome might be used in this role. All three transgenic strains examined here express the EGFP gene; two (EGFP and autoHermes) have this gene placed under the actin5C promoter control and so express this gene in all mitotically active tissues throughout development The third strain (pBacMOS) has expression of this gene localized to the anal papillae of larvae and the adult brain. One strain (autoHermes) contains an autonomous Hermes element, and autonomous transposable elements may be one mechanism by which beneficial transgenes are mobilized through mosquito populations. Autonomous Hermes elements have been shown to transpose via cut-and-paste transposition in the somatic nuclei of a transgenic strain of Ae. aegypti, but as yet no evidence of germ-line mobility of these elements in these strains has been found (21).
The results reported here show how three transgenic lines of Ae. aegypti compare to nontransformed mosquitoes with respect to demographic parameters. These data do not, however, permit the specific source of fitness costs to be identified. There are three major sources of fitness costs associated with the possession of a transgene. First, insertional mutagenesis resulting from the integration of the transgene into the host's genome could result in the partial or complete disruption of gene function. Second, the expression of the transgene may be detrimental to the organism. Finally, the process of creating transgenic insects involves at least two generations of brother/sister matings and results in significant inbreeding depression. The three transgenic strains used in this study were created independently and are unlikely to have the transgenes inserted into precisely the same region of the Aedes genome. Both Hermes insertions into the genome involve the transposable element and the flanking plasmid DNA integrating into the genome (9, 21), whereas insertion of pBacMOS was more precise and involved only the piggyBac element (10). The autoHermes element has been shown to be mobile in somatic nuclei in our experimental Ae. aegypti strain during subsequent generations (21), and the insertion of this element by cut-and-paste transposition into multiple somatic nuclei during development might be predicted to reduce the viability and fitness of these mosquitoes, because inevitably genes critical for development and metabolism will be affected by autoHermes insertion either in or near them. Fitness issues related to insertion placement need further investigation.
In our studies, we have placed the EGFP gene under the control of two different promoters, the actin5C promoter of D. melanogaster and the 3xP3 synthetic promoter that directs expression to the adult brain and the larval anal papalliae. If the presence of EGFP in the cell is a strong contributor to observed declines in fitness, it might be expected that strains expressing EGFP in all mitotically active tissues throughout development might be more adversely affected than strains in which expression is confined to a few tissues during specific life stages. This was not observed, because neither the EGFP nor autoHermes strains was consistently less fit for any of the parameters measured when compared to the pBacMOS strain. We believe it unlikely that decreased fitness observed for the pBacMOS element is due to mobilization of endogenous mariner elements by the Mos transposase located in this piggyBac element. In Ae. aegypti, the Mos transposase located on this element cross-mobilizes D. melanogaster mariner elements at a very low frequency, 0.007% (10). Although we cannot definitively rule out crossmobilization of Aedes mariner elements, no crossmobilization of mariner elements by heterologous transposases has been reported and, if it were the case here, it might be predicted to be at an even lower frequency than reported for D. melanogaster mariner mobilization in Ae. aegypti (10).
Inbreeding depression can also reduce mosquito fitness (23). This phenomenon has not been studied in transgenic mosquitoes, which undergo severe inbreeding during the first few generations after microinjection as laboratory colonies are established. As for other insect species that are genetically transformed using transposable elements, each transgenic line arises from a single fertilized zygote containing transgenic gametes. The transgenic strains used in this study all arose from the Orlando strain of Ae. aegypti that had been maintained in our laboratories for many years without any invigoration from field material. Outcrosses of the transgenic strains were always made to the Orlando strain, and so at no stage was any heterozygosity at new loci introduced to the transgenic strains. The degree to which inbreeding depression can reduce the fitness of transgenic mosquitoes is currently not known but needs to be evaluated in future studies. Additionally, we did not measure the mating competitiveness of transgenic males or the efficiency with which larvae acquire food resources, both of which could conceivably affect fitness estimates under competitive conditions.
Our results show that transgenesis in Ae. aegypti can come at a substantial cost to fitness. An argument for using genetic transformation in insects, and in mosquitoes in particular, is that the changes made are small and very specific, unlike previous genetic manipulation technologies in insects that relied on chromosomal translocations with or without chemical-induced mutagenesis, with consequent dramatic declines in fitness and mating competitiveness. Genetic transformation by transposable elements remains, in comparison, a very precise technology; however, we suggest that it cannot be assumed that even the simplest insertions into the mosquito genome will have no cost to fitness. Our results support the previous findings that transgenic strains of An. stephensi containing Minos-based transposable elements cannot outcompete wild-type strains (8). Our results are also consistent with previous studies on the impact of P element insertion on the fitness of D. melanogaster. Somatic movement of P has been shown to reduce the life span, fitness, mating ability, and locomotion of D. melanogaster (6, 24), whereas other studies have indicated that remobilization of P elements in this species is mainly responsible for the decline in fitness and viability seen in progeny resulting from dysgenic matings (25).
Although it is certainly not necessary to subject all transgenic lines of mosquitoes to the analyses described here, it is important to apply these types of tests to strains that are proposed for release programs. It is also important to determine the molecular and genetic basis of the decrease in fitness so that these effects can be reduced or eliminated. These types of studies will enhance the ability to develop gene vectors that will have minimal effects on fitness and so make transgenic mosquitoes for control programs more rapid, facilitating greater certainty for success. There may always be some cost to fitness when using transgenic strains. If so, the fitness costs of transgenesis need to be established so the magnitude of a desired gene-driving system needed to overcome this can be determined. These opposing forces need to be characterized and quantified if genetic control programs using transgenic mosquitoes are to be feasible.
Acknowledgments
This work was supported in part by grants from the University of California Mosquito Research Program (Award No. 00-011-1-1 to M.S.H.) and the National Institutes of Health (Award AI45741 to P.W.A.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: EGFP, enhanced GFP.
References
- 1.Adelman, Z. N., Jasinskiene, N. & James, A. A. (2002) Mol. Biochem. Parasitol. 121, 1–10. [DOI] [PubMed] [Google Scholar]
- 2.Collins, F. H., Kamau, L., Ranson, H. A. & Vulule, J. M. (2000) Bull. World Health Org. 78, 1412–1423. [PMC free article] [PubMed] [Google Scholar]
- 3.Asman, S. M., Nelson, R. L., MacDonald, P. T., Milby, M. M., Reeves, W. C., White, K. D. & Fine, P. E. M. (1979) Mosquito News 39, 248–258. [Google Scholar]
- 4.Terwedow, H. A., Asman, S. M., MacDonald, P. T., Nelson, R. L. & Reeves, W. C. (1977) Ann. Entomol. Soc. Am. 70, 849–854. [Google Scholar]
- 5.Tiedje, J. M., Colwell, R. K., Grossmna, Y. L., Hodson, R. E., Lenski, R. E., Mack, R. N. & Regal, P. J. (1989) Ecology 70, 298–315. [Google Scholar]
- 6.McKay, T. F. C. (1989) Genome 31, 284–295. [DOI] [PubMed] [Google Scholar]
- 7.Woodruff, R. C. (1992) Genetica 86, 143–154. [DOI] [PubMed] [Google Scholar]
- 8.Catterucia, F., Godfray, H. C. J. & Crisanti, A. (2003) Science 299, 1225–1227. [DOI] [PubMed] [Google Scholar]
- 9.Pinkerton, A. C., Michel, K., O'Brochta, D. A. & Atkinson, P. W. (2000) Insect Mol. Biol. 9, 1–10. [DOI] [PubMed] [Google Scholar]
- 10.Wilson, R., Orsetti, J., Klocko, A. D., Aluvihare, C., Peckham, E., Atkinson, P. W., Lehane, M. J. & O'Brochta, D. A. (2003). Insect Biochem. Mol. Biol. 33, 853–863. [DOI] [PubMed] [Google Scholar]
- 11.Berghammer, A. J., Klingler, M. & Wimmer, E. A. (1999) Nature 402, 370–371. [DOI] [PubMed] [Google Scholar]
- 12.Carey, J. R. (1993) Applied Demography for Biologists with Special Emphasis on Insects (Oxford Univ. Press, New York).
- 13.Birch, L. C. (1948) J. Anim. Ecol. 17, 15–26. [Google Scholar]
- 14.Miller, R. G. (1974) Biometrika 61, 1–16. [Google Scholar]
- 15.Efron, B. (1981) Biometrika 68, 589–599. [Google Scholar]
- 16.Meyer, J. S., Ingersoll, C. G., McDonald, L. L. & Boyce, M. S. (1986) Ecology 67, 1156–1166. [Google Scholar]
- 17.Shao, J. & Tu, D. (1995) The Jackknife and Bootstrap (Springer, New York).
- 18.Braig, H. R. & Yan, G. (2002) in Genetically Engineered Organisms, eds. Letourneau, D. K. & Burrows, B. E. (CRC, Boca Raton, FL), pp. 251–314.
- 19.Giumond, N., Bideshi, D. K., Pinkerton, A. C., Atkinson, P. W. & O'Brochta, D. A. (2003) Mol. Genet. Genom. 268, 779–790. [DOI] [PubMed] [Google Scholar]
- 20.Lozovsky, E. R., Nurminsky, D., Wimmer, E. A. & Hartl, D. L. (2002) Genetics 160, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brochta, D. A., Sethuraman, N., Wilson, R., Hice, R. H., Pinkerton, A. C., Leveseque, C. S., Bideshi, D. K., Jasinskiene, N., Coates, C. J., James, A. A., et al. (2003) J. Exp. Biol. 206, 3823–3834. [DOI] [PubMed] [Google Scholar]
- 22.Taylor, C., Toure, Y. T., Carnahan, J., Norris, D. E., Dolo, G., Traore, S. F., Edillo, F. E. & Lanzaro, G. C. (2001) Genetics 157, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armbruster, P., Hutchinson, R. A. & Linvell, T. (2000) Proc. R. Soc. London Ser. B 267, 1939–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodruff, R. C., Thompson, J. N., Jr., Barker J. S. & Huai, H. (1999) Genetica 107, 261–269. [PubMed] [Google Scholar]
- 25.Eanes, W. F., Wesley, C., J. Hey, J., Houle, D. & Ajioka, J. W. (1988) Genet. Res. Camb. 52, 17–26. [Google Scholar]