Abstract
Background
Several vector systems have been developed to express any gene desired to be studied in Bacillus subtilis. Among them, the transcriptionally regulated promoters involved in carbohydrate utilization are a research priority. Expression systems based on Bacillus promoters for xylose, maltose, and mannose utilization, as well as on the heterologous E. coli lactose promoter, have been successfully constructed. The promoter of the mtlAFD operon for utilization of mannitol is another promising candidate for its use in expression vectors. In this study, we investigated the regulation of the mtl genes in order to identify the elements needed to construct a strong mannitol inducible expression system in B. subtilis.
Results
Regulation of the promoters of mtlAFD operon (PmtlA) and mtlR (PmtlR) encoding the activator were investigated by fusion to lacZ. Identification of the PmtlA and PmtlR transcription start sites revealed the σA like promoter structures. Also, the operator of PmtlA was determined by shortening, nucleotide exchange, and alignment of PmtlA and PmtlR operator regions. Deletion of the mannitol-specific PTS genes (mtlAF) resulted in PmtlA constitutive expression demonstrating the inhibitory effect of EIICBMtl and EIIAMtl on MtlR in the absence of mannitol. Disruption of mtlD made the cells sensitive to mannitol and glucitol. Both PmtlA and PmtlR were influenced by carbon catabolite repression (CCR). However, a CcpA deficient mutant showed only a slight reduction in PmtlR catabolite repression. Similarly, using PgroE as a constitutive promoter, putative cre sites of PmtlA and PmtlR slightly reduced the promoter activity in the presence of glucose. In contrast, glucose repression of PmtlA and PmtlR was completely abolished in a ΔptsG mutant and significantly reduced in a MtlR (H342D) mutant.
Conclusions
The mtl operon promoter (PmtlA) is a strong promoter that reached a maximum of 13,000 Miller units with lacZ as a reporter on low copy plasmids. It is tightly regulated by just one copy of the mtlR gene on the chromosome and subject to CCR. CCR can be switched off by mutations in MtlR and the glucose transporter. These properties and the low costs of the inducers, i.e. mannitol and glucitol, make the promoter ideal for designing regulated expression systems.
Keywords: PTS, PRD, Activator, Catabolite repression, Mannitol operon
Background
For many years, Bacillus subtilis has been vastly investigated not only as the model of Gram-positive bacterium, but also as an expression platform for homologous and heterologous gene expression. Efficient secretion of enzymes such as proteases, lipases, and amylases along with its safe use and completely sequenced genome is promising for industrial applications. However, several bottlenecks exist that limit the expression potential, e.g. low level of heterologous protein production, instability of the plasmids, inclusion body formation, and production of extracellular proteases [1]. So far, several expression systems have been developed in order to overcome the low production categorized to inducer-specific promoters, growth-phase promoters and autoinducible promoters [2]. Among them, systems based on Pspac [3] and Pxyl [4] have been widely used. Recently, two other systems were developed based on PmanP [5] and Pglv [6] promoters.
B. subtilis utilizes many carbohydrates via the phosphoenolpyruvate-dependent phosphotransferase system (PTS). In a PTS pathway, the sugar is translocated across the cytoplasmic membrane and concomitantly phosphorylated by a sugar-specific permease, i.e. Enzyme II (EII). The phosphate is provided by phosphoenolpyruvate (PEP), which is synthesized during glycolysis. The phosphate transfer from PEP to sugar is catalyzed by PTS Enzyme I, phosphocarrier protein HPr, and EIICBA. The latter is responsible for the uptake of the sugars. In addition to uptake and phosphorylation, the PTS is involved in the regulation of catabolic operons. Hereby, the PTS ensures the hierarchical utilization of carbon sources, a phenomenon known as carbon catabolite repression (CCR) [7].
The CCR of sugar operons is dependent on phosphorylated intermediates of the PTS. Despite the fact that the phosphate transfer cascade is similar in both Gram-positive and Gram-negative bacteria, there are some outstanding differences in the regulation of CCR. While in Gram-negative bacteria, such as Escherichia coli, the phosphorylation status of the cytoplasmic EIIA domain for glucose (EIIAGlc) plays the main role, it is HPr in low G+C Gram-positive bacteria, such as B. subtilis, that is dominant. In B. subtilis, HPr shows two phosphorylation sites. Phosphorylation at His15 is needed for the transfer reaction to EIIA component, whereas the phosphorylation at Ser46 plays a regulatory role [8]. HPr (Ser46~P) (or Crh (Ser46), which shares 45% identity in amino acid sequence with HPr, but contains only the serine regulatory site) is able to interact with catabolite control protein A (CcpA) in order to form a complex. Afterwards, this complex binds to so-called cre (catabolite-responsive element) sites in the promoter region of the relevant genes and leads to positive or negative regulation of the genes depending on the location of cre (for review see [9-14]). In addition to CcpA-dependent carbon catabolite repression, there are also CcpA-independent systems in B. subtilis [15,16]. For instance, trehalose and sucrose are known to be regulated by the inducer exclusion mechanism. In these systems the transporter lacks the specific EIIA domains. Therefore, the phosphate is transferred from the EIIA domain of EIICBAGlc to the EIIB domains of the trehalose and sucrose transporters. Glucose competes with the phosphorylation of the EIIB domains and hereby reduces sucrose and trehalose transport [17,18]. A second, more important CcpA independent CCR occurs via PTS regulation domains (PRDs) in transcriptional antiterminators and activators. PRD containing antiterminators have two PRDs in addition to a RNA binding site. One PRD (PRDII) is phosphorylated by HPr (His15) and the other (PRDI) is phosphorylated by the specific PTS transport system, which is regulated by the antiterminator. For antiterminator activity, usually the PRDI has to be dephosphorylated and the PRDII phosphorylated, which is the case in the presence of the specific sugar and absence of glucose. PRD containing activators are similar, but contain a DNA binding site instead of the RNA binding site, and they have additional domains like the mannose activator, which contains an EIIA and EIIB domain like the PTS transporters [16,19-22].
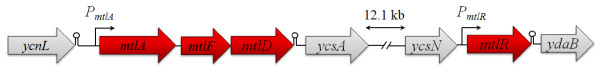
Mannitol consumption by B. subtilis is another example for regulation by PRD containing activators. The polyol is taken up by the mannitol system as one of the 17 PTS in B. subtilis, which were identified by genome sequencing and protein annotations. Four genes are needed: mtlA (encoding enzyme IICBMtl), mtlF (encoding enzyme IIAMtl), mtlD (encoding mannitol-1-phosphate dehydrogenase), and mtlR (encoding the transcriptional activator) (Figure 1). The first genes comprise the mtl operon, whereas mtlR is located at 14.4 kb downstream from the mtl operon [23-27]. Previously, it was shown that transcription of the mannitol operon is activated by MtlR [28]. The MtlR protein consists of a DNA binding domain and two PRD domains as well as EIIAMtl-like and EIIBGat-like domains. Recent studies of MtlR have indicated that phosphorylation of the PRDII domain of MtlR by HPr (His15), as well as the absence of phosphorylation of EIIAMtl-like and EIIBGat-like domains, respectively, have a stimulatory effect on the activity, whereas the dephosphorylation of PRDI reduced the MtlR activity 5 to 25% [29]. Besides mannitol, the transcription of the mtl operon is induced by glucitol, a non-PTS sugar. Also, mtlD is required for the assimilation of glucitol in B. subtilis [28]. Basically, glucitol is taken up by a H+-symporter (encoded by gutP) without any chemical modification and oxidized to fructose by glucitol dehydrogenase (gutB). In addition to the GutP transporter, it was observed that glucitol could be weakly taken up by EIICBAMtl and phosphorylated to glucitol 6-phosphate [30].
Figure 1.
Genetic map of mtl operon. Organization of the mannitol PTS encoding genes consisting of mtlAFD and its regulator mtlR in B. subtilis 168 is depicted.
In this study, we characterize the transcriptional activity of both PmtlA and PmtlR depending on the presence of mannitol and other sugars as well as the influence of various mutations on the regulation of the two mannitol promoters.
Methods
Strains, media and growth condition
Bacterial strains used in this study are listed in Table 1. Transformants of E. coli and B. subtilis were selected on LB agar [31], supplemented with ampicillin (100 μg ml-1) or spectinomycin (100 μg ml-1) depending on the plasmid antibiotic marker. Unless otherwise specified, knock-out B. subtilis mutants were selected on LB agar, containing erythromycin (5 μg ml-1) or chloramphenicol (5 μg ml-1). Mutations were confirmed by cultivating the mutants in modified Spizizen salts medium [32]. Trisodium citrate, glucose, and trace elements were replaced by 0.02% (w/v) casamino acids. For tryptophan auxotrophic B. subtilis 168 and its derivatives, 50 μg ml-1 tryptophan was added. The carbon source was either 1% (w/v) sterile-filtered mannitol, glucitol or glucose. All of the strains were incubated at 37°C under a shaking condition at 200 rpm.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant structure | Source, reference, or construction |
|---|---|---|
| E. coli | ||
| JM109 | recA1, endA1, gyrA96, thi, hsdR17, supE44, relA, λ-, Δ(lac-proAB), [F', traD36, proAB, lacIqZΔM15] | [69] |
| B. subtilis | ||
| 3NA | spo0A3 | [70] |
| 168 | trpC2 | Bacillus Genetic Stock Center |
| KM12 | spo0A3 ΔmtlAF::ermC | 3NA transformed with pKAM5 |
| KM13 | spo0A3 ΔmtlAFD::ermC | 3NA transformed with pKAM6 |
| KM15 | spo0A3 ΔmtlR::ermC | 3NA transformed with pKAM4 |
| KM37 | spo0A3 mtlD::ermC | 3NA transformed with pKAM14 |
| KM39 | spo0A3 ΔgutRBPydjE::cat | 3NA transformed with pKAM13 |
| KM40 | spo0A3 ΔmtlAFD::ermC ΔgutRBPydjE::cat | KM13 transformed with pKAM13 |
| KM162 | spo0A3 ΔmtlR::ermC, hisI', spc | KM15 transformed with pHM30 |
| KM163 | spo0A3 mtlR-H342D | KM162 transformed with pKAM66 |
| MW312 | spo0A3 (ptsG-pstH')::ermC | 3NA transformed with pMW312.2 |
| MW373 | spo0A3 ΔptsG | MW312 transformed with pMW373.3 |
| TQ303 | spo0A3 ΔccpA::ermC | [5] |
| TQ432 | trpC2 ptsH-H15A amyE::cat | [5] |
| Plasmids | ||
| pHM30 | hisF, hisI', spc, yvcA, yvcB, bla | [37] |
| pHM31 | hisF, hisI, yvcA, yvcB, bla | [37] |
| pKAM1 | spc, ter- PmtlA (-181/-1)α -lacZ-ter, repA | This study |
| pKAM4 | 'ycsN-ermC-ydaB', bla, spc | This study |
| pKAM5 | 'ycnL-ermC-PmtlA-mtlD', bla, spc | This study |
| pKAM6 | 'ycnL-ermC-ycsA', bla, spc | This study |
| pKAM9 | spc, ter- PmtlA (-161/-1) -lacZ-ter, repA | This study |
| pKAM12 | spc, ter- PmtlA' (-161/-32)-lacZ-ter, repA | This study |
| pKAM13 | 'ydjC-cat-pspA', bla, spc | This study |
| pKAM14 | 'mtlD::ermC, bla, spc | This study |
| pKAM18 | spc, ter-PmtlR-lacZ-ter, repA | This study |
| pKAM27 | spc, ter- PmtlA (-161/-32)*[cttta→gaaat] -lacZ-ter, repA | This study |
| pKAM43 | spc, ter- 'PmtlA (-157/-32) -lacZ-ter, repA | This study |
| pKAM44 | spc, ter- PmtlA' (-161/-52) -lacZ-ter, repA | This study |
| pKAM45 | spc, ter- PmtlA (-161/-32)*[ctttc→gaaag] -lacZ-ter, repA | This study |
| pKAM48 | spc, ter- 'PmtlA (-159/-32) -lacZ-ter, repA | This study |
| pKAM49 | spc, ter- 'PmtlA (-155/-32) -lacZ-ter, repA | This study |
| pKAM52 | spc, ter- PmtlA (-161/-32)*[ttcaa→aagtt] -lacZ-ter, repA | This study |
| pKAM57 | spc, ter- 'PmtlA (-153/-32) -lacZ-ter, repA | This study |
| pKAM58 | spc, ter- 'PmtlA (-149/-32) -lacZ-ter, repA | This study |
| pKAM59 | spc, ter- 'PmtlA (-151/-32) -lacZ-ter, repA | This study |
| pKAM66 | hisF, hisI, mtlR-H342-D, yvcA, yvcB, bla | This study |
| pKAM84 | spc, ter- PmtlA (-161/-32)*[gtcct→cagga] -lacZ-ter, repA | This study |
| pKAM88 | spc, ter- PgroE - (crePmtlA) -UTRPmtlR-lacZ-ter, repA | This study |
| pKAM89 | spc, ter- PgroE - (crePmtlR) -UTRPmtlR-lacZ-ter, repA | This study |
| pKAM90 | spc, ter- PgroE -(creacsA) -UTRPmtlR-lacZ-ter, repA | This study |
| pKAM91 | spc, ter- PgroE - (cremtlA) -UTRPmtlR-lacZ-ter, repA | This study |
| pKAM92 | spc, ter- PmtlA (-161/-32)*[ccaaa→ggttt] -lacZ-ter, repA | This study |
| pKAM101 | spc, ter- PgroE -UTRPmtlR-lacZ-ter, repA | This study |
| pMW312.2 | glcT'-ermC-ptsHI', bla, spc | This study |
| pMW363.1 | ter, spc, manP'-cat-yjdB-yjdA', bla | Laboratory Stock |
| pMW373.3 | 'glcT-PptsGHI-ptsH', bla, spc | This study |
| pSUN279.2 | ter-PmanR-manR-PmanP-lacZ-ter, repA, oripUC18, oripBS72, bla, spc | [5] |
| pSUN338.3 | yvcL-ermC-yvcN, bla, spc | [5] |
| pSUN356.7 | manR'-ermC-manA', bla, spc | [5] |
α The numbers are showing the position of the sequence with respect to mtlA start codon.
For induction of the mtl promoters, 85 ml LB medium in 500 ml Erlenmeyer flasks were inoculated by an overnight culture in a dilution of 1:50 and incubated at 37°C under shaking condition (200 rpm). Aliquots of 8 ml with an OD600 of 0.4 were divided in a 100 ml Erlenmeyer flask and subsequently induced by addition of different carbohydrates to a final concentration of 0.2% (w/v) each, i.e. mannitol, mannitol + glucose, glucose, mannitol + xylose, xylose, glucitol, glucitol + glucose. Cultures were harvested 1 h after addition of sugars and used for enzyme activity studies. As a defined medium, Spizizen salts medium (SSM) (14 g of K2HPO4, 6 g of KH2PO4, 2 g of (NH4)2SO4, 1 g of trisodium citrate, 0.2 g of MgSO4.7H2O, and 10 g of glycerol per liter) supplemented by trace elements (1.5 mg of CaCl2.2H2O, 50.1 mg of FeCl3.6H2O, 60.3 mg of Na2-EDTA, 0.54 mg of ZnSO4.7H2O, 0.3 mg of MnSO4.H2O, 0.48 mg of CuSO4.5H2O, 0.54 mg of CoCl2.6H2O per liter) was also applied in induction studies with the same procedure. All of the experiments were repeated at least 3 times and mean values were used for comparison.
Materials
D-mannitol was purchased from Serva Electrophoresis GmbH (Article No. 28410; Heidelberg, Germany) and D-glucitol (D-sorbitol; Cat. No. 85532) obtained from Sigma-Aldrich Chemie GmbH (Taufkirchen, Germany). Other chemicals were purchased from Merck (Darmstadt, Germany), Sigma-Aldrich (Taufkirchen, Germany) or Fluka (Buchs, Germany). DNA modifying enzymes were obtained from Roche Applied Science (Mannheim, Germany) or New England Biolabs GmbH (Frankfurt, Germany). Oligonucleotides were synthesized by Eurofins MWG Operons (Ebersberg, Gemany).
DNA manipulation and transformation
Standard molecular techniques including E. coli transformation were carried out according to Sambrook et al. [33]. B. subtilis was naturally transformed using "Paris Method" [32,34]. PCRs were performed using Pfu DNA polymerase from Promega GmbH (Mannheim, Germany) on a MiniCycler from Biozym and the DNA constructs were sequenced by GATC Biotech (Konstanz, Germany). DNA preparation kits from Qiagen (Hilden, Germany) were applied for chromosomal DNA or plasmid extraction according to the manufactures' instruction. DNA sequencing reagents (AutoRead sequencing kit) were obtained from GE Healthcare (Munich, Germany).
Construction of expression vectors
The oligonucleotides used in this study are listed in Table 2. Chromosomal DNA of B. subtilis 168 was applied as the template for PCR reactions. The promoter region of the mannitol operon (PmtlA) was amplified using s5526/s5527 (plasmid pKAM1). Shortening of the 5'-end of the PmtlA region was performed by s6209/s5527 (pKAM9). For construction of pKAM12, oligonucleotides s6209/s6213 were applied to remove the wild type Shine-Dalgarno of PmtlA. Additionally, the untranslated region (UTR) of PmtlA in pKAM12 was further shortened by s6209/s6727 (pKAM44). Moreover, gradual shortening of PmtlA 5' was performed by s6792/s6213 (pKAM48), s6726/s6213 (pKAM43), s6793/s6213 (pKAM49), s6829/s6213 (pKAM57), s6210/s6213 (pKAM59), and s6830/s6213 (pKAM58). Mutation of the base pairs between -35 box and MtlR binding site of PmtlA was carried out by s7091/s6213 (pKAM92), s7065/s6213 (pKAM84), and s6688/s6213 (pKAM27) oligonucleotides. Construction of pKAM45 was performed by fusion PCR. Primary PCRs s6209/s6728 and s6729/s6213 were followed by final PCR s6209/s6213 using primary PCR products as a template. Plasmid pKAM52 was also constructed in the same way using s6209/s6800 and s6801/s6213 PCR products as a template in the final PCR by s6209/s6213 oligonucleotides. Amplification of the promoter region of mtlR (PmtlR) was carried out using s5799/s6392 (pKAM18). All of the amplified promoter fragments were double digested by NheI and AflII in order to be fused to lacZ as the reporter gene in pSUN279.2 [5], a derivative of pMTLBS72 [35]. Plasmid pMTLBS72 is a B.subtilis/E.coli shuttle vector containing a pBR322 origin of replication for E. coli and pBS72 origin of replication for B. subtilis. Replicon pBS72, which has been isolated from B. subtilis, is a stable theta replicating plasmid with low copy number in B. subtilis (6 copies per chromosome) [36].
Table 2.
Oligonucleotides used in this study
| Name | Sequence (5'→3') | Purpose |
|---|---|---|
| Expression vectors | ||
| s5526 | AAAAAAGCTAGCGGCTCCTGAAACCAGGAG | Amplification of PmtlA |
| s5527 | AAAAACTTAAGATATAAACCCTCCCTGTTTTG | Amplification of PmtlA |
| s5799 | AAGCTAGCTACGATATTCCATAAAAAGC | Amplification of PmtlR |
| s6209 | AAAAAGCTAGCTTTTTATTTTTAAAAAATTGTCACAGTCA | Shortening upstream of PmtlA |
| s6210 | AAAAAAGCTAGCTAAAAAATTGTCACAGTCATGTGC | Shortening upstream of PmtlA |
| s6213 | AAAAAACTTAAGTAAGATACAAAAATATGTTCAGAGA | Shortening downstream of PmtlA |
| s6392 | AAAAAACTTAAGAGCCAATCTTGATGTGCGG | Amplification of PmtlR |
| s6688 | AAAAAAGCTAGCTTTTTATTTTTAAAAAATTGTCACAGTCATGTGCCAAAGTCCTGAAATCTTTCAA TTGTATAGGGACTG | Mutation between -35 and MtlR binding site |
| s6726 | AAAAAAGCTAGCTATTTTTAAAAAATTGTCACAGT | Shortening upstream of PmtlA |
| s6727 | AAAAAACTTAAGAGAGAATGATGCTTCCCTTTG | Shortening downstream of PmtlA |
| s6728 | ATACAATTCTTTCTAAAGAGGACTTTGGCACATG | Mutation between -35 and MtlR binding site |
| s6729 | CCTCTTTAGAAAGAATTGTATAGGGACTGTAAGCGT | Mutation between -35 and MtlR binding site |
| s6792 | AAAAAAGCTAGCTTTATTTTTAAAAAATTGTCACA | Shortening upstream of PmtlA |
| s6793 | AAAAAAGCTAGCTTTTTAAAAAATTGTCACAGTC | Shortening upstream of PmtlA |
| s6800 | CGCTTACAGTCCCTATACAAAACTTAGTAAAGAGGACTTTGGCAC | Mutation between -35 and MtlR binding site |
| s6801 | CCAAAGTCCTCTTTACTAAGTTTTGTATAGGGACTGTAAGCG | Mutation between -35 and MtlR binding site |
| s6829 | AAAAAAGCTAGCTTTAAAAAATTGTCACAGTCAT | Shortening upstream of PmtlA |
| s6830 | AAAAAAGCTAGCAAAAATTGTCACAGTCATGTG | Shortening upstream of PmtlA |
| s7065 | AAAAAAGCTAGCTTTTTATTTTTAAAAAATTGTCACAGTCATGTGCCAAACAGGACTTTACTTTCAA TTGTATAGGG | Mutation between -35 and MtlR binding site |
| s7091 | AAAAAAGCTAGCTTTTTATTTTTAAAAAATTGTCACAGTCATGTGGGTTTGTCCTCTTTACTTTCAA TTGTATA | Mutation between -35 and MtlR binding site |
| s7098 | AAAAAAGCTAGCAGCTATTGTAACATAATCGGT | Fusion of PgroE-(cre)-UTRPmtlR |
| s7189 | CCTTAAAACGCTTACAGCAATTCTTATAATAAAGAATCTCC | Fusion of PgroE-(crePmtlA)-UTRPmtlR |
| s7190 | TGCTGTAAGCGTTTTAAGGAAACCTCTCTATATCCTCTA | Fusion of PgroE-(crePmtlA )-UTRPmtlR |
| s7191 | CCATAAAACGCTTTCAACAATTCTTATAATAAAGAATCTCC | Fusion of PgroE-(crePmtlR)-UTRPmtlR |
| s7192 | TGTTGAAAGCGTTTTATGGAAACCTCTCTATATCCTCTA | Fusion of PgroE-(crePmtlR )-UTRPmtlR |
| s7193 | CCTGGTAACGCTTTCAACAATTCTTATAATAAAGAATCTCC | Fusion of PgroE-(creacsA)-UTRPmtlR |
| s7194 | TGTTGAAAGCGTTACCAGGAAACCTCTCTATATCCTCTA | Fusion of PgroE-(creacsA )-UTRPmtlR |
| s7195 | CCTGTTCACGCTTTCAGCAATTCTTATAATAAAGAATCTCC | Fusion of PgroE-(cremtlA)-UTRPmtlR |
| s7196 | TGCTGAAAGCGTGAACAGGAAACCTCTCTATATCCTCTA | Fusion of PgroE-(cremtlA )-UTRPmtlR |
| s7237 | GTAGAGGATATAGAGAGGTTTCCCAATTCTTATAATAAAGAATCTCC | Fusion of PgroE-UTRPmtlR |
| s7238 | GGAGATTCTTTATTATAAGAATTGGGAAACCTCTCTATATCCTCTAC | Fusion of PgroE-UTRPmtlR |
| Integration vector | ||
| s5069 | AAAAAAGAATTCGATATCAGATCTACGCGTTAACCCGGGC | Amplification of ermC |
| s5070 | AAAAAACAATTGAATCGATTCACAAAAAATAGG | Amplification of ermC |
| s5621 | AAAAAAGGCGCCTGGATTACCGTCTCATCG | Amplification of glcT |
| s5622 | AAAAAAGGATCCAACCGCTTCCGCCTCATGAA | Amplification of glcT |
| s5623 | GTGTTAGTACGCCGTGCTT | Amplification of ptsHI |
| s5624 | GTCGCAATCATAGGGAACAT | Amplification of ptsHI |
| s5809 | AAAAAAGATATCAACGCCCTTGCCCTTTC | Amplification of ycsN |
| s5810 | AAAAAAAGATCTGCATCAGCTGGTAAACTGAT | Amplification of ycsN |
| s5812 | AAAAAAAGGCCTAACACAAATGTTGTTTCTGC | Amplification of ydaB |
| s5860 | AAAAAAACTAGTACCTGCATGGCACACGT | Amplification of ydaB |
| s5866 | AAAAAAGGATCCATAAGAATTGACCTCCTCT | Amplification of glcT-PptsGHI |
| s5867 | AAAAAAGGATCCTAAGGGTGTTAGTACGCCGT | Amplification of ptsHI |
| s5918 | AAAGATCTAACCAGGAGCCTTTTTATTTT | Amplification of PmtlA |
| s5919 | CGAAATGTAAGGCGATCATATATAAACCCTCCCTGTT | Amplification of PmtlA |
| s5920 | AACAGGGAGGGTTTATATATGATCGCCTTACATTTCG | Amplification of mtlD |
| s5921 | AAGATATCGACCGTAAACAGCTTCCGTT | Amplification of mtlD |
| s5994 | AAACTAGTAAGAAACTTAATCAATAACCGAC | Amplification of ycsA |
| s5995 | AAAGGCCTTCTCGATTCCGCTATAATCAG | Amplification of ycsA |
| s6067 | CCTGAAAGAAACACCATGCCCGAAC | Amplification of ycnL |
| s6068 | AAGATATCGAAAGAAACACCATGCCCGAAC | Amplification of ycnL |
| s6079 | AAAAAAACTAGTCTTTGGCACATGACTGTGACA | Amplification of ycnL |
| s6080 | AAAGATCTCTTTGGCACATGACTGTGACA | Amplification of ycnL |
| s6302 | AAAAAAGAATTCGGTATCTATCTTTTATGCCAA | Amplification of ydjC |
| s6303 | AAAAAAGCTAGCTACGTAGTTCTGTCAGCAATC | Amplification of ydjC |
| s6304 | AAAAAACTTAAGATCATTGAAGATGTTTCTTGA | Amplification of pspA |
| s6305 | AAAAAACATATGCAGCAATTTGATTCGCCGC | Amplification of pspA |
| s6344 | AAAAAAGATATCGATCGCCTTACATTTCGGTGC | Amplification of mtlD |
| s6345 | AAAAAACATATGTTAAAATGATGGCGTGCAACG | Amplification of mtlD |
| s6865 | GCTGACGGCCGGCTCCAGATCT GCA ATC AAG CCT TCA TAT AA | Amplification of mtlR-H342D |
| s6866 | TTATATGAAGGCTTGATTGCAGATC TGG AGC CGG CCG TCA GC | Amplification of mtlR-H342D |
| s6867 | AAAAAA ACTAGT TTA CAG TAT GTT TTT TTC TTT CAT | Amplification of mtlR-H342D |
| s6949 | AAAAAA CCCGGG TAC GAT ATT CCA TAA AAA GC | Amplification of mtlR-H342D |
| Primer extension | ||
| s5959 | Cy5-GCTGCAAGGCGATTAAGTTGG | Hybridized to lacZ |
| s5960 | Cy5-CCAGTCACGACGTTGTAAAAC | Hybridized to lacZ |
Fusion of the cre sites of PmtlA, PmtlR, the internal cre site of mtlA, and cre site of acsA to PgroE was performed by fusion PCR. In this way, the primary PCRs s7098/s7189 and s7190/s6392 (pKAM88), s7098/s7191 and s7192/s6392 (pKAM89), s7098/s7193 and s7194/s6392 (pKAM90), s7098/s7195 and s7196/s6392 (pKAM91) and s7098/s7237 and s7238/s6392 (pKAM101) were followed by final PCR s7098/s6392. The final products PgroE-(cre)-UTRPmtlR were double digested by NheI/AflII and ligated into pSUN279.2.
Construction of mtl and gut mutant strains
Deletion of mtlR, mtlAF, and mtl operon was performed by homologous recombination, replacing the gene of interest by erythromycin resistance gene via double crossover. Integration vector pSUN338.3 [5] was used as the parental vector for construction of the integration vectors in this study. To delete mtlR, downstream flanking gene of mtlR, namely ydaB, was amplified by s5860/s5812 and cloned into the vector, digested by SpeI/StuI. Into the resulting vector the upstream gene ycsN of mtlR, amplified with oligonucleotides s5809/s5810, was inserted between the EcoRV/BglII sites resulting in the cassette of 'ycsN-ermC-ydaB' (pKAM4). Deletion of the mtlAF was performed by the fusion of the PmtlA and mtlD. Fusion PCR was carried out applying s5918/s5919 and s5920/s5921 for the primary reactions, and the final PCR fragment using s5918/s5921 was digested by BglII/EcoRV in order to clone the fragment upstream and opposite direction of erythromycin in pSUN338.3. Amplification of ycnL, the upstream flanking gene of mtl operon, by s6067/s6079 was performed and the product fragment was cloned between the StuI/SpeI sites. The resulting plasmid named pKAM5 contained the 'ycnL-ermC-PmtlA-mtlD' cassette. Disruption of the mtlD was performed by amplification of mtlD, utilizing s6344/s6345 and cloning the resulting fragment with EcoRV/NdeI digestion into pSUN338.3. Erythromycin gene was amplified by s5069/s5070 and blunt-cloned into the plasmid by disruption of mtlD' via HincII restriction site (plasmid pKAM14). Transcription of the erythromycin resistance gene was in opposite direction to the mtl operon transcription after integration into the chromosome by double crossover. Deletion of the mtl operon was performed through the construction of pKAM6. Amplification of the upstream gene, ycnL, was performed by s6068/s6080, followed by cloning between EcoRV/BglII restriction sites of pSUN338.3. Downstream flanking gene, ycsA, was amplified by s5994/s5995 and cloned via SpeI/StuI sites. The final sequence 'ycnL-ermC-ycsA' was used for integration into the chromosome. Deletion of the gut operon was done by replacing the complete operon of glucitol (ΔgutRBPydjE::cat) by a chloramphenicol resistance gene. For this purpose, plasmid pMW363.1 (a derivative of pSUN338.3 harboring chloramphenicol resistance) was used as the parental plasmid. Amplification of the upstream flanking gene of gut operon, namely ydjC, was performed by s6302/s6303, followed by cloning into EcoRI/NheI cut sites. As the downstream flanking gene, pspA was amplified by s6304/s6305 and cloned into the plasmid through AflII/NdeI digestion. The resulting plasmid, named pKAM13, was used for the deletion of gut operon in B. subtilis 3NA.
Prior to transformation of B. subtilis 3NA with plasmids pKAM4, pKAM5, pKAM6, pKAM13, and pKAM14, the DNA was linearized by PacI in order to prevent the single crossover integration. The transformants were selected on LB plates supplemented with erythromycin or chloramphenicol, depending on the plasmid. Additionally, the selected mutants were counter-selected on LB plates containing spectinomycin in order to remove the single crossover mutants. Finally, the deletion on the chromosome was confirmed by PCR using the relevant primers. All of the mutants were cultured in minimal medium harboring 1% of either mannitol or glucitol as the sole carbon source in order to confirm the disability of growth with mannitol.
Markerless deletion of ptsG mutant
Construction of the ΔptsG mutant was carried out in two steps leading to a markerless mutant. The first step included a disruption of ptsG and a 5' part of ptsH by the integration vector pMW312.2. This plasmid was a derivative of pSUN356.7 [5]. Using oligonucleotides s5621/s5622, the upstream region of ptsG (glcT) was amplified. The amplified fragment was cut by KasI/BamHI and ligated into pSUN356.7, which was digested by KasI/BglII. The resulting vector was subsequently cut by PstI. Next, the amplified fragment with s5623/s5624, harboring the downstream region of ptsG ('ptsHI), was integrated. Thereby, the integration vector, pMW312.2, was constructed. Naturally competent B. subtilis 3NA cells were transformed by pMW312.2 and selected on LB agar plates supplemented with erythromycin and 0.5% (w/v) glucitol. The growth of the resulting strain was checked on minimal medium agar plates containing 0.5% (w/v) mannose as the sole carbon source. The deleted mutant (named MW312) could not grow on the mannose minimal medium due to the incapability of producing HPr (encoded by ptsH) and enzyme I (encoded by ptsI). In the second step, a new integration vector was constructed, containing the intact ptsG-ptsHI promoter region fused to the intact ptsHI region. For this purpose, the upstream region, glcT-PptsGHI was amplified by s5621/s5866 and the downstream sequence, ptsHI region, was amplified by s5867/s5624. The fragments were cut by KasI/BamHI and BamHI/PstI, respectively, and integrated via a 3-fragment-ligation into KasI/PstI cut pMW312.2. The resulting integration vector pMW373.3 was used for transformation of naturally competent MW312 cells. For the selection procedure, minimal mannose medium was applied and the mannose positive transformants were counter-selected for erythromycin sensitivity. Finally, the chromosomal DNA of ΔptsG (strain MW373) was extracted and checked by the relevant primers.
Markerless integration of PmtlR-mtlR-H342D into the chromosome
Mutation of the histidine residue of mtlR to aspartate was carried out using fusion PCR. In this way, primary PCRs by oligonucleotides s6949/s6865 and s6866/s6867 were followed by final PCR using s6949/s6867 and PCR products of primary PCRs as the template. The resulting fragment PmtlR-mtlR-H342D was digested by XmaI/SpeI and cloned into pHM31 via XmaI/NheI restriction sites (pKAM66). To integrate the mtlR allele into the chromosome of B. subtilis KM15 (ΔmtlR), an integration system based on histidine auxotrophy was applied [37]. Briefly, B. subtilis KM15 was transformed by pHM30 harboring truncated hisI and spectinomycin resistant genes. The transformants were selected on LB-spectinomycin agar (strain KM162). Afterwards, strain KM162 was transformed by pKAM66 harboring intact hisI and PmtlR-mtlR-H342D and the transformants were selected on Spizizen salts agar supplemented with 1% glycerol. The transformants were then validated on LB-spectinomycin agar and Spizizen salts agar with no citrate and supplemented by 0.5% mannitol as the sole carbon source. Finally, the integration was confirmed by PCR (strain KM163).
β-galactosidase assay
The β-galactosidase activity was measured by using o-nitrophenyl-β-galactopyranoside (ONPG) as a substrate according to Miller assay [32,38] with the modification of Sun & Altenbuchner [5].
Primer extension
Determination of the transcription start site (TSS) of PmtlA and PmtlR was performed by primer extension method. For this purpose, two Cy5 5'-labeled oligonucleotides, i.e. s5959 and s5960 hybridizing to nucleotides 69 to 89 and 31 to 51 downstream of lacZ start codon, were designed. B. subtilis 3NA harboring pKAM1 containing PmtlA-lacZ and pKAM18 containing PmtlR-lacZ were cultivated in LB medium and induced at OD600 of 0.4 by 0.8% mannitol. Total RNA was extracted 1 h after addition of mannitol at 37°C and 200 rpm using the Qiagen RNeasy mini kit (Hilden, Germany). Avian myeloblastosis virus reverse transcriptase and T7 polymerase (Roche, Mannheim, Germany) were applied for reverse transcription and DNA sequencing, respectively. The generated cDNAs and sequencing fragments were analyzed on a denaturing polyacrylamide sequencing gel (GE Healthcare).
Results
B. subtilis PmtlA activity
To characterize the promoter of the mtl operon (PmtlA), the complete sequence from the ycnL stop codon upstream of the mtlA promoter down to the start codon of mtlA was amplified by PCR and transcriptionally fused to the lacZ reporter gene in the E. coli/B. subtilis shuttle vector pSUN279.2 (Figure 2A). With this new plasmid pKAM1, competent B. subtilis 3NA cells were transformed. The resulting strain was cultivated in LB broth and the mtlA promoter induced by adding 0.2% mannitol. In this way, the highest β-galactosidase activity was observed about one hour after the addition of mannitol (data not shown). Therefore, all further β-galactosidase activity measurements were performed at that time. The addition of the inducer mannitol resulted in a 21-fold induction, from 44 to 926 Miller units (M.U.).
Figure 2.
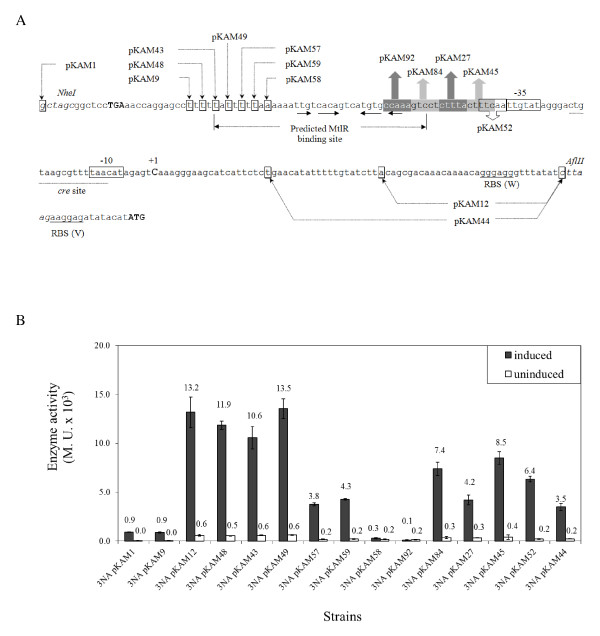
Promoter of mtl operon fused to lacZ (A) and β-galactosidase activity of the constructs (B). (A) Upstream sequence of lacZ in plasmid pKAM1 and deletion derivatives are shown. The intervening sequence between stop codon of ycnL (bold capital letter) and mtlA start codon was obtained from B. subtilis and was inserted between NheI and AflII site of the vector. It includes the promoter elements of mtl operon (PmtlA). The promoter -10 and -35 putative boxes are enclosed by rectangles, while the single C residue (bold capital letter) is the transcription start site. The determined cre site (underline) has an overlap with the -10 box. Ribosomal binding sites of the promoter (W) and vector (V) are underlined, and the lacZ start codon is marked in bold capital letters. The first bases of the shortened promoters are shown by boxes and the arrows. (B) Activity of different constructs of PmtlA in B. subtilis 3NA containing the wild type promoter (pKAM1), the 5' shortened PmtlA, i.e., pKAM9, pKAM43, pKAM48, pKAM49, pKAM57, pKAM58, and pKAM59, as well as 3' shortened PmtlA, i.e. pKAM12 and pKAM44, were induced by 0.2% of mannitol at OD600 of 0.4. β-galactosidase activity of the cells was measured after 1 h of induction. Plasmids pKAM27, pKAM45, pKAM52, pKAM84, and pKAM92 contain the complementary base pairs in comparison to a wild type promoter in the discriminated region between the reported MtlR binding site and -35 box.
The mtl operon is activated by MtlR. So far, the regulator binding site, -10 and -35 boxes of the PmtlA were reported by alignment of the mtlA promoter regions from Geobacillus stearothermophilus and B. subtilis [28]. Hence, shortening of the promoter region as used in pKAM1 was carried out for the determination of the boundaries of PmtlA. In the first step, a deletion from the 5'-end of the promoter was performed removing 20 bps. Determining β-galactosidase activity of this construct, namely pKAM9, showed that this deletion had no effect on the promoter activity (Figure 2B). The deleted sequence embraced the transcription terminator region of the ycnL gene. Afterwards, pKAM9 was used to delete 31 bps from the 3'-end of PmtlA containing the mtlA wild type ribosomal binding site. Induction studies with this newly shortened plasmid (pKAM12) by mannitol showed a remarkable increase of β-galactosidase activity (about 13 to 15-fold) in the basal expression level, as well as under fully induced conditions, compared to pKAM1. This increase might be either due to the shortened untranslated region of mRNA or the presence of two ribosomal binding sites in pKAM1, one from the vector pSUN279.1 and one from mtlA. The latter one is deleted in pKAM12 as depicted in Figure 2A. Further deletion of the next 20 bps from the 3'-end reduced the activity drastically (pKAM44). Therefore, starting from pKAM12, a series of 6 deletions were made at the 5'-end of PmtlA to define the MtlR binding site more precisely. Each construct had two bps more deleted at the 5'-end, giving the plasmids pKAM48, pKAM43, pKAM49, pKAM57, pKAM59, and pKAM58. Deletion of the first two bps of the proposed MtlR binding site (pKAM49) had almost no influence on promoter activity. By deletion of the next 2 bps (pKAM57) and 4 bps (pKAM59), the promoter was still inducible, but showed about a 3-fold reduced activity. Finally, deletion of the next two bps (pKAM58) had a drastic effect and led to a complete loss of promoter activity (Figure 2B). To define the 3'-end of the MtlR binding site, the region between the -35 promoter sequence and the proposed MtlR binding site was exchanged by a complementary DNA sequence. This was done in steps of 5 bps each leading to the plasmids pKAM52, pKAM45, pKAM27, and pKAM84. In all plasmids the promoter was still inducible but β-galactosidase activity was reduced. Finally, exchange of the 5 bps in the palindromic sequence of MtlR binding site in pKAM92 inactivated the PmtlA.
Studying PmtlR activity
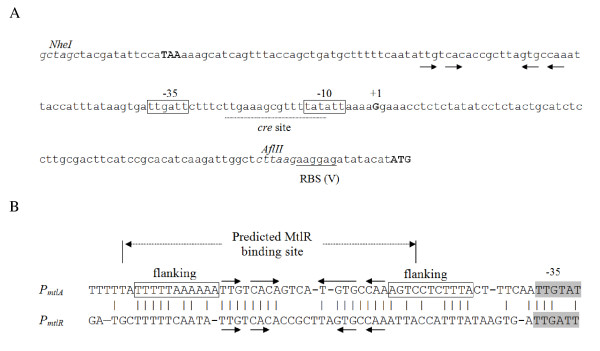
The promoter region of mtlR, amplified by PCR, included the stop codon of the gene ycsN upstream of mtlR and ended at 15 bps upstream of mtlR start codon (Figure 3A). The fragment without the putative mtlR ribosomal binding site was placed in front of the lacZ in pSUN279.1 (pKAM18). Subsequently, B. subtilis 3NA was transformed by pKAM18 in order to monitor the PmtlR activity. Similar to PmtlA, mannitol was able to induce PmtlR. However, PmtlR showed a remarkably lower activity. The β-galactosidase activity in the non-induced cells was 23 M.U. and was raised 4.1-fold to 95 M.U. in 1 h (Table 3). Obviously, MtlR is able to autoinduce its own synthesis. When MtlR is able to induce its own synthesis, there should be a MtlR binding site upstream of PmtlR similar to PmtlA. Indeed, when the upstream sequences of both promoters were aligned, a 42 bps sequence was identified at a distance of 7 bps to the -35 promoter sequences, which share high sequence identity (Figure 3B).
Figure 3.
Promoter of mtlR fused to lacZ (A) and alignment of PmtlA and PmtlR operators (B). (A) Upstream sequence of lacZ in pKAM18. The promoter region of mtlR locating downstream of ycsN stop codon (bold capital letter) and 8 bps upstream of the mtlR start codon is placed between NheI and AflII sites of the vector pSUN279.2. The -10 and -35 boxes (rectangles), as well as transcription start site (G residue; bold capital letter), and lacZ start codon (bold capital letters) are shown. The cre sequence overlaps the Pribnow box (underlined). (B) Alignment of the DNA sequences 55 bps upstream of -35 boxes of PmtlA and PmtlR including the MtlR binding sites.
Table 3.
Activity of PmtlA (pKAM12) and PmtlR (pKAM18) in B. subtilis 3NA and mutants thereof
| B. subtilis | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Treatment (0.2%) | 3NA pKAM12 | KM12 pKAM12 | KM15 pKAM12 | KM37 pKAM12 | KM163 pKAM12 | 3NA pKAM18 | KM12 pKAM18 | KM15 pKAM18 | KM37 pKAM18 |
| Mannitol | 13178 ± 1563 | 13862 ± 1342 | 92 ± 49 | 10801 ± 3290 | 13722 ± 1231 | 95 ± 25 | 243 ± 34 | 16 ± 1 | 156 ± 39 |
| Mannitol + Glucose | 5920 ± 894 | 10289 ± 1131 | 61 ± 46 | 1921 ± 967 | 11795 ± 729 | 28 ± 18 | 143 ± 14 | 5 ± 1 | 53 ± 21 |
| Glucose | 482 ± 86 | 10493 ± 1569 | 57 ± 42 | 1021 ± 379 | 6208 ± 583 | 14 ± 4 | 135 ± 39 | 5 ± 1 | 24 ± 4 |
| Mannitol + Xylose | 13693 ± 1343 | 14084 ± 2081 | 81 ± 45 | 11762 ± 3814 | 12499 ± 2112 | 99 ± 30 | 214 ± 33 | 15 ± 2 | 148 ± 49 |
| Xylose | 555 ± 120 | 14046 ± 2391 | 84 ± 52 | 2834 ± 943 | 1879 ± 76 | 25 ± 3 | 217 ± 41 | 15 ± 1 | 46 ± 7 |
| Glucitol | 8009 ± 947 | 12519 ± 1012 | 81 ± 53 | 11841 ± 3728 | 12425 ± 3297 | 51 ± 26 | 192 ± 43 | 10 ± 1 | 143 ± 42 |
| Glucitol + Glucose | 4559 ± 555 | 10743 ± 663 | 65 ± 51 | 1492 ± 611 | 11555 ± 2364 | 26 ± 22 | 142 ± 36 | 6 ± 1 | 44 ± 13 |
| Uninduced | 594 ± 83 | 15056 ± 2420 | 95 ± 59 | 2838 ± 933 | 2389 ± 645 | 23 ± 4 | 265 ± 71 | 16 ± 1 | 47 ± 10 |
The strains were induced by 0.2% (w/v) of sugars, or no inducer was added (control). β-galactosidase activity was measured 1 h after addition of inducer. Activity was calculated as Miller units. Mutations: KM12 (ΔmtlAF); KM13 (ΔmtlAFD); KM15 (ΔmtlR); KM37 (mtlD::ermC); KM163 (mtlR-H342D).
Determination of the transcriptional start sites of PmtlA and PmtlR
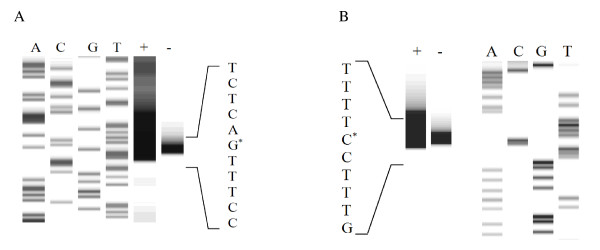
In order to determine the transcription start sites (TSS) of PmtlA and PmtlR, primer extension experiments were performed. B. subtilis 3NA containing the plasmid pKAM1 and pKAM18, respectively, was cultivated in LB liquid medium and RNA was extracted from cells induced by mannitol and from non-induced cells. For designing of the Cy5-5'-labeled primers, the sequence of lacZ, which is identical in the two vectors, was chosen. Using primer extension technique, TSS of the PmtlA was identified at a C residue locating 92 bps upstream of the lacZ start codon, namely 72 bps upstream of mtlA (Figure 4A). Accordingly, -10 (TAACAT) and -35 (TTGTAT) boxes were determined resembling the house-keeping sigma factor (σA) binding site. Besides, one cre site (CTGTAAGCGTTTTAA) within the sequence of the PmtlA was found having 2 mismatches (underlined) compared to the consensus sequence (WTGNAARCGNWWWCA) (Figure 2A) [39]. The cre site has an overlap with the -10 box of PmtlA. Likewise, the transcription start site of PmtlR was identified to be a single G residue (Figure 4B). Accordingly, -10 box (TATATT), -35 box (TTGATT), along with cre site (TTGAAAGCGTTTTAT), were determined (Figure 3A). Also in this case, the cre site overlaps with the Pribnow box and has 2 mismatches compared to the cre consensus sequence (underlined).
Figure 4.
Primer extension. (A) Primer extension of PmtlA in pKAM12. The procedure has been explained in Methods. A, C, G, and T represent the dideoxynucleotide triphosphates used for the pKAM1 sequencing, and the + and - display the primer extension reaction of induced and uninduced sample, respectively. (B) Primer extension of PmtlR in pKAM18 and DNA sequencing reaction.
Induction and catabolite repression of PmtlA and PmtlR
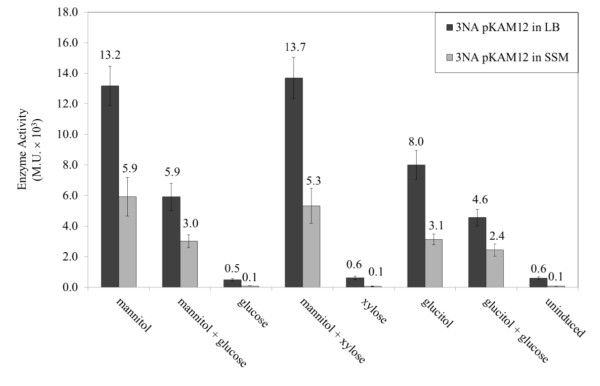
Induction and catabolite repression of PmtlA and PmtlR on the plasmids pKAM12 and pKAM18 in B. subtilis 3NA was investigated by cultivating the strains in LB liquid medium, as well as in Spizizen salts medium, with the addition of either mannitol, glucose, the non-PTS sugars glucitol and xylose, alone or in combination of two sugars (Figure 5). After one hour of induction, promoter activity was monitored by β-galactosidase activity. Although the promoter activity was almost doubled in LB medium, the PmtlA was less inducible in LB compared to SSM caused by a higher basal activity. However, comparison of the minimal and rich medium showed no significant difference in the glucose repression in PmtlA (ratio mannitol/mannitol + glucose). Therefore, further measurements were performed in LB medium due to a higher activity of the promoter and a shorter lag phase of growth. As shown in Table 3, both promoters were induced by mannitol and to a lesser extent by glucitol, whereas xylose had no effect. The presence of glucose repressed promoter activity during induction by mannitol and glucitol; as a result, PmtlA activity dropped at least twofold and PmtlR activity about threefold. This means that both promoters underlie catabolite repression.
Figure 5.
β-galactosidase activity of B. subtilis 3NA pKAM12. β-galactosidase activity of B. subtilis 3NA pKAM12 induced in LB medium compared to Spizizen salts medium (SSM) 1 h after addition of inducer.
Deletion of mtl and gut genes
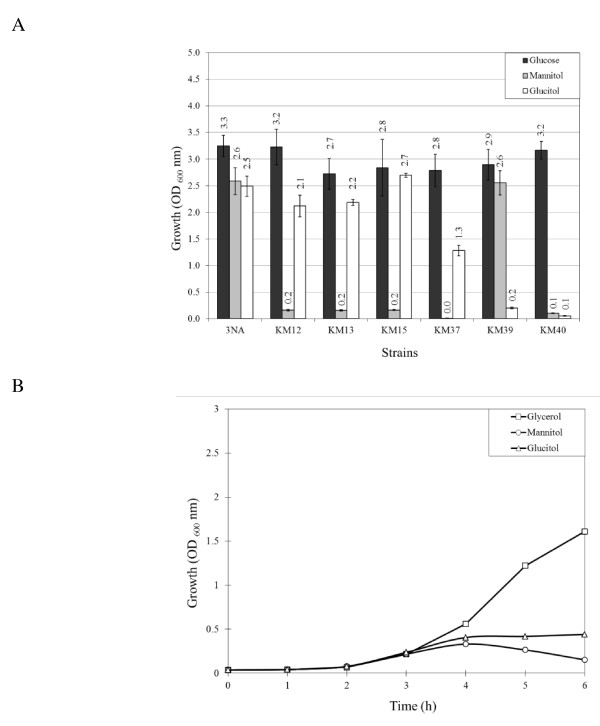
Genes involved in mannitol utilization, i.e. mtlA, mtlF, mtlD, and mtlR, were replaced or disrupted by an erythromycin resistance gene in order to investigate their probable influence on PmtlA activity, as well as on cell growth, with mannitol or glucitol as carbon source. The mutants, i.e. KM12 (ΔmtlAF::ermC), KM13 (ΔmtlAFD::ermC), KM15 (ΔmtlR::ermC) and KM37 (mtlD::ermC) were used to inoculate minimal medium with 1% mannitol to an optical density of 0.01 (OD600) and incubated for 16 h. Very weak growth of bacteria to an OD600 of maximum 0.2 was detected with KM13, KM12, and KM15 strains; however, KM37 strictly showed no growth (Figure 6A). Additionally, the mtl mutants were cultured in minimal medium with 1% glucitol as the sole carbon source. The mutants KM12, KM13, and KM15 grew similar to the wild type. However, KM37 showed half of the growth of the wild type. This phenomenon was reported by Watanabe et al. [28] at first.
Figure 6.
Growth of mtl and gut mutants (A) and growth curve of mtlD::ermC mutant (B). (A) Growth of the B. subtilis mtl and gut mutants in minimal medium. Optical density at 600 nm of the strains KM12 (ΔmtlAF), KM13 (ΔmtlAFD), KM15 (ΔmtlR), KM37 (mtlD::ermC), KM39 (ΔgutRBPydjE), KM40 (ΔgutRBPydjE ΔmtlAFD), as well as of B. subtilis 3NA (wild type), was measured after 16 h incubation at 37°C on a rotary shaker with 200 rpm. Minimal medium was supplemented by 1% of either mannitol, glucitol or glucose (control) as the sole carbon source. (B) The toxicity effect of the mannitol and glucitol in KM37. Strain KM37 was grown in LB medium containing 1% glycerol, mannitol, or glucitol and the growth curve was monitored at intervals of 1 h.
As described, KM37 was the only strain that strictly showed no growth in the presence of mannitol as the sole carbon source and a reduced growth with glucitol. Consequently, in another effort the growth of KM37 in LB medium enriched by either 1% glycerol (as control), mannitol or glucitol was monitored. As depicted in Figure 6B, tracking the growth curve of KM37 at 1-h intervals revealed that the presence of mannitol and glucitol retarded the growth of the cells in the exponential phase. In fact, the maximal cell density observed with KM37 was 0.3 OD600 with mannitol and 0.4 OD600 with glucitol after 6 h incubation. By microscopy, we discovered that the cells became very swollen during incubation probably due to the accumulation of mannitol 1-phosphate. The same phenomenon was observed in the presence of glucitol, where probably glucitol 6-P is formed.
Besides the mtl operon, the complete gut operon, namely gutRBPydjE, was replaced by the chloramphenicol resistance gene in order to investigate the probable interaction of the two degradation pathways. The gut genes were deleted in B. subtilis 3NA as well as in KM13 (ΔmtlAFD::ermC). The latter strain was used as the control in which no growth should be observed for both sugars. Growth of KM39 (ΔgutRBPydjE::cat) in minimal medium containing either 1% glucitol or 1% mannitol as the sole carbon source was strongly reduced in the case of glucitol, whereas for mannitol it was not affected. In the double mutant KM40 growth on either glucitol or mannitol was strongly reduced as expected (Figure 6A).
Activity of PmtlA and PmtlR in mtl mutants
The activity of PmtlA in the absence of mannitol PTS components was investigated to determine if these components had an effect on the promoter activity. The strain KM12 lacks mtlA and mtlF encoding the EIICBMtl and EIIAMtl, respectively. Strain KM12 was transformed by pKAM12 and β-galactosidase activities were determined in the presence or absence of mannitol, glucitol, or xylose of the resulting strain. The results indicated that the activity of PmtlA was constitutive (Table 3). In other words, the promoter showed highest activity regardless of the presence or absence of the inducer. Obviously, the components of mannitol transporter had an inhibitory effect on the PmtlA activity in the absence of mannitol. However, glucose repression was functional to some extent, e.g. 1.3-fold reduction of β-galactosidase activity by mannitol and 1.2-fold by glucitol when glucose was added.
Next, the mtlD deficient mutant KM37 was transformed by pKAM12. The deficiency of the mannitol 1-phosphate dehydrogenase not only increased the basal expression level of PmtlA (4.7-fold), but also enhanced the glucose repression from 2.2-fold in the wild type to 5.6-fold in the mutant. As expected, addition of mannitol or glucitol inhibited the growth during the 1 h of induction. However, the cells grew normally when glucose was added (data not shown). In accordance with pKAM12, similar results were obtained with pKAM18 in KM12 and KM37 mutants (Table 3). Surprisingly, PmtlA was equally induced by glucitol and mannitol (approx. 11,000 M.U.). Finally, deletion of the mtlR encoding the activator of the operon drastically decreased the activity of the PmtlA to about 95 M.U. (KM15 pKAM12, Table 3). This value is six times lower compared to the uninduced promoter in a wild type strain. No remarkable difference was observed between the different sugars tested, although addition of glucose reduced the activity slightly (1.2 to 1.5- fold). Finally, KM15 (ΔmtlR::ermC) was transformed by pKAM18 harboring PmtlR. In this mutant the promoter was no longer inducible. This confirms the assumption that PmtlR is an autoregulatory promoter. The maximum activity was reduced to 16 M.U., which was approximately the basal activity of PmtlR in the wild type strain, and the presence of glucose led to a further reduction.
Phosphorylation of MtlR by HPr (His15~P)
It is known that PEP-dependent phosphorylation of HPr (His15) plays an essential role in the activity of PRD containing regulators such as ManR [5] and LevR [40]. Therefore, strain TQ432 harboring ptsH-H15A mutation was transformed by pKAM12. Results of induction of the TQ432 pKAM12 by different sugars are shown in Table 4. Similar to ΔmtlR mutant, no induction was observed by mannitol or glucitol. Moreover, no glucose repression was observed on the basal expression level due to a slow metabolism of the glucose. These results were in line with the in vitro studies of Joyet et al. [29] in which phosphorylation of PRDII domain of MtlR was observed in the presence of HPr (H15~32P).
Table 4.
Activity of PmtlA (pKAM12) and PmtlR (pKAM18) in B. subtilis 3NA, and CCR mutants
| B. subtilis | |||||||
|---|---|---|---|---|---|---|---|
| Treatment (0.2%) | 3NA pKAM12 | TQ303 pKAM12 | MW373 pKAM12 | TQ432 pKAM12 | 3NA pKAM18 | TQ303 pKAM18 | MW373 pKAM18 |
| Mannitol | 13178 ± 1563 | 6120 ± 1701 | 11301 ± 628 | 61 ± 7 | 83 ± 3 | 93 ± 4 | 77 ± 17 |
| Mannitol + Glucose | 5920 ± 894 | 1636 ± 437 | 11788 ± 550 | 56 ± 7 | 19 ± 1 | 38 ± 3 | 73 ± 17 |
| Glucose | 482 ± 86 | 287 ± 158 | 398 ± 137 | 58 ± 8 | 12 ± 2 | 29 ± 1 | 25 ± 5 |
| Mannitol + Xylose | 13693 ± 1343 | 4982 ± 628 | 10533 ± 712 | 51 ± 7 | 84 ± 8 | 86 ± 2 | 74 ± 16 |
| Xylose | 555 ± 120 | 387 ± 32 | 417 ± 54 | 44 ± 9 | 25 ± 3 | 33 ± 3 | 25 ± 5 |
| Glucitol | 8009 ± 947 | 3489 ± 854 | 5211 ± 720 | 43 ± 5 | 40 ± 11 | 68 ± 4 | 41 ± 9 |
| Glucitol + Glucose | 4559 ± 555 | 774 ± 255 | 5324 ± 432 | 37 ± 3 | 17 ± 8 | 31 ± 2 | 40 ± 10 |
| Uninduced | 594 ± 83 | 501 ± 61 | 433 ± 115 | 59 ± 7 | 23 ± 4 | 33 ± 1 | 25 ± 6 |
Expression of lacZ by PmtlA (pKAM12) and PmtlR (pKAM18) in B. subtilis 3NA as well as in CCR affected mutants, such as TQ303 (ΔccpA), TQ432 (ptsH-H15A), and MW373 (ΔptsG) were measured as described in the legend of Table 3.
Activity of PmtlA and PmtlR in CCR deficient mutants
By transforming B. subtilis TQ303, which lacks the ccpA gene, a central element of the catabolite repression system, the effect of catabolite repression on PmtlA was investigated with plasmid pKAM12. This mutant grew slower than the wild type and showed a longer lag phase. In contrast to our expectations, the ccpA mutant demonstrated a similar or even slightly stronger catabolite repression than the wild type (3.7 versus 2.2-fold) when mannitol together with glucose were added (Table 4). The higher rate of repression might be caused by the fact that the β-galactosidase activity after 1 h induction with mannitol alone was about threefold lower than in the wild type, whereas the basal activity of PmtlA in the ccpA mutant resembled the wild type.
In addition to PmtlA, regulation of PmtlR was considered by the transformation of TQ303 mutant with pKAM18. The fully induced PmtlR by mannitol showed about the same activity as in the wild type in contrast to PmtlA. By adding of mannitol together with glucose, the β-galactosidase activity was less reduced compared to the wild type strain. On the other hand, the basal expression level in LB and LB with glucose or xylose was slightly increased at about the same amount as the catabolite repression was reduced (Table 4).
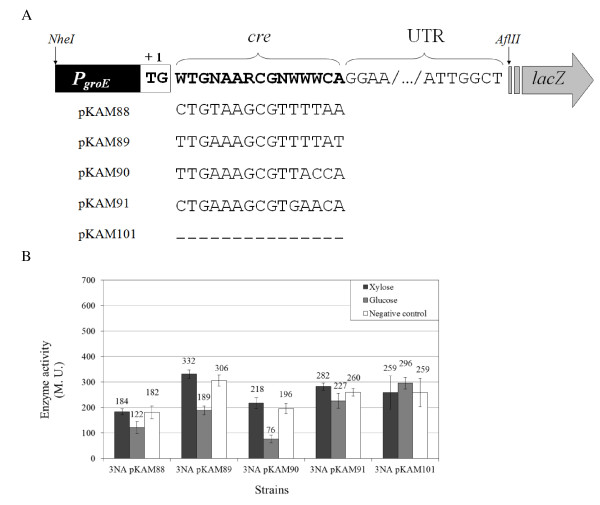
Function of cre sites downstream of promoter
In order to clarify whether the cre sites of the PmtlA and PmtlR are functional, they were inserted downstream of the transcription start site of a constitutive promoter. Insertion of cre site at the +1 transcriptional start site inhibits the promoter clearance in a road block mechanism and RNA polymerase (RNAP) can be stalled when the HPr (S46~P)-CcpA binds to DNA [41]. In this case, the promoter of the groESL operon (PgroE) was fused to an untranslated region of PmtlR (UTRPmtlR) (Figure 7A). In addition to the cre sites of PmtlA and PmtlR, another cre site, locating inside the mtlA gene (+1156 to +1169), has been reported [22]. Comparison of this cre site with the cre consensus sequence revealed only one mismatch (Figure 7A). As a positive control, the functional cre site of acsA (acetyl-CoA synthetase) was applied [42,43], while direct fusion of PgroE-UTRPmtlR was used as the negative control. The constructs PgroE-(cre)-UTRPmtlR were fused to lacZ in pSUN279.2. Transformation of B. subtilis 3NA by pKAM88 (crePmtlA), pKAM89 (crePmtlR), pKAM90 (creacsA), and pKAM91 (cremtlA) was carried out. Afterwards, the transformants were grown in LB and at the OD600 of 0.4 either glucose (0.2%), xylose (0.2%), or no sugar was added. For all 4 promoters containing a putative cre site a reduction of β-galactosidase activity was observed by the addition of glucose. For the cre site inside the mtlA it was only 1.2-fold. The cre sites of PmtlA and PmtlR reduced the activity of PgroE 1.5- to 1.6-fold, whereas the cre site of acsA repressed the promoter expression 2.6-fold.
Figure 7.
Fusion of cre sites to PgroE (A) and repression of the constructs by glucose (B). (A) Insertion of the cre sites of PmtlA (pKAM88), PmtlR (pKAM89), and of the internal cre site of mtlA (pKAM91), as well as the cre site of acsA, between PgroE and PmtlR untranslated region. As negative control, PgroE was directly fused to PmtlR untranslated region (pKAM101). (B) Catabolite repression of B. subtilis 3NA pKAM88, pKAM89, pKAM90, pKAM91, and pKAM101 in the presence and absence of glucose (PTS) and xylose (non-PTS).
Deletion of ptsG relieves glucose repression of PmtlA and PmtlR
As demonstrated before, the putative cre sequences had weak effects on the catabolite repression of PmtlA and PmtlR. Thus, we hypothesized that the catabolite repression in this system is mainly due to phosphorylation and dephosphorylation of one of the PRD domains of MtlR via HPr and maybe via the IICBAGlc transporter. The ptsG encoding the transporter of glucose is the first gene of an operon, followed by ptsH and ptsI [26,44]. Disruption or deletion of ptsG by an antibiotic resistance gene may result in the loss of expression of ptsH and ptsI encoding the HPr and Enzyme I proteins, respectively. Therefore, a two-step method was developed. In the first step ptsG was replaced by an erythromycin resistance gene. These mutants were unable to grow on mannose and other PTS sugars. In the second step, the antibiotic resistance gene was replaced by the promoter PptsGHI in which the selection was based on regaining the ability to grow on mannose. The resulting strain MW373 was transformed by pKAM12 and pKAM18 and incubated in the presence of different sugars (Table 4). MW373 harboring pKAM12 was still inducible by mannitol and glucitol, but showed identical β-galactosidase activity when glucose was simultaneously added. Similarly, deletion of the ptsG led to a loss of the glucose repression in PmtlR (Table 4).
Catabolite repression through dephosphorylation of MtlR (H342D)
As shown, glucose repression was completely abolished in ΔptsG mutant. The absence of EIICBAGlc in the cell could affect the level of fructose 1,6-bisphosphate in the cell and finally the CcpA-dependent CCR. But, on the other hand the weakness of the cre sites to repress the promoter activity led us to hypothesize that the EIICBAGlc interacts with other regulatory components of mannitol PTS. Lately, it is shown that the mutation of His 342 to Asp in PRDII domain of MtlR reduced the catabolite repression [29]. Therefore, it is assumed that the dephosphorylation of MtlR by glucose PTS transporter leads to a catabolite repression. In order to confirm this observation in our construct, integration of the gene encoding the mutant MtlR (H342D) into the genome of ΔmtlR strain was performed by the markerless integration system based on histidine auxotrophy (see Methods). Transformation of the mtlR-H342D strain, namely KM163, by pKAM12 was followed by induction studies. As shown in table 3, only a slight catabolite repression was observed in this mutant, although the basal activity was increased due to the mutation of the MtlR. Surprisingly, glucose as the repressor of the system increased the activity of the PmtlA. This phenomenon was also observed by other PTS sugars such as mannose and sucrose, although fructose showed no influence (data not shown).
Discussion
In this study, regulation of the mtl operon and its activator was investigated by fusion of the promoter region of mtlAFD and mtlR to lacZ. Identification of transcription start sites revealed that PmtlA and PmtlR have the σA promoter structure. Shortening the upstream region of PmtlA showed that the two first base pairs of the predicted binding site by Watanabe et al. [28] have no effect on the PmtlA activity. In fact, these two nucleotides are not part of the highly conserved sequence found between PmtlR and PmtlA operator regions, and therefore might not belong to the binding site. Changing the base pairs between the predicted MtlR binding site and -35 showed a reduction in the activity of the promoter, although the inducibility of the promoter remained intact. Generally, transcriptional activators in their active conformation bind as homodimers or homomultimers to their operators [45,46]. Presumably, the mutations between operator and -35, as well as shortening the 5'-end of the MtlR binding site, reduce the binding affinity of the MtlR monomer to the affected operator site, while binding of the second monomer to the intact operator site stabilizes the activator-operon complex. Consequently, mutation in one of the distal or proximal flanking sequences of the operator changes the stability of RNAP-promoter complex mediated by the activator, and thereby affects the overall expression of the promoter [46]. This could explain the inducible but lower activity of the PmtlA with a shortened MtlR binding site. Furthermore, alignment of the operator region of PmtlA and PmtlR indicated that the short palindromic sequence in the operator is enclosed by conserved flanking sequences, consisting of 11 base pairs (Figure 3B). In fact, the MtlR binding site seems to be longer than the predicted sequence extending towards the -35 box. Previously, DNA footprint studies also indicated that the operator of the mannitol promoter in G. stearothermophilus extends near to -35 box [47]. Apparently, the distance between regulator and RNAP binding sites is shorter than the distance found in promoters regulated by class I type activators, which is -61 to -91 bps. Consequently, MtlR is likely a class II type activator in which the regulator makes a direct contact to domain 4 of σ70 [48-53].
Activity of PmtlA and PmtlR was monitored in mtl mutants by the presence of mannitol as the inducer, glucitol as a nonspecific inducer, and glucose as the effector of carbon catabolite repression. In the mtlR mutant, both PmtlA and PmtlR activities were nearly abolished. In contrast to most prokaryotic transcriptional regulators, PRD containing activators do not bind effector molecules and activity is controlled by phosphorylation and dephosphorylation. Accordingly, expression of PmtlA and PmtlR was also abolished in a HPr (H15A) mutant. These results were in line with recent in vitro studies on MtlR indicating that phosphate transfer from HPr (H15) to PRDII of MtlR is essential for stimulation of MtlR activity. This is similar to other PTS transcriptional activators [29]. On the other hand, deletion of the mannitol-specific PTS genes (mtlAF) resulted in a constitutive activity of PmtlA and PmtlR indicating an inhibitory effect of the EIICBMtl and EIIAMtl on PmtlA and PmtlR. MtlR belongs to the LevR-type regulator. Four such transcription activators are known in B. subtilis (LevR, LicR, ManR, and MtlR) [16]. The inhibitory effect of the specific transporters on their activators has been shown already for EIILev [54,55], EIICel [56], and EIIMan [5]. MtlR phosphorylation studies by Joyet et al. [29] have also indicated that the phosphorylation of MtlR by mannitol transporter inactivates the regulator.
The replacement of mtlD with erythromycin resistance gene made the cells sensitive to glucitol as well as mannitol. So far, it is clear that mtlD not only plays a vital role in the consumption of mannitol, but also is necessary for assimilation of glucitol. Induction of PmtlA in the gutRBPydjE deleted mutant showed that the gut system components, especially GutR, have no effect on PmtlA activity (data not shown). Northern hybridization studies, as well as induction studies, proved the ability of glucitol to induce the mtl operon [28]. A weak transport of glucitol by the mannitol specific PTS-transporter in B. subtilis was observed by Chalumeau et al. [30]. This could be due to a relaxed specificity of the PTS transporters in which the transporter may transport more than one sugar. So far, it is observed that PtsG takes up sucrose and salicin in addition to glucose, and the β-glucoside permease (BglP) is capable of a weak uptake of glucose [57,58]. Thus, the inducibility of PmtlA and PmtlR by glucitol is likely due to the uptake of glucitol by mannitol transporter where EIICBMtl and EIIAMtl dephosphorylate the Cys419 and H599 residues of MtlR, and activates the regulator. Previously, the weak activity of mannitol 1-phosphate dehydrogenase with sorbitol 6-phosphate was detected by Horwitz and Kaplan [59]. It can be assumed that a small amount of glucitol in the growth media is not taken up by the glucitol transporter, but is phosphorylated to glucitol 6-phosphate by EIICBAMtl during transport. Afterwards, mannitol 1-phosphate dehydrogenase oxidizes glucitol 6-phosphate to fructose 6-phosphate. Therefore, deletion of the mannitol dehydrogenase should lead to an accumulation of the non-metabolized glucitol 6-phosphate and finally to the killing of the cell.
In addition to mtl deficient mutants, catabolite repression of PmtlA and PmtlR was investigated in CcpA-dependent CCR mutants. Expression of PmtlA in the ΔccpA mutant was dramatically reduced, although the catabolite repression was increased. In fact, the ΔccpA mutant grew slower than the wild type strain due to pleiotropic roles of CcpA in the nitrogen regulation, branched chain amino acid synthesis as well as tricarboxylic acid cycle [60,61]. In contrast, catabolite repression of PmtlR was reduced in the ΔccpA strain; however, the PmtlR activity was not completely relieved from CCR. Obviously, no exact interpretation was obtained by investigation of general trans elements of CCR. Therefore, we focused on the cis element of CCR, which is the cre site of PmtlA and PmtlR. The influence of cre sites from PmtlA, PmtlR, acsA, and mtlA were compared by using the constitutive promoter of groESL operon. In the presence of glucose the cre sites of PmtlA and PmtlR significantly repressed the expression of lacZ from PgroE, whereas the cre site located inside the mtlA gene had almost no effect. Therefore, it seems that two weak cre sites, located in the regulator and operon promoter work together to repress the mannitol genes, whereas in the similar B. subtilis mannose system, a strong cre site in the promoter of the regulatory gene (PmanR) is the only cis element for CcpA-dependent CCR [5]. These results were in line with the previous attempts in mtl operon representing the CcpA-dependent CCR in mannitol PTS [62,63].
So far, it is shown that the glucose repression is abolished when ptsG was disrupted [64,65]. In order to see the probable effect of the glucose PTS transporter, ptsG was deleted from the genome in a way that PptsGHI was fused to ptsHI. As expected, activity of the PmtlA and PmtlR in the ΔptsG mutant was identical in the presence and absence of glucose. The reason why we did not see any effect of the cre/CcpA system in the presence of glucose can be explained by the fact that HPr kinase is only active in the presence of glycolytic intermediates. Due to the deletion of ptsG no glucose is taken up by the EIICBAGlc. From the second glucose transporter (GlcU) it is known that this gene (glcU) is expressed 3 h after sporulation under the control of σG [66,67]. Finally, expression of glcP (glucose/mannose:H+ symporter), a third glucose permease depends on glucose 6-phosphate produced by glucose PTS [65,68]; therefore, reduction of fructose 1,6-bisphosphate pool in the cell eliminates the cre/CcpA dependent CCR. Since CcpA-dependent CCR was completely abolished only in ΔptsG and ptsH-H15A mutants, CcpA dependent CCR might not play the main role in the mannitol regulation. The strong effect of glucose might be explained by a hierarchy in the affinity of HPr (H15~P) to EIICBAGlc versus EIIAMtl and PRDII domain of MtlR. Alternatively, EIICBAGlc might also inactivate MtlR directly by dephosphorylation of PRDII domain. Mutation of the histidine 342 to aspartate located in PRDII domain of MtlR supported the assumption of dephosphorylation of MtlR by glucose PTS components. In this mutant addition of PTS sugars such as glucose, mannose, and sucrose (except fructose) increased the activity of the promoter. This could be due to dephosphorylation of the EIIA and EIIB domains of MtlAF/or MtlR by the specific transporters of glucose, mannose, and sucrose: However, further experiments are necessary to clarify this effect. Overall, catabolite repression in mtl system is mainly functional at posttranslational level by PtsG and at the transcription initiation level by CcpA dependent CCR, which switch off the expression of mannitol uptake system.
Conclusions
Characterization and optimization of the promoter region of mtl operon, as well as its regulator, resulted in construction of a highly inducible expression system in B. subtilis based on mannitol as a cheap inducer. Activity of PmtlA and PmtlR in several mutants revealed that this system is mainly regulated by the phosphorylation and dephosphorylation state of the specific regulator. This accelerates the further manipulation of the strain to enhance the expression system.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KMH designed and performed the experiments and analyzed the data. MW constructed the ptsG mutant and wrote the corresponding section in Materials. JA supervised and coordinated the project. KMH wrote the paper which was later revised and corrected by JA. All authors approved the final version of the manuscript.
Contributor Information
Kambiz Morabbi Heravi, Email: kambiz_morabbi@yahoo.com.
Marian Wenzel, Email: marian.wenzel@iig.uni-stuttgart.de.
Josef Altenbuchner, Email: josef.altenbuchner@iig.uni-stuttgart.de.
Acknowledgements
We are very grateful to Dr. Tianqi Sun for providing the plasmids and strains, and also for her helpful suggestions during this study. We thank Silke Weber for her technical assistance. This study was supported by the German Academic exchange service (DAAD), and the materials were partially provided by Lonza AG, Switzerland. This work was supported by the German Research Foundation (DFG) within the funding programme OpenAccess Publishing.
References
- Li WF, Zhou XX, Lu P. Bottlenecks in the expression and secretion of heterologous proteins in Bacillus subtilis. Res Microbiol. 2004;155:605–610. doi: 10.1016/j.resmic.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Schumann W. Production of recombinant proteins in Bacillus subtilis. Adv Appl Microbiol. 2007;62:137–189. doi: 10.1016/S0065-2164(07)62006-1. [DOI] [PubMed] [Google Scholar]
- Yansura DG, Henner DJ. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L, Mogk A, Schumann W. A xylose inducible Bacillus subtilis integration vector and its application. Gene. 1996;181:71–76. doi: 10.1016/S0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- Sun T, Altenbuchner J. Characterization of a mannose utilization system in Bacillus subtilis. J Bacteriol. 2010;192:2128–2139. doi: 10.1128/JB.01673-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Zhang W, Zhang X, Cen P. Construction and characterization of a novel maltose inducible expression vector in Bacillus subtilis. Biotechnol Lett. 2006;28:1713–1718. doi: 10.1007/s10529-006-9146-z. [DOI] [PubMed] [Google Scholar]
- Warner JB, Lolkema JS. CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol Rev. 2003;67:475–490. doi: 10.1128/MMBR.67.4.475-490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Küster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Molecular Microbiology. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Stülke J, Hillen W. Coupling physiology and gene regulation in bacteria: The phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften. 1998;85:583–592. doi: 10.1007/s001140050555. [DOI] [PubMed] [Google Scholar]
- Kotrba P, Inui M, Yukawa H. Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J Biosci Bioeng. 2001;92:502–517. doi: 10.1263/jbb.92.502. [DOI] [PubMed] [Google Scholar]
- Titgemeyer F, Hillen W. Global control of sugar metabolism: a Gram-positive solution. Antonie Van Leeuwenhoek. 2002;82:59–71. doi: 10.1023/A:1020628909429. [DOI] [PubMed] [Google Scholar]
- Dahl MK. CcpA-independent carbon catabolite repression in Bacillus subtilis. J Mol Microbiol Biotechnol. 2002;4:315–321. [PubMed] [Google Scholar]
- Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem. 2009;73:245–259. doi: 10.1271/bbb.80479. [DOI] [PubMed] [Google Scholar]
- Dahl MK. Enzyme IIGlc contributes to trehalose metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1997;148:233–238. doi: 10.1111/j.1574-6968.1997.tb10294.x. [DOI] [Google Scholar]
- Sutrina SL, Reddy P, Saier MH, Reizer J. The glucose permease of Bacillus subtilis is a single polypeptide chain that functions to energize the sucrose permease. J Biol Chem. 1990;265:18581–18589. [PubMed] [Google Scholar]
- Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD - a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- Stülke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- van Tilbeurgh H, Declerck N. Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr Opin Struct Biol. 2001;11:685–693. doi: 10.1016/S0959-440X(01)00267-6. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Galinier A, Masuda S. In: Bacillus subtilis and its closest relatives: from genes to cells. Sonenshein AL, Hoch JA, Losick R, editor. Washington, D.C: ASM Press; 2002. Carbohydrate uptake and metabolism; pp. 129–150. [Google Scholar]
- Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A. et al. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology. 2009;155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Wipat A. Sequencing and functional analysis of the genome of Bacillus subtilis strain 168. FEBS Lett. 1996;389:84–87. doi: 10.1016/0014-5793(96)00524-8. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V. et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Reizer J, Bachem S, Reizer A, Arnaud M, Saier MH, Stülke J. Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology. 1999;145:3419–3429. doi: 10.1099/00221287-145-12-3419. [DOI] [PubMed] [Google Scholar]
- Reizer J, Sutrina SL, Wu LF, Deutscher J, Reddy P, Saier MH. Functional interactions between proteins of the phosphoenolpyruvate-sugar phosphotransferase systems of Bacillus subtilis and Escherichia coli. Journal of Biological Chemistry. 1992;267:9158–9169. [PubMed] [Google Scholar]
- Watanabe S, Hamano M, Kakeshita H, Bunai K, Tojo S, Yamaguchi H. et al. Mannitol-1-phosphate dehydrogenase (MtlD) is required for mannitol and glucitol assimilation in Bacillus subtilis: Possible cooperation of mtl and gut operons. J Bacteriol. 2003;185:4816–4824. doi: 10.1128/JB.185.16.4816-4824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyet P, Derkaoui M, Poncet S, Deutscher J. Control of Bacillus subtilis mtl operon expression by complex phosphorylation-dependent regulation of the transcriptional activator MtlR. Mol Microbiol. 2010;76:1279–1294. doi: 10.1111/j.1365-2958.2010.07175.x. [DOI] [PubMed] [Google Scholar]
- Chalumeau H, Delobbe A, Gay P. Biochemical and genetic study of D-glucitol transport and catabolism in Bacillus subtilis. J Bacteriol. 1978;134:920–928. doi: 10.1128/jb.134.3.920-928.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Adams JN, Ting RC. Transduction of lactose-utilizing ability among strains of E. coli and S. dysenteriae and the properties of the transducing phage particles. Virology. 1960;12:348–390. doi: 10.1016/0042-6822(60)90161-6. [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons; 1990. [Google Scholar]
- Sambrook J, Fritsch EF, Manniatis T. Molecular cloning: a laboratory manual. NY: Cold spring Harbor Laboratory; 1989. [Google Scholar]
- Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagodich AV, Cherva EA, Shtaniuk YV, Prokulevich VA, Fomichev YK, Prozorov AA. et al. Construction of a vector system for molecular cloning in Bacillus subtilis and Escherichia coli. Mol Biol (Mosk) 2005;39:306–309. doi: 10.1007/s11008-005-0043-7. [DOI] [PubMed] [Google Scholar]
- Titok MA, Chapuis J, Selezneva YV, Lagodich AV, Prokulevich VA, Ehrlich SD. et al. Bacillus subtilis soil isolates: plasmid replicon analysis and construction of a new theta-replicating vector. Plasmid. 2003;49:53–62. doi: 10.1016/S0147-619X(02)00109-9. [DOI] [PubMed] [Google Scholar]
- Motejadded H, Altenbuchner J. Integration of a lipase gene into the Bacillus subtilis chromosome: Recombinant strains without antibiotic resistance marker. Iran J Biotechnol. 2007;5:105–109. [Google Scholar]
- Miller JH. Experiments in molecular genetics. New York: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 2000;28:1206–1210. doi: 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stülke J, Martin-Verstraete I, Charrier V, Klier A, Deutscher J, Rapoport G. The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon. Journal of Bacteriology. 1995;177:6928–6936. doi: 10.1128/jb.177.23.6928-6936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F. Repression of transcription initiation in bacteria. J Bacteriol. 1999;181:2987–2991. doi: 10.1128/jb.181.10.2987-2991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Turinsky AJ, Henkin TM. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalieckas JM, Wray LV, Fisher SH. Expression of the Bacillus subtilis acsA gene: Position and sequence context affect cre-mediated carbon catabolite repression. J Bacteriol. 1998;180:6649–6654. doi: 10.1128/jb.180.24.6649-6654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorec M, Postma PW. Cloning and nucleotide sequence of the ptsG gene of Bacillus subtilis. Mol Gen Genet. 1992;234:325–328. doi: 10.1007/BF00283853. [DOI] [PubMed] [Google Scholar]
- Krauss Gerhard. Biochemistry of signal transduction and regulation. Weinheim: Wiley-VCH; 2005. [Google Scholar]
- Ishihama A. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- Henstra SA, Tuinhof M, Duurkens RH, Robillard GT. The Bacillus stearothermophilus mannitol regulator, MtlR, of the phosphotransferase system - A DNA-binding protein, regulated by HPr and IICBmtl-dependent phosphorylation. J Biol Chem. 1999;274:4754–4763. doi: 10.1074/jbc.274.8.4754. [DOI] [PubMed] [Google Scholar]
- Barnard A, Wolfe A, Busby S. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr Opin Microbiol. 2004;7:102–108. doi: 10.1016/j.mib.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Browning DF, Busby SJW. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Ghosh T, Bose D, Zhang XD. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol Rev. 2010;34:611–627. doi: 10.1111/j.1574-6976.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- van Hijum SAFT, Medema MH, Kuipers OP. Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiol Mol Biol Rev. 2009;73:481–509. doi: 10.1128/MMBR.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Verstraete I, Debarbouille M, Klier A, Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990;214:657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- Martin-Verstraete I, Charrier V, Stülke J, Galinier A, Erni B, Rapoport G. et al. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol Microbiol. 1998;28:293–303. doi: 10.1046/j.1365-2958.1998.00781.x. [DOI] [PubMed] [Google Scholar]
- Tobisch S, Glaser P, Kruger S, Hecker M. Identification and characterization of a new beta-glucoside utilization system in Bacillus subtilis. J Bacteriol. 1997;179:496–506. doi: 10.1128/jb.179.2.496-506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling O, Herzberg C, Hertrich T, Vorsmann H, Jessen D, Hubner S. et al. Keeping signals straight in transcription regulation: specificity determinants for the interaction of a family of conserved bacterial RNA-protein couples. Nucleic Acids Research. 2006;34:6102–6115. doi: 10.1093/nar/gkl733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stülke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Molecular Microbiology. 1997;25:65–78. doi: 10.1046/j.1365-2958.1997.4351797.x. [DOI] [PubMed] [Google Scholar]
- Horwitz SB, Kaplan NO. Hexitol dehydrogenases of Bacillus subtilis. J Biol Chem. 1964;239:830–838. [PubMed] [Google Scholar]
- Moreno MS, Schneider BL, Maile RR, Weyler W, Saier MH. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Reizer J, Fischer C, Galinier A, Saier MH, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca GL, Chung YJ, Barabote RD, Weyler W, Schilling CH, Saier MH. Catabolite repression and activation in Bacillus subtilis: Dependency on CcpA, HPr, and HprK. J Bacteriol. 2005;187:7826–7839. doi: 10.1128/JB.187.22.7826-7839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem S, Faires N, Stülke J. Characterization of the presumptive phosphorylation sites of the Bacillus subtilis glucose permease by site-directed mutagenesis: implication in glucose transport and catabolite repression. FEMS Microbiol Lett. 1997;156:233–238. doi: 10.1111/j.1574-6968.1997.tb12733.x. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Chauvaux S, Choi P, Saier MH. Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: Identification of a novel hexose:H+ symporter. J Bacteriol. 1998;180:498–504. doi: 10.1128/jb.180.3.498-504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H, Bassias J, Jankovic I, Bruckner R. Identification of a gene in Staphylococcus xylosus encoding a novel glucose uptake protein. J Bacteriol. 1999;181:4929–4936. doi: 10.1128/jb.181.16.4929-4936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Nicholson WL, Neitzke KD, Setlow P, Freese E. Sigma-G RNA polymerase controls forespore-specific expression of the glucose dehydrogenase operon in Bacillus subtilis. Nucleic Acids Res. 1989;17:999–1017. doi: 10.1093/nar/17.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaoka T, Ochi K. Glucose uptake pathway-specific regulation of synthesis of neotrehalosadiamine, a novel autoinducer produced in Bacillus subtilis. J Bacteriol. 2007;189:65–75. doi: 10.1128/JB.01478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 Vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Michel JF, Millet J. Physiological studies on early-blocked sporulation mutants of Bacillus subtilis. J Appl Bacteriol. 1970;33:220–227. doi: 10.1111/j.1365-2672.1970.tb05246.x. [DOI] [PubMed] [Google Scholar]