Abstract
Background
The cancer stem cell model has been proposed based on the linkage between human embryonic stem cells and human cancer cells. However, the evidences supporting the cancer stem cell model remain to be collected. In this study, we extensively examined the expression of human embryonic stem cell-associated signatures including core genes, transcription factors, pathways and microRNAs in various cancers using the computational biology approach.
Results
We used the class comparison analysis and survival analysis algorithms to identify differentially expressed genes and their associated transcription factors, pathways and microRNAs among normal vs. tumor or good prognosis vs. poor prognosis phenotypes classes based on numerous human cancer gene expression data. We found that most of the human embryonic stem cell- associated signatures were frequently identified in the analysis, suggesting a strong linkage between human embryonic stem cells and cancer cells.
Conclusions
The present study revealed the close linkage between the human embryonic stem cell associated gene expression profiles and cancer-associated gene expression profiles, and therefore offered an indirect support for the cancer stem cell theory. However, many interest issues remain to be addressed further.
Background
The development of human embryonic stem cell (hESC) is controlled by specific signatures, including specific transcription factors (TFs), pathways, microRNAs (miRNAs) and core genes. These signatures determine the self-renewal or differentiation fate of hESCs. Cancer is one of the developmental diseases. The initiation, proliferation and metastasis of cancer are often associated with the abnormalities of developmental signatures. Like hESCs, cancer cells are endowed with the ability to self-renew and proliferate indefinitely.
Based on accumulated evidence linking cancer cells to hESCs, some researchers proposed cancer stem cell (CSC) hypothesis [1]. A CSC is defined as "a cell within a tumor that possesses the capacity to self-renew and to cause the heterogeneous lineages of cancer cells that comprise the tumor [2] ". This hypothesis suggests that a small percentage of hESC-like CSCs are responsible for initiating and replenishing the tumor, and the dormant CSCs may account for cancer metastasis, chemoresistance and recurrence so that they become potential targets for improved cancer therapies. One type of evidence supporting the CSC model is the identification of surface markers of cancer-initiating cells (CICs; also known as cancer stem cells) in various human tumor types. Dick et al reported that only a subset of cells were able to transplant AML into recipient mice [3,4]. These tumorigenic cells were defined as CD34+CD38-, indicating a presence of CD34 proteins and a lack of CD38 proteins on their surface [5]. Dirks et al successfully isolated CSCs (CD133+ cells) from different phenotypes of brain tumors [6,7]. The CSCs were also identified in a list of the other tumor types including breast tumors [8], melanoma [9], ovarian cancer [10,11], prostate cancer [12], pancreatic cancer [13,14], sarcoma [15] and colon cancer [16,17]. Although the CSC theory is supported by some experimental evidences, much contention exists over whether these evidences are sufficiently valid or merely are some artifacts [18-21].
Some other types of evidence seems to lend support to the CSC theory, although they are not direct or absolutely convincing. For example, hESCs share cellular and molecular phenotypes with tumor cells and cancer cell lines [22]. Human induced pluripotent stem cells (HiPSCs) were first derived with four transcription factors: OCT4, SOX2, MYC and KLF4 [23] or OCT4, SOX2, NANOG, and LIN28 [24]. All these transcription factors have been reported to be highly expressed in various types of cancer [25-29]. Furthermore, silencing of tumor suppressor gene p53 significantly increased the reprogramming efficiency of human somatic cells [30]. Activation of telomerase is in part responsible for long lifespan of stem cells as well as anti-apoptosis of cancer cells [13,31-34]. Cell cycle regulation plays a critical role in both stem cells and cancer cells [35-39].
The linkage between hESC-specific gene expression profiles and cancer-specific gene expression profiles may provide evidence in support of the CSC model. To this end, many studies have identified hESC-associated gene expression signatures (hESCGESs) [40-44], and several studies have examined the expression of hESCGESs in human cancer [45-49]. In [45], the authors provided first clinical evidence for the implication of a "glioma stem cell" or "self-renewal" phenotype in treatment resistance of glioblastoma. In [46], the authors found the hESCGESs that distinguished primary from metastatic human germ cell tumors. In [47], the authors identified a subset of hESC-associated transcription regulators that were highly expressed in poorly differentiated tumors. In [48], the authors revealed that an increased expression of some hESCGESs identified poorly differentiated lung adenocarcinoma. In [49], the authors compared the expression of pluripotency factors OCT4, SOX2, KLF4 and MYC in 40 human tumor types to that of their normal tissue counterparts using publicly available gene expression data, and found significant overexpression of at least one out of them in 18 out of the 40 cancer types investigated. Furthermore, they found that these genes were associated with tumor progression or bad prognosis. All together, these studies revealed that "stemness" gene expression signatures were associated with tumor malignancies, and therefore might be informative molecular predictors of cancer therapy outcome [50].
In this study, we investigated the linkage between hESCGESs and tumor malignancies by an extensive examination of the expression of hESCGESs in various human tumor types. We used 51 publicly available gene expression datasets, which involve 23 human tumor types [51].
Methods
Identification of human stem cell-associated gene expression signatures
The self-renewal and differentiation of hESCs are controlled by hESC-specific signal molecules in a signaling-specific manner. Through a substantial survey of related literatures, we collected four types of hESCGESs: genes, pathways, TFs and miRNAs.
We collected 24 hESC-associated gene sets which were classified into five groups (Table 1 and Additional file 1, Table S1).
Table 1.
hESC-associated gene sets
| Group | Gene set | #Genes | Description |
|---|---|---|---|
| hESC exp1 | 379 | Overexpressed in hESCs according to 5 or more out of 20 profiling studies | |
| hESC exp2 | 40 | Overexpressed in hESCs according to a meta-analysis of 8 profiling studies | |
| hESC exp3 | 48 | Overexpressed in hESCs in at least 10 studies | |
| hESC exp4 | 30 | Underexpressed in hESCs in at least 6 studies | |
| hESC exp5 | 189 | TF genes in hESCs | |
| hESC expressed | hESC exp6 | 44 | Highly expressed in undifferentiated hESCs |
| hESC exp7 | 994 | High connectivity (≥ 500) in the global co-expression networks of hESCs | |
| hESC exp8 | 22 | Candidate hESC markers | |
| hESC exp9 | 27 | Differentially upregulated in hESCs | |
| hESC exp10 | 31 | The most abundant transcripts expressed in hESCs | |
| hESC exp11 | 1, 000 | Significantly identified in hESC lines by SAM analysis | |
| Nanog targets | 988 | Genes activated by Nanog in hESCs | |
| Oct4 targets | 290 | Genes activated by Oct4 in hESCs | |
| NOS targets | Sox2 targets | 734 | Genes activated by Sox2 in hESCs |
| NOS targets | 179 | Overlap of three above sets | |
| NOS TFs | 37 | Transcription regulators in NOS targets set | |
| NOS co-bound | 353 | Genes co-bound by Nanog, Oct4 and Sox2 in hESCs | |
| Suz12 targets | 1, 040 | Genes bound by Suz12 | |
| Polycomb targets | Eed targets | 1, 066 | Genes bound by Eed |
| H3K27 bound | 1, 121 | Genes bound by H3K27 | |
| PRC2 targets | 654 | Overlap of three above sets | |
| Myc targets | Myc targets1 | 230 | E-box-containing genes regulated by c-Myc in cultured cell lines |
| Myc targets2 | 775 | Genes commonly regulated by c-Myc and Max in a Burkitt's lymphoma cell line | |
| P53 targets | 35 | Common gene lists regulated by p53 in mouse and human | |
A number of developmental signal pathways, such as Wnt, Notch, Hedgehog and Bmi-1, are necessary for regulation of stem cell self-renewal and differentiation. We identified 54 signal pathways as the hESC-associated pathway signatures (Table 2).
Table 2.
hESC-associated signal pathways, TFs and miRNAs
| Pathway | TF | miRNA |
|---|---|---|
| Activin | TP53 | miR-143 |
| AKT | MYC | miR-145 |
| ALK | GATA4 | miR-187 |
| ATM | SMAD1 | miR-296 |
| BMI1 | ESRRB | miR-301 |
| BMP | SOX2 | miR-21 |
| Cell cycle | NANOG | let-7a |
| EGF/EGFR | KLF4 | miR-371 |
| ERBB2 | MYB | miR-372 |
| ERK | MYCN | miR-373 |
| FGF | ZFX | miR-367 |
| Glycolysis | STAT3 | miR-302a |
| Hippo | ZIC3 | miR-302a* |
| IGF | ZFP42 | miR-302b |
| JAK/STAT | SALL4 | miR-302b* |
| c-KIT | REST | miR-302c |
| Lefty | TCF3 | miR-302c* |
| LIF | HOXB1 | miR-302d |
| MAPK | HAND1 | miR-200c |
| MEK/ERK | POU5F1 | miR-222 |
| NF-κB | SRY | |
| NHEJ/HR a | TBX5 | |
| Nodal | E2F4 | |
| Notch | GATA6 | |
| p53 | PAX6 | |
| PRC2 | TCF4 | |
| PDGF | FOXD3 | |
| PI3K | CNOT3 | |
| PTEN | ZEB2 | |
| RAS | ESX1L | |
| RTK | ||
| Hedgehog (SHH) | ||
| Smad | ||
| Stat3 | ||
| Telomerase | ||
| TGFβ | ||
| VEGF | ||
| WNT | ||
a NHEJ: non-homologous DNA end-joining; HR: homologous recombination
We identified 189 key TFs involved in regulation of hESC self-renewal and differentiation including three core TFs OCT4, SOX2 and NANOG with essential roles in the transcriptional control of the regulatory circuitry underlying pluripotency [43,52]. Table 2 lists 30 "critical" TFs. The complete TF list is presented in Additional file 2, Table S2.
Recent research indicates that miRNAs have an important role in regulating stem cell self-renewal and differentiation [53]. We identified 114 hESC-associated miRNAs. Table 2 lists one part of them. The complete miRNA list is presented in Additional file 3, Table S3.
Identification of tumor-associated gene expression signatures
We identified differentially expressed genes among normal vs. tumor or good prognosis vs. poor prognosis phenotypes classes using univariate F-test for unpaired samples or t-test for paired samples at 0.05 significance level. This procedure was implemented with the class comparison between groups of arrays tool in BRB-ArrayTools, an integrated package developed by Simon et al for the visualization and statistical analysis of DNA microarray gene expression data [54]. The software can be freely downloaded from the website: http://linus.nci.nih.gov/BRB-ArrayTools.html.
We identified important pathways, TFs and miRNAs by analyzing gene sets for differential expression among pre-defined classes. The pre-defined phenotypes classes in the class comparison algorithm involved two types: normal vs. tumor and good prognosis vs. poor prognosis. The latter is concerned with tumor subtypes which exhibit different clinical outcome such as metastasis or not, relapse or disease free, drug or radio therapy sensitive or resistance etc., and different tumor progression grades. The LS or KS permutation test and Efron-Tibshirani's GSA maxmean test were used to determine the significant gene sets at 0.05 significance level. The pathways (BioCarta) related to the significant gene sets were identified. The TFs were identified by the gene sets, in each of which all genes were experimentally verified to be targets of the same transcription factor. Each miRNA potentially targeting all the genes in one of the gene sets was identified. The identification of important pathways, TFs and miRNAs was performed with the gene set expression class comparison tool in BRB-ArrayTools.
In addition, we used the survival analysis tool in BRB-ArrayTools to find genes, pathways, TFs and miRNAs related to survival for the partial datasets which provided related data. All the executive parameters were identical to those used in the class comparison.
We compared the identified gene sets, pathways, TFs and miRNAs to those in hESCGESs, and found their overlaps, respectively.
Materials
We analyzed 51 human gene expression datasets involving 23 tumor types (Table 3). For each dataset, we carried out class comparison and/or survival analysis algorithm to identify informative genes, pathways, TFs and miRNAs. A total of 75 class comparison and survival analysis were carried out (Table 4). All the references relevant to Table 1, Table 2, Table 3 and Table 4 are presented in Additional file 4.
Table 3.
Fifty-one human tumor gene expression datasets
| Tumor Type | # Datasets |
|---|---|
| Bladder Cancer | 1 |
| Brain Cancer | 6 |
| Breast Cancer | 5 |
| Colon Cancer | 1 |
| Cervical Cancer | 1 |
| Embryonal Cancer | 1 |
| Esophageal Cancer | 1 |
| Gastric Cancer | 2 |
| Head and Neck Cancer | 3 |
| Leukemia | 3 |
| Liver Cancer | 1 |
| Lung Cancer | 2 |
| Lymphoma | 4 |
| Medulloblastoma | 1 |
| Melanoma | 2 |
| Mesothelioma | 1 |
| Ovarian Cancer | 1 |
| Pancreatic Cancer | 1 |
| Prostate Cancer | 5 |
| Renal Cancer | 4 |
| Soft Tissue Sarcoma | 2 |
| Thyroid Cancer | 1 |
| Uterine Leiomyoma | 2 |
Table 4.
Summary of the algorithms performed for all datasets
| Algorithms | #Algorithms performed | |
|---|---|---|
| Class comparison | normal vs. tumor | 31 |
| good prognosis vs. poor prognosis | 38 | |
| Survival analysis | 6 | |
Results
Overlaps between hESCGESs genes and tumor-associated genes
In the total of 75 class comparisons and survival analyses, we identified 72 sets of differentially expressed genes significant at 0.05 threshold level (Additional file 5, Table S4). We analyzed the overlap between each of the 72 gene sets and each of the 24 hESC-associated gene sets. We found that they have considerable overlaps. For example, all the 379 genes in the hESC exp1 gene sets of Table 1 appeared in at least one of the 72 differentially expressed gene sets (DEGSs). Among them, 308 genes appeared in 10 or more DEGSs, and 120 genes appeared in 20 or more DEGSs. The most frequently overlapping gene was MTHFD2 (methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase), which occurred in one half the 72 DEGSs. The second most overlapping genes are MCM4 and MCM6 (34 overlaps), two members of the gene family encoding the mini chromosome maintenance complex. All the 40 genes in the hESC exp2 gene sets of Table 1 also occurred in at least one DEGS, and 26 genes occurred in no less than 10 DEGSs. Among them, MYBL2, a member of the MYB family of transcription factor genes involved in cell cycle progression, most frequently occurred in the DEGSs (31 times).
Table 5 gives the number of the genes which have 10 or more overlaps and the top 10 overlapping genes in each of the 24 hESC-associated gene sets, suggesting that a large proportion of the hESC-associated genes are also related to cancer. Gene function enrichment analysis suggests that a substantial portion of the genes listed in Table 5 are involved in cell cycle regulation, DNA damage repair and replication, apoptosis, development and differentiation, cell adhesion and TF activity (Table 6).
Table 5.
Overlaps between the 24 hESC-associated gene sets and the 72 differentially expressed gene sets
| Gene sets | #Genes with 10 or more overlaps b | Top 10 overlapping genes |
|---|---|---|
| hESC exp1 | 308 (81%) | MTHFD2, MCM4, MCM6, LGALS8, PPP2R1B, RFC4, GART, BUB1, LCK, PTPN2 |
| hESC exp2 | 26 (65%) | MYBL2, EPHA1, ORC2, DTYMK, PRKD3, NCAPH, ETV4, DSCC1, CDC25A, PWP2 |
| hESC exp3 | 38 (79%) | BUB1, DLGAP5, SLC16A1, USP9X, HSPA4, TERF1, PSIP1, PLA2G16, UGP2, BMPR1A |
| hESC exp4 | 29 (97%) | SPARC, COL1A2, COL3A1, COL1A1, CD47, COL5A2, KRT18, KRT8, LUM, COL6A3 |
| hESC exp5 | 135 (71%) | TCF4, STAT1, GATA3, MAF, MYC, MYBL2, ILF3, SMAD4, FUBP1, GATA2 |
| hESC exp6 | 28 (64%) | CD9, IL6ST, PTEN, EDNRB, KIT, NR5A2, IFITM2, CRABP2, NFYC, PODXL |
| hESC exp7 | 863 (87%) | GNAS, FN1, SPARC, MCM6, MCM4, TOP2A, COL1A2, COL3A1, RFC4, RAB31 |
| hESC exp8 | 11 (50%) | FAS, CKS1B, GJA1, NPM1, TGIF1, HMGA1, DNMT3A, ERH, SOX2, DNMT3B |
| hESC exp9 | 21 (78%) | PCNA, CKS1B, FAS, TERF1, GJA1, CCNB1, NPM1, FZD7, SFRP1, HMGA1 |
| hESC exp10 | 30 (97%) | PGK1, BAK1, HNRNPA1, EEF1A1, PPIA, GAPDH, GJA1, TMED2, EEF1B2, NPM1 |
| hESC exp11 | 749 (75%) | PDE4DIP, FGFR2, CKS2, MTHFD2, SOX4, SLC2A3, STAT1, MAPK1, PSMB2, MCM4 |
| Nanog targets | 711 (72%) | CALD1, FGFR2, CKS2, TNPO1, KIAA0101, SPARC, TOP2A, ARHGAP1, B2M, VCAN |
| Oct4 targets | 211 (73%) | FGFR2, TCF4, KIAA0101, TOP2A, PPP2R1B, TCF12, UBE2D3, RAB5A, HMGB2, PTPN2 |
| Sox2 targets | 542 (74%) | FGFR2, TOP2A, ARHGAP1, VCAN, PPP2R1B, UBE2D3, CBX3, RAB5A, PIK3R3, H2AFX |
| NOS targets | 132 (74%) | FGFR2, TOP2A, PPP2R1B, UBE2D3, RAB5A, PTPN2, SET, FGFR1, BUB3, ADD3 |
| NOS TFs | 28 (76%) | STAT3, MYST3, IFI16, MLLT10, FOXO1, PHF17, ZFP36L1, TAF12, HHEX, ZEB2 |
| NOS co-bound | 199 (56%) | FGFR2, TOP2A, PPP2R1B, UBE2D3, RAB5A, PTPN2, SET, FGFR1, BUB3, BMI1 |
| Suz12 targets | 512 (49%) | PDE4DIP, BCL2, GNAS, PTGER3, CD44, RAB31, CYP1B1, EPHB1, GATA3, HLF |
| Eed targets | 510 (48%) | PDE4DIP, BCL2, PTGER3, PRKCB, CD44, INPP4A, CYP1B1, EPHB1, GATA3, HLF |
| H3K27 bound | 557 (50%) | PDE4DIP, BCL2, PTGER3, PRKCB, EPHB1, GATA3, NTRK2, CD47, GPD1L, NCAM1 |
| PRC2 targets | 397 (61%) | PDE4DIP, BCL2, PTGER3, EPHB1, GATA3, NTRK2, NCAM1, CACNA1D, GATA2, GPM6B |
| Myc targets1 | 204 (89%) | BCL2, MCM4, TGFB3, CCND2, APC, MUC1, TCF12, ENO1, APP, CSTB |
| Myc targets2 | 645 (83%) | CKS2, PPP2R1B, TCF12, UBE2D3, CDC25B, H2AFX, HLA-A, PDK3, PRKDC, TMF1 |
| P53 targets | 27 (77%) | GATM, MYH11, TP53, BTG2, INPP5D, MDM2, MYB, SPARCL1, PLK4, FMO5 |
b The percentage of the overlapping gene number relative to the total gene number for each of the 24 hESC-associated gene sets is given in parenthesis.
Table 6.
Functional categories of the genes listed in Table 5
| Functional categories | Representative genes |
|---|---|
| Cell cycle regulation | ORC2, NCAPH, DSCC1, CDC25A, CDC25B, CKS2, CKS1B, BUB3, CCND2, CCNB1, DLGAP5 |
| DNA repair/replication | TOP2A, MCM4, MCM6, RFC4, PCNA, PPIA, HMGB2, H2AFX, PRKDC |
| Apoptosis | FAS, BAK1, IFI16, PHF17, BTG2, PSMB2, BCL2, APC, TP53 |
| Development/differentiation | ETV4, FGFR1, FGFR2, VCAN, STAT3, HHEX, EPHB1, NTRK2, GPM6B, TGFB3, BMI1, SOX2, DNMT3B, SOX4 |
| Cell adhesion | VCAN, CD9, CD44, CD47, NCAM1, COL6A3, PODXL, FN1 |
| TF activity | GATA2, TMF1, STAT1, STAT3, MAF, FUBP1, MYB, NFYC, TGIF1, SOX2, ZEB2, TAF12, ZFP36L1, MLLT10, MYST3, TCF4, TCF12, ETV4, MYBL2 |
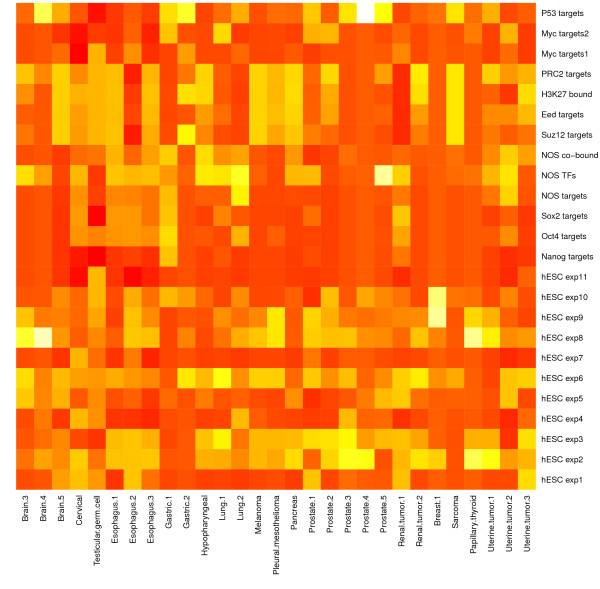
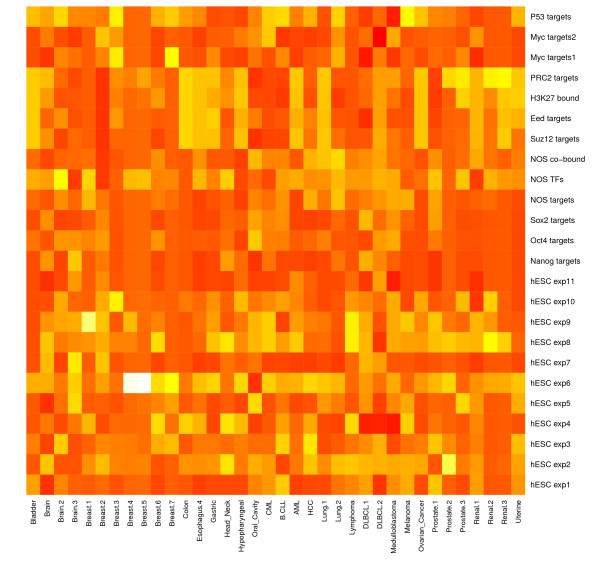
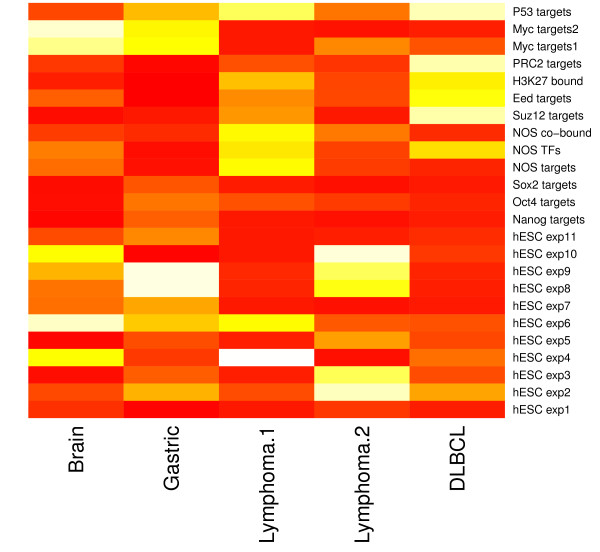
We carried out significance analyses of the overlapping gene sets between each of the 72 DEGSs and each of the 24 hESC-associated gene sets based on the hypergeometric test. Three heatmaps of hypergeometric p-values are presented in Figure 1, Figure 2 and 3, which visualize the significance of the overlap between the hESC-associated gene sets and the DEGSs among normal vs. tumor, good prognosis vs. poor prognosis phenotypes classes, and survival analysis, respectively (the detailed description of all the datasets related to each figure is provided in Additional file 6). These figures show that the targets of three core hESC-associated TF OCT4, SOX2 and NANOG have significant overlaps with most of the DEGSs. Two gene sets targeted by MYC also shows significant overlaps with most of the DEGSs. These results suggest that key hESC-associated gene expression signatures have important implications in pathogenesis of cancer.
Figure 1.
Significance of overlap between hESC and tumor gene sets by normal vs. tumor class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure 2.
Significance of overlap between hESC and tumor gene sets by good vs. poor prognosis class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure 3.
Significance of overlap between hESC and tumor gene sets by survival analysis. The detailed description of all the datasets is provided in Additional file 6.
Overlaps between hESCGESs pathways and tumor-associated pathways
In the total of 75 class comparison and survival analyses, we identified 68 groups of pathways significant at 0.05 threshold level. Among the 54 hESC-associated signal pathways signatures, 26 pathways appeared at least in eight different groups and the other 28 pathways didn't appear in any group. The most frequent identified pathway was the Cell Cycle pathway, which appeared for 57 times (84% occurrence rate), and the next one was the MAPK pathway which was identified for 50 times (74% occurrence rate). Table 7 lists all the 26 pathways and their occurrence frequencies in the 68 groups of pathways significant in the cancer datasets. These pathways have been proven to play important roles in both maintenance of hESC function and tumorigenesis.
Table 7.
Twenty-six hESC-associated pathways frequently identified in tumors
| Pathway | Frequency |
|---|---|
| Cell Cycle | 57 |
| MAPK | 50 |
| IGF | 31 |
| EGF/EGFR | 30 |
| ERK | 29 |
| SHH | 28 |
| AKT | 26 |
| RAS | 25 |
| NF-κB | 22 |
| Telomerase | 22 |
| p53 | 21 |
| WNT | 20 |
| PRC2 | 19 |
| ALK | 16 |
| NOTCH | 15 |
| ATM | 14 |
| VEGF | 14 |
| PDGF | 13 |
| ERBB2 | 11 |
| JAK/STAT | 11 |
| PI3K | 11 |
| PTEN | 11 |
| TGFβ | 11 |
| MEK | 10 |
| STAT3 | 9 |
| Glycolysis | 8 |
Clearly, the Cell Cycle pathway plays an extremely important role in regulation of the self-renewal and pluripotency process of hESCs [55-59]. The undifferentiated hESCs have a short G1 phase, and therefore show rapid cell cycle characteristic relative to differentiated somatic cells. The unorthodox G1/S phase transition feature in the hESC cell cycle is associated with the deregulated proliferation and differentiation blockades of tumor cells [39,60-65]. The MAPK (Mitogen-Activated Protein Kinase) pathway regulates both the early embryonic development and the embryonic stem cell commitment from early steps of the process to mature differentiated cells [66]. The role of MAPK pathway in cancer is prominent as cancer can be perceived as a disease of communication between and within cells. The statistical significance analysis also shows that both the Cell Cycle pathway and MAPK pathway have important association with a majority of tumor types (see Additional file 7, Figure S1, Additional file 8, Figure S2 and Additional file 9, Figure S3).
The importance of IGF signaling pathway for maintenance of hESCs has been proven [67-70]. This signaling pathway appears to play a crucial role in cancer and can be of potential interest in cancer therapy [71-77]. The ERK pathway is active in the undifferentiation status of hESCs. Its activation is critical in maintenance of hESC self-renewal [78-81]. On the other hand, there has been accumulating evidence of ERK pathway (RAF-MEK-ERK signaling cascade) in oncogenesis to make it an attractive target for drug development [82].
Interestingly, almost all the widely-recognized hESC-associated pathways such as SHH, WNT, PRC2, Notch, PTEN and TGFβ have important linkage with cancer (see Table 7). The SHH (Sonic Hedgehog) signaling pathway is one of the key regulators of human embryonic development [83-87]. Activation of the pathway leads to an increased risk of the development of cancerous malignancies [87-94]. The WNT signaling pathway is a network of a number of proteins acting as a critical regulator of hESCs [43,56,59,69,79,84,85,95-103]. However, the deregulation of the pathway has been closely associated with cancer [83,86,90,94,103-114]. The PRC2 (Polycomb Repressive Complex 2) pathway is involved in control of the developmental regulators in hESCs [50,56,115-118]. The expression of PRC2 components is upregulated in various cancers such as melanoma, lymphoma, and breast and prostate cancer. The Notch signaling pathway plays a key role in the normal development of hESCs and many other cell types depending on the expression level and cellular context of the Notch receptors [84,85,101,119]. Its deregulation potentially contributes to cancer development in several different ways [111,120-126]. The PTEN (PhosphaTase and Tensin Homolog) acts as a tumor suppressor gene involved in regulation of the cell cycle, preventing cells from growing and dividing too rapidly. This pathway is also critical for stem cell maintenance [59,69,83]. The TGFβ (Transforming Growth Factor β) signaling pathway is of central importance to the self renewal of hESCs [43,59,69,79,84,85,96,98-102,115,127,128]. This signal pathway is involved in a wide range of cellular processes in both the adult organism and the developing embryo. It plays a role in both tumor suppression and tumor progression depending on cellular context [129-132].
Additional two important pathways involved in both hESCs function and tumorigenesis are p53 and telomerase pathways. They were identified for 21 and 22 times in our 68 class comparison or survival analysis (see Table 7). The p53 pathway can maintain the homeostasis of self-renewal and differentiation of hESCs [133-135]. Inactivation of this pathway in several cancer types may correlates with hESC-specific signatures [22,136,137]. Telomerase enzyme levels or activity has shown to be highly expressed in embryonic stem cells [79]. On the other hand, telomerase is reactivated and serves to maintain telomere length in most advanced cancers [34].
Taken together, the high overlap between hESCGESs pathways and tumor-associated pathways reveals that there exist common mechanisms underlying cancerous malignancies and "stemness" of hESCs.
Overlaps between hESCGESs TFs and tumor-associated TFs
We identified 73 groups of targets of TFs significant at 0.05 threshold level. Among the 189 hESC-associated TF signatures, 42 TFs appeared at least in three different groups and the others didn't show in any group. The most frequently identified TF was MYC with 56% occurrence rate (41 occurrences), and the next one was MYB with 51% occurrence rate (37 occurrences). The complete 42 TFs accompanying with their occurrence frequencies are presented in Table 8.
Table 8.
Forty-two hESC-associated TFs frequently identified in tumors
| TF | Frequency |
|---|---|
| MYC | 41 |
| MYB | 37 |
| SP1 | 33 |
| TP53 | 33 |
| E2F4 | 33 |
| TFAP2A | 32 |
| E2F1 | 32 |
| JUN | 30 |
| SMAD1 | 27 |
| TAL1 | 24 |
| NFKB1 | 23 |
| STAT3 | 23 |
| MYBL2 | 22 |
| ETS1 | 21 |
| ETS2 | 20 |
| POU2F1 | 20 |
| POU2F2 | 18 |
| STAT1 | 18 |
| WT1 | 18 |
| ETV4 | 18 |
| HOXA9 | 17 |
| SMAD3 | 17 |
| E2F2 | 17 |
| SP3 | 16 |
| LEF1 | 14 |
| NFKB2 | 14 |
| POU5F1 | 13 |
| PAX6 | 11 |
| STAT4 | 11 |
| SMAD4 | 10 |
| GLI1 | 10 |
| PAX3 | 9 |
| PAX2 | 8 |
| MYBL1 | 8 |
| PAX5 | 7 |
| SMAD2 | 7 |
| GLI2 | 6 |
| PAX8 | 6 |
| STAT2 | 6 |
| GLI3 | 3 |
| POU3F2 | 3 |
| E2F3 | 3 |
From Table 8, we can see a number of "stemness" TFs identified as informative in tumors. Evidently, MYC is one of the most important TFs in both hESCs and Cancer cells [22,23,44,48,49,52,56,116,138-140]. MYC represses differentiation and maintains the self-renewal of mouse and human pluripotent stem cells [138,141]. MYC regulatory networks may account for most of the transcriptional similarity between embryonic stem cells and cancer cells [139]. The statistical significance analysis also shows that MYC plays an important role in most of the tumor types analyzed (see Additional file 10, Figure S4, Additional file 11, Figure S5 and Additional file 12, Figure S6).
Another extremely important TF is POU5F1 (OCT4), which is necessary for induction of pluripotent stem cells from human somatic cells [23,24]. OCT4 constitutes the core transcriptional regulatory circuitry in hESCs in combination with SOX2 and NANOG essentially responsible for the early development and propagation of undifferentiated hESCs [43,44,52,56,58,59,79,84,97,116,117,119,142,143]. OCT4 expression appears to be important in maintaining the undifferentiated state of embryonal carcinoma [86,144], as well as in other cancers [27,145].
Our analysis results suggest that several families of hESC-associated TFs like MYB, E2F, PAX, SMAD, STAT, POU, SP and GLI, are related to cancer (Table 8). For example, three members of the TF family MYB: MYB, MYBL1 and MYBL2, appear to be closely associated with cancer (Table 8). In fact, a substantial number of studies have revealed that they had important roles in regulation of stem cell self-renewal and differentiation [146,147], and the development of cancer [148,149]. E2F plays a crucial role in control of the cell cycle progression and regulating the expression of genes required for G1/S transition [150], and therefore is important for stem cell self-renewal and differentiation. The members of the family E2F1, 2, 3 and E2F4 have been reported to be associated with cancer [151]. PAX plays an essential role in regulating cell proliferation and self-renewal, resistance to apoptosis, migration of embryonic precursor cells, and the coordination of specific differentiation programs during embryonic development [59], as well as the development of cancer [152]. SMAD regulates cell proliferation and differentiation by activating downstream TGFß gene transcription. Its members play important roles in hESC fate determination [98], and cancerous pathogenesis [153]. STAT regulates cell growth, survival and differentiation via activation by JAK (Janus kinase). This pathway is critical for regulation of stem cell self-renewal and differentiation [101]. Deregulation of this pathway is frequently observed in various tumor types [154]. POU mainly regulate the development of an organism, and are also involved in various cancers [155]. SP1 and SP3 are two members of the TF family SP (Specificity Protein) which binds GC-rich DNA sequences. Their roles in hESCs and cancer cells have been widely recognized [26]. GLI encompasses three members: GLI1, GLI2 and GLI3, all of which mediate the Hedgehog pathway and therefore are involved in hESC fate determination and cancerous pathogenesis [87].
In summary, the substantial overlap between the TFs involved in hESC fate determination and the TFs involved in cancerous pathogenesis suggests that hESCs and cancer cells may share essential regulatory mechanisms.
Overlaps between hESCGESs miRNAs and Tumor-associated miRNAs
We identified 67 groups of miRNA targets significant at 0.05 threshold level. Among the 114 hESC-associated miRNA signatures, 102 miRNAs appeared at least in eight different groups and the other 12 miRNAs didn't show in any group. The most frequently identified miRNA was miR-29c, which occurred for 34 times (51% occurrence rate), and the next one was miR-200b which occurred for 30 times (45% occurrence rate). Table 9 lists 50 miRNAs whose occurrence frequencies are no less than 20. The complete 102 miRNAs accompanying with their occurrence frequencies are presented in Additional file 13, Table S5.
Table 9.
Fifty hESC-associated miRNAs frequently identified in tumors
| miRNA | Frequency |
|---|---|
| miR-29c | 34 |
| miR-200b | 30 |
| miR-19b | 29 |
| miR-29a | 29 |
| miR-29b | 29 |
| let-7a | 28 |
| miR-520f | 28 |
| miR-21 | 27 |
| miR-302c | 27 |
| miR-302d | 27 |
| miR-494 | 27 |
| miR-518b | 27 |
| miR-519c | 27 |
| miR-520a | 27 |
| miR-200c | 26 |
| miR-26a | 26 |
| miR-302a | 26 |
| miR-30d | 26 |
| miR-124a | 25 |
| miR-16 | 25 |
| miR-19a | 25 |
| miR-302b | 25 |
| miR-374 | 25 |
| miR-518c | 25 |
| miR-519b | 25 |
| miR-15a | 24 |
| miR-15b | 24 |
| miR-18a | 24 |
| miR-20b | 24 |
| miR-301 | 24 |
| miR-520b | 24 |
| miR-520c | 24 |
| miR-130a | 23 |
| miR-18b | 23 |
| miR-369-3p | 23 |
| miR-520d | 23 |
| miR-520e | 23 |
| miR-103 | 22 |
| miR-154 | 22 |
| miR-20a | 22 |
| miR-525 | 22 |
| miR-93 | 22 |
| miR-17-5p | 21 |
| miR-302c* | 21 |
| miR-470 | 21 |
| miR-515-5p | 21 |
| miR-517c | 21 |
| miR-106a | 20 |
| miR-146b | 20 |
| miR-96 | 20 |
Notably, there is a broad range of overlap between "stemness" miRNAs and oncogenic miRNAs. Most of the important "stemness" miRNAs are presented in Table 9 or Table S5. The miR-302 cluster miRNAs (miR-302a, miR-302a*, miR-302b, miR-302b*, miR-302c, miR-302c*, miR-302d) have been shown to regulate important cellular functions in hESCs, including cell proliferation and chromatin structure, and have been consistently reported to be overexpressed in hESCs [156]. All the seven members of this group appear in Table S5, and five of them are also presented in Table 9, indicative of their close linkage with cancer. Some literatures have reported the relatedness between miRNA-302 family and tumorigenecity [157-160]. Another group of miR-200 family miRNAs (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) have been revealed to be hESC-specific, and upregulated in hESCs [156,161,162]. Three of them are presented in Table S5 and miR-200b and miR-200c are also listed in Table 9 with relatively high frequencies (30 and 26, respectively), strongly indicating their association with cancer. In fact, this miRNA family plays an important role in cancerous pathogenesis [163-165]. The miRNA-520 cluster on chromosome 19 was highly expressed in undifferentiated hESCs, and might be closely involved in hESC function [156,166]. Its eight members miRNA-520a-h show in Table S5 and six members miRNA-520a-f also show in Table 9, suggesting that the miRNA family has tight connection with cancer. Many studies have revealed the relatedness between its members and cancer [167-170]. The miR-518b, miR-518c, miR-519b and miR-519c have been consistently reported to be overexpressed in undifferentiated hESCs [156,166,171,172]. Our analysis outcomes suggest that they may be closely involved in the development of cancer (Table 9). This finding is supported by some studies [173,174]. In addition, the other miRNA families shown in Table 9 like miRNA-29, 19, 15, 20 and let-7 have been revealed to be involved in both hESC fate determination and cancerous pathogenesis [53,96,161,175].
The statistical significance analysis shows that some "stemness" miRNAs like miR-29 family member miR-29a, miR-29b and miR-29c are associated with a broad spectrum of tumor types (see Additional file 14, Figure S7, Additional file 15, Figure S8 and Additional file 16, Figure S9).
Taken together, a number of miRNAs play crucial roles in both hESC fate determination and tumorigenicity.
Discussion
Although the evidence strongly supporting the CSC theory remains insufficient, and the fundamental experimental evidence for CSCs based on mouse xenograft models are controversial [21], the CSC model is attractive for it provides reasonable explanation of the development mechanisms underlying cancer, as well as a promise of improved cancer therapies. Therefore, any proof in favor of the CSC theory is valuable in the biology of cancer.
In this study, we provided an indirect evidence for the CSC theory using the computational biology approach. We found a strong linkage between hESCs and cancer cells by an examination of the similarity between the hESC-specific gene expression profiles and cancer-specific gene expression profiles. The hESC-specific gene expression signatures including genes, pathways, TFs and miRNAs were generally differentially expressed among normal vs. tumor phenotypes, or among cancer subtypes with distinct clinical outcomes. The genes important in regulation of hESC self-renewal and differentiation such as SOX2 and MYB, were also closely involved in tumorigenicity. The signal pathways such as the Cell Cycle, MAPK, SHH, WNT, PRC2, Notch, PTEN and TGFβ involved in the hESC fate determination were also strongly associated with cancer genesis, progression and prognosis. The typical hESC-specific TFs like OCT4 and c-Myc (also known as MYC), appeared to be important in control of the undifferentiated state of cancer cells. The miRNAs overexpressed in undifferentiated hESCs like miRNA-302, 200 and 520 cluster miRNAs, were closely involved in the development of cancer.
Generally speaking, the cell cycle regulation mechanism mostly underlies the commonality between hESCs and cancer cells. Differing from somatic cells, hESCs have an abbreviated G1 phase in cell cycle, which is critical for maintenance of hESC self-renewal and pluripotency. The abbreviated G1 phase is also largely responsible for the uncontrolled proliferation of tumor cells which escape from the programmed cell death during the G1 phase [62]. In fact, the hESC-associated signatures most frequently identified in tumors are mainly involved in regulation of cell cycle (see Table 6, Table 7, Table 8 and Table 9). Among them, the TF c-Myc is the core signature connecting hESCs with cancer cells. c-Myc binds genic and intergenic regions to regulate the expression of thousands of genes and noncoding RNAs throughout the genome [138]. c-Myc is involved in the cell cycle regulation by directly regulating cell cycle regulators [44,116,138], or regulating miRNAs which inhibit cell cycle regulators [96,138]. The role of c-Myc in linking hESCs with cancer has been recognized [138,139].
Here we identified differentially expressed genes at 0.05 significance level. A more stringent significance threshold of 0.001 would be more statistically reasonable if considering corrections of multiple hypotheses. Because the numbers of significant pathways, TFs and miRNAs identified by analyses of gene sets would be small for a majority of datasets if the significance threshold of 0.001 were used under which the number of differentially expressed genes were still often substantial, we selected the 0.05 significance level for all the differentially expressed analyses in order to keep consistency.
One limitation of this study was that the analyses were mainly based on the computational biology approach which needs experimental validation to corroborate these findings. In addition, some finer analyses such as grouping the overlaps of gene signatures between hESCs and tumors according to different tumor categories, separating the differentially expressed genes into the overexpressed and underexpressed genes etc., may contribute to a better understanding of the similarities between hESCs and tumor cells in gene expression profiles. Another limitation of this study was that we identified tumor-associated gene expression signatures based on whole tumor samples which might be derived from the majority of tumor cells, not necessarily from the minority of CSCs so that the overlapping signatures identified between hESCs and tumors might not be able to provide a strong support for the CSC model. If the tumor-associated gene expression signatures were identified by comparison between isolated CSCs versus non-CSCs fraction of the same tumor, the same results would be more reliable in support of the CSC model. These issues could be addressed in future research.
A further problem is the intertwined relationships between stem cell, cancer and ageing [176]. Cancer is actually an age-related disease as the incidence of cancer grows exponentially with ageing. Meanwhile, ageing is mostly caused by a decline in the replicative function of stem cell [177], and in turn aging has effects on the function of stem cell [178]. Thus, an in-depth investigation of the molecular mechanisms that connect stem cell, cancer and ageing will be necessary for postponing ageing and overcoming cancer.
Conclusions
The present results revealed the close linkage between the hESC-specific gene expression profiles and cancer-specific gene expression profiles, and therefore offered an indirect support for the CSC theory. However, many interest issues remain to be addressed further.
Availability of supporting data
The 51 human cancer gene expression datasets are available at the following website: http://linus.nci.nih.gov/~brb/DataArchive_New.html. All the other datasets supporting the results of this article are included within the article and its additional files.
Competing interests
The author declares that he has no competing interests.
Authors' contributions
XW designed and performed the research. XW wrote the manuscript.
Supplementary Material
Table S1. 24 hESC-associated gene sets.
Table S2. 189 hESC-associated transcription factors.
Table S3. 114 hESC-associated microRNAs.
Supplementary references. The list of references for the hESC-associated gene expression signatures identified and human tumor gene expression datasets used.
Table S4. 72 differentially expressed gene sets.
Figure S1. Significance of overlap between hESC and tumor pathways by normal vs. tumor class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S2. Significance of overlap between hESC and tumor pathways by good vs. poor prognosis class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S3. Significance of overlap between hESC and tumor pathways by survival analysis. The detailed description of all the datasets is provided in Additional file 6.
Figure S4. Significance of overlap between hESC and tumor TFs by normal vs. tumor class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S5. Significance of overlap between hESC and tumor TFs by good vs. poor prognosis class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S6. Significance of overlap between hESC and tumor TFs by survival analysis. The detailed description of all the datasets is provided in Additional file 6.
Table S5. 102 miRNAs identified at least in eight different groups.
Figure S7. Significance of overlap between hESC and tumor miRNAs by normal vs. tumor class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S8. Significance of overlap between hESC and tumor miRNAs by good vs. poor prognosis class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S9. Significance of overlap between hESC and tumor miRNAs by survival analysis. The detailed description of all the datasets is provided in Additional file 6.
Acknowledgements
The author thanks Dr. Richard Simon for his critical reading of this manuscript and very valuable comments which helped the author to improve the manuscript.
References
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CHM, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE, Herlyn M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Zhang S, Balch C, Chan MW, Lai H-C, Matei D, Schilder JM, Yan PS, Huang THM, Nephew KP. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68(11):4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65(8):3025–3029. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4(2):141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Gibbs CP, Kukekov VG, Reith JD, Tchigrinova O, Suslov ON, Scott EW, Ghivizzani SC, Ignatova TN, Steindler DA. Stem-like cells in bone sarcomas: implications for tumorigenesis. Neoplasia. 2005;7(11):967–976. doi: 10.1593/neo.05394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nature medicine. 2009;15(9):1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science (New York, N Y) 2007;317(5836):337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- Kennedy JA, Barabe F, Poeppl AG, Wang JCY, Dick JE. Comment on "Tumor growth need not be driven by rare cancer stem cells". Science (New York, N Y) 2007;318(5857):1722. doi: 10.1126/science.1149590. author reply 1722. [DOI] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11(4):268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, N Y) 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16(4):318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Lambertini C, Pantano S, Dotto GP. Differential control of Notch1 gene transcription by Klf4 and Sp3 transcription factors in normal versus cancer-derived keratinocytes. PLoS One. 2010;5(4):e10369. doi: 10.1371/journal.pone.0010369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29(14):2153–2159. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- Ji J, Zheng P-S. Expression of Sox2 in human cervical carcinogenesis. Hum Pathol. 2010;41(10):1438–1447. doi: 10.1016/j.humpath.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Chiou S-H, Yu C-C, Huang C-Y, Lin S-C, Liu C-J, Tsai T-H, Chou S-H, Chien C-S, Ku H-H, Lo J-F. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14(13):4085–4095. doi: 10.1158/1078-0432.CCR-07-4404. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JCI. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. Telomeres, stem cells, senescence, and cancer. J Clin Invest. 2004;113(2):160–168. doi: 10.1172/JCI20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehezkel S, Rebibo-Sabbah A, Segev Y, Tzukerman M, Shaked R, Huber I, Gepstein L, Skorecki K, Selig S. Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics. 2011;6(1):63–75. doi: 10.4161/epi.6.1.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA. 1994;91(21):9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31(1):9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Medina R, Lengner CJ, Mandeville M, Qiao M, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Reprogramming the pluripotent cell cycle: restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. J Cell Physiol. 2011;226(5):1149–1156. doi: 10.1002/jcp.22440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5(2):141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neganova I, Zhang X, Atkinson S, Lako M. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene. 2009;28(1):20–30. doi: 10.1038/onc.2008.358. [DOI] [PubMed] [Google Scholar]
- Collins K, Jacks T, Pavletich NP. The cell cycle and cancer. Proc Natl Acad Sci USA. 1997;94(7):2776–2778. doi: 10.1073/pnas.94.7.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266(5192):1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang AX, Joshi BH, Ginis I, Thies RS, Amit M, Lyons I, Condie BG, Itskovitz-Eldor J, Rao MS, Puri RK. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103(8):2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RAR, Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13(6):601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- Sperger JM, Chen X, Draper JS, Antosiewicz JE, Chon CH, Jones SB, Brooks JD, Andrews PW, Brown PO, Thomson JA. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci USA. 2003;100(23):13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou M-F, de Tribolet N, Regli L, Wick W, Kouwenhoven MCM, Hainfellner JA, Heppner FL, Dietrich P-Y, Zimmer Y, Cairncross JG, Janzer R-C, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related "self-renewal" signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- Santagata S, Ligon KL, Hornick JL. Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am J Surg Pathol. 2007;31(6):836–845. doi: 10.1097/PAS.0b013e31802e708a. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40(5):499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan KA, Chen G, Kalemkerian GP, Wicha MS, Beer DG. An embryonic stem cell-like signature identifies poorly differentiated lung adenocarcinoma but not squamous cell carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15(20):6386–6390. doi: 10.1158/1078-0432.CCR-09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenhals M, Kassambara A, De Vos J, Hose D, Moreaux J, Klein B. Embryonic stem cell markers expression in cancers. Biochem Biophys Res Commun. 2009;383(2):157–162. doi: 10.1016/j.bbrc.2009.02.156. [DOI] [PubMed] [Google Scholar]
- Glinsky GV. "Stemness" genomics law governs clinical behavior of human cancer: implications for decision making in disease management. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26(17):2846–2853. doi: 10.1200/JCO.2008.17.0266. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer Inform. 2008;6:9–15. doi: 10.4137/cin.s448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10(2):116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Informatics. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23(7):837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, Mongan NP. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18(7):1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DAG, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Molecular and cellular biology. 2008;28(20):6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Peterson H, Chavez L, Kahlem P, Lehrach H, Vilo J, Adjaye J. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS One. 2010;5(5):e10709. doi: 10.1371/journal.pone.0010709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li H, Liu Y, Mattson MP, Rao MS, Zhan M. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PLoS One. 2008;3(10):e3406. doi: 10.1371/journal.pone.0003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends in cell biology. 2002;12(9):432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Miyazaki H, Shiozaki A, Niisato N, Ohsawa R, Itoi H, Ueda Y, Otsuji E, Yamagishi H, Iwasaki Y, Nakano T, Nakahari T, Marunaka Y. Chloride ions control the G1/S cell-cycle checkpoint by regulating the expression of p21 through a p53-independent pathway in human gastric cancer cells. Biochem Biophys Res Commun. 2008;366(2):506–512. doi: 10.1016/j.bbrc.2007.11.144. [DOI] [PubMed] [Google Scholar]
- Massague J. G1 cell-cycle control and cancer. Nature. 2004;432(7015):298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- Michalides RJAM, van de Brekel M, Balm F. Defects in G1-S cell cycle control in head and neck cancer: a review. Head Neck. 2002;24(7):694–704. doi: 10.1002/hed.10109. [DOI] [PubMed] [Google Scholar]
- Niklinska W, Laudanski J, Niklinski J. G1 phase of the cell cycle control and lung cancer: biological and clinical implications. Minireview. Neoplasma. 2001;48(3):157–163. [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1(3):222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- Binetruy B, Heasley L, Bost F, Caron L, Aouadi M. Concise review: regulation of embryonic stem cell lineage commitment by mitogen-activated protein kinases. Stem Cells. 2007;25(5):1090–1095. doi: 10.1634/stemcells.2006-0612. [DOI] [PubMed] [Google Scholar]
- Wang L, Schulz TC, Sherrer ES, Dauphin DS, Shin S, Nelson AM, Ware CB, Zhan M, Song C-Z, Chen X, Brimble SN, McLean A, Galeano MJ, Uhl EW, D'Amour KA, Chesnut JD, Rao MS, Blau CA, Robins AJ. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110(12):4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, Ramos-Mejia V, Rouleau A, Yang J, Bosse M, Lajoie G, Bhatia M. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448(7157):1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- Brandenberger R, Khrebtukova I, Thies RS, Miura T, Jingli C, Puri R, Vasicek T, Lebkowski J, Rao M. MPSS profiling of human embryonic stem cells. BMC Dev Biol. 2004;4:10. doi: 10.1186/1471-213X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celil AB, Campbell PG. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. The Journal of biological chemistry. 2005;280(36):31353–31359. doi: 10.1074/jbc.M503845200. [DOI] [PubMed] [Google Scholar]
- Velcheti V, Govindan R. Insulin-like growth factor and lung cancer. J Thorac Oncol. 2006;1(7):607–610. [PubMed] [Google Scholar]
- Izycki T, Chyczewska E, Naumnik W, Talalaj J, Panek B, Ossolinska M. Serum levels of IGF-I and IGF-II in patients with lung cancer during chemotherapy. Exp Oncol. 2004;26(4):316–319. [PubMed] [Google Scholar]
- Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol. 2011;226(4):1118–1127. doi: 10.1002/jcp.22433. [DOI] [PubMed] [Google Scholar]
- Guvakova MA, Surmacz E. Tamoxifen interferes with the insulin-like growth factor I receptor (IGF-IR) signaling pathway in breast cancer cells. Cancer Res. 1997;57(13):2606–2610. [PubMed] [Google Scholar]
- Anisimov VN. Insulin/IGF-1 signaling pathway driving aging and cancer as a target for pharmacological intervention. Exp Gerontol. 2003;38(10):1041–1049. doi: 10.1016/s0531-5565(03)00169-4. [DOI] [PubMed] [Google Scholar]
- Shukla V, Coumoul X, Vassilopoulos A, Deng C-X. IGF signaling pathway as a selective target of familial breast cancer therapy. Curr Mol Med. 2008;8(8):727–740. doi: 10.2174/156652408786733694. [DOI] [PubMed] [Google Scholar]
- Vanamala J, Reddivari L, Radhakrishnan S, Tarver C. Resveratrol suppresses IGF-1 induced human colon cancer cell proliferation and elevates apoptosis via suppression of IGF-1R/Wnt and activation of p53 signaling pathways. BMC Cancer. 2010;10:238. doi: 10.1186/1471-2407-10-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HB, Kim JS, Kwon H-J, Nam KH, Youn HS, Sok D-E, Lee Y. Basic fibroblast growth factor activates ERK and induces c-fos in human embryonic stem cell line MizhES1. Stem Cells Dev. 2005;14(4):395–401. doi: 10.1089/scd.2005.14.395. [DOI] [PubMed] [Google Scholar]
- Liu N, Lu M, Tian X, Han Z. Molecular mechanisms involved in self-renewal and pluripotency of embryonic stem cells. J Cell Physiol. 2007;211(2):279–286. doi: 10.1002/jcp.20978. [DOI] [PubMed] [Google Scholar]
- Levenstein ME, Ludwig TE, Xu R-H, Llanas RA, VanDenHeuvel-Kramer K, Manning D, Thomson JA. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells. 2006;24(3):568–574. doi: 10.1634/stemcells.2005-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak P, Dvorakova D, Koskova S, Vodinska M, Najvirtova M, Krekac D, Hampl A. Expression and potential role of fibroblast growth factor 2 and its receptors in human embryonic stem cells. Stem Cells. 2005;23(8):1200–1211. doi: 10.1634/stemcells.2004-0303. [DOI] [PubMed] [Google Scholar]
- Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283(2):125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3(12):895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J. Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2007;25(2):500–510. doi: 10.1634/stemcells.2006-0426. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13(14):4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- Sell S. Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol. 2004;51(1):1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn N-s, Jackson KW, Suri P, Wicha MS. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer research. 2006;66(12):6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Guicherit OM, Zaharian BI, Xu Y, Chai L, Wichterle H, Kon C, Gatchalian C, Porter JA, Rubin LL, Wang FY. Identification of a small molecule inhibitor of the hedgehog signaling pathway: effects on basal cell carcinoma-like lesions. Proc Natl Acad Sci USA. 2003;100(8):4616–4621. doi: 10.1073/pnas.0732813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Yue W, Wei B, Wang N, Li T, Guan L, Shi S, Zeng Q, Pei X, Chen L. Sonic hedgehog pathway is essential for maintenance of cancer stem-like cells in human gastric cancer. PLoS One. 2011;6(3):e17687. doi: 10.1371/journal.pone.0017687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimeault M, Hauke R, Mehta PP, Batra SK. Recent advances in cancer stem/progenitor cell research: therapeutic implications for overcoming resistance to the most aggressive cancers. J Cell Mol Med. 2007;11(5):981–1011. doi: 10.1111/j.1582-4934.2007.00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher FC. Hedgehog signaling and therapeutics in pancreatic cancer. Carcinogenesis. 2011;32(4):445–451. doi: 10.1093/carcin/bgq280. [DOI] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, de Sauvage FJ. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391(6662):90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- Wicking C, McGlinn E. The role of hedgehog signalling in tumorigenesis. Cancer Lett. 2001;173(1):1–7. doi: 10.1016/s0304-3835(01)00676-0. [DOI] [PubMed] [Google Scholar]
- Yanai K, Nakamura M, Akiyoshi T, Nagai S, Wada J, Koga K, Noshiro H, Nagai E, Tsuneyoshi M, Tanaka M, Katano M. Crosstalk of hedgehog and Wnt pathways in gastric cancer. Cancer Lett. 2008;263(1):145–156. doi: 10.1016/j.canlet.2007.12.030. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10(1):55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Mallanna SK, Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344(1):16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez L, Bais AS, Vingron M, Lehrach H, Adjaye J, Herwig R. In silico identification of a core regulatory network of OCT4 in human embryonic stem cells using an integrated approach. BMC genomics. 2009;10:314. doi: 10.1186/1471-2164-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132(6):1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Besser D. Expression of nodal, lefty-a, and lefty-B in undifferentiated human embryonic stem cells requires activation of Smad2/3. The Journal of biological chemistry. 2004;279(43):45076–45084. doi: 10.1074/jbc.M404979200. [DOI] [PubMed] [Google Scholar]
- Rao RR, Stice SL. Gene expression profiling of embryonic stem cells leads to greater understanding of pluripotency and early developmental events. Biol Reprod. 2004;71(6):1772–1778. doi: 10.1095/biolreprod.104.030395. [DOI] [PubMed] [Google Scholar]
- Rho J-Y, Yu K, Han J-S, Chae J-I, Koo D-B, Yoon H-S, Moon S-Y, Lee K-K, Han Y-M. Transcriptional profiling of the developmentally important signalling pathways in human embryonic stem cells. Hum Reprod. 2006;21(2):405–412. doi: 10.1093/humrep/dei328. [DOI] [PubMed] [Google Scholar]
- Richards M, Tan S-P, Tan J-H, Chan W-K, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22(1):51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Carethers JM. Intersection of transforming growth factor-beta and Wnt signaling pathways in colorectal cancer and metastasis. Gastroenterology. 2009;137(1):33–36. doi: 10.1053/j.gastro.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411(6835):349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- Skotheim RI, Lind GE, Monni O, Nesland JM, Abeler VM, Fossa SD, Duale N, Brunborg G, Kallioniemi O, Andrews PW, Lothe RA. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res. 2005;65(13):5588–5598. doi: 10.1158/0008-5472.CAN-05-0153. [DOI] [PubMed] [Google Scholar]
- Hadjihannas MV, Behrens J. CIN By WNT: growth pathways, mitotic control and chromosomal instability in cancer. Cell Cycle. 2006;5(18):2077–2081. doi: 10.4161/cc.5.18.3282. [DOI] [PubMed] [Google Scholar]
- Terry S, Yang X, Chen M-W, Vacherot F, Buttyan R. Multifaceted interaction between the androgen and Wnt signaling pathways and the implication for prostate cancer. J Cell Biochem. 2006;99(2):402–410. doi: 10.1002/jcb.20983. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Shigematsu H, Nakajima T, Kubo R, Motohashi S, Sekine Y, Shibuya K, Iizasa T, Hiroshima K, Nakatani Y, Gazdar AF, Fujisawa T. Synchronous alterations of Wnt and epidermal growth factor receptor signaling pathways through aberrant methylation and mutation in non small cell lung cancer. Clin Cancer Res. 2007;13(20):6087–6092. doi: 10.1158/1078-0432.CCR-07-0591. [DOI] [PubMed] [Google Scholar]
- Zardawi SJ, O'Toole SA, Sutherland RL, Musgrove EA. Dysregulation of Hedgehog, Wnt and Notch signalling pathways in breast cancer. Histol Histopathol. 2009;24(3):385–398. doi: 10.14670/HH-24.385. [DOI] [PubMed] [Google Scholar]
- Li C, Zhang Y, Lu Y, Cui Z, Yu M, Zhang S, Xue X. Evidence of the cross talk between Wnt and Notch signaling pathways in non-small-cell lung cancer (NSCLC): Notch3-siRNA weakens the effect of LiCl on the cell cycle of NSCLC cell lines. J Cancer Res Clin Oncol. 2011;137(5):771–778. doi: 10.1007/s00432-010-0934-4. [DOI] [PubMed] [Google Scholar]
- Chen X, Stoeck A, Lee SJ, Shih I-M, Wang MM, Wang T-L. Jagged1 expression regulated by Notch3 and Wnt/b-catenin signaling pathways in ovarian cancer. Oncotarget. 2010;1(3):210–218. doi: 10.18632/oncotarget.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Nakajima K, Kawamoto K, Kikuno N, Kawakami K, Yamamura S, Ueno K, Majid S, Saini S, Ishii N, Dahiya R. Wnt antagonist gene DKK2 is epigenetically silenced and inhibits renal cancer progression through apoptotic and cell cycle pathways. Clin Cancer Res. 2009;15(18):5678–5687. doi: 10.1158/1078-0432.CCR-09-0558. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mah N, Prigione A, Wolfrum K, Andrade-Navarro MA, Adjaye J. A transcriptional roadmap to the induction of pluripotency in somatic cells. Stem Cell Rev. 2010;6(2):282–296. doi: 10.1007/s12015-010-9137-2. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh Y-H, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung W-K, Clarke ND, Wei C-L, Ng H-H. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K-i, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136(14):2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol Ther. 2002;1(5):466–476. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- Strizzi L, Hardy KM, Seftor EA, Costa FF, Kirschmann DA, Seftor REB, Postovit L-M, Hendrix MJC. Development and cancer: at the crossroads of Nodal and Notch signaling. Cancer Res. 2009;69(18):7131–7134. doi: 10.1158/0008-5472.CAN-09-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28(3):339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Zhang X-m, Wang J-x, Lei X-g, Cheng H, Wang L-l, Yao G-y. [Regulation and mechanism of Notch signaling pathway in small cell lung cancer] Zhonghua Bing Li Xue Za Zhi. 2010;39(2):95–99. [PubMed] [Google Scholar]
- Efferson CL, Winkelmann CT, Ware C, Sullivan T, Giampaoli S, Tammam J, Patel S, Mesiti G, Reilly JF, Gibson RE, Buser C, Yeatman T, Coppola D, Winter C, Clark EA, Draetta GF, Strack PR, Majumder PK. Downregulation of Notch pathway by a gamma-secretase inhibitor attenuates AKT/mammalian target of rapamycin signaling and glucose uptake in an ERBB2 transgenic breast cancer model. Cancer Res. 2010;70(6):2476–2484. doi: 10.1158/0008-5472.CAN-09-3114. [DOI] [PubMed] [Google Scholar]
- Rose SL. Notch signaling pathway in ovarian cancer. Int J Gynecol Cancer. 2009;19(4):564–566. doi: 10.1111/IGC.0b013e3181a12ed2. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, Ali S, Abbruzzese JL, Gallick GE, Sarkar FH. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69(6):2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118(Pt 19):4495–4509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–267. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Akhurst RJ, Derynck R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001;11(11):S44–51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- Yang G, Yang X. Smad4-mediated TGF-beta signaling in tumorigenesis. Int J Biol Sci. 2010;6(1):1–8. doi: 10.7150/ijbs.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10(6):415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, Wu J, Ding M, Deng H. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem. 2007;282(8):5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- Grandela C, Pera MF, Grimmond SM, Kolle G, Wolvetang EJ. p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res. 2007;1(2):116–128. doi: 10.1016/j.scr.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Sabapathy K, Klemm M, Jaenisch R, Wagner EF. Regulation of ES cell differentiation by functional and conformational modulation of p53. Embo J. 1997;16(20):6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(52):22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17(2):211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Dalton S. Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen Med. 2010;5(6):947–959. doi: 10.2217/rme.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS One. 2008;3(12):e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80(1):9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo ABH, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K, Fleck RA, Ford A, Gertow K, Gertsenstein M, Gokhale PJ, Hamilton RS, Hampl A, Healy LE, Hovatta O, Hyllner J, Imreh MP, Itskovitz-Eldor J, Jackson J, Johnson JL, Jones M, Kee K, King BL, Knowles BB, Lako M, Lebrin F, Mallon BS, Manning D, Mayshar Y, McKay RDG, Michalska AE, Mikkola M, Mileikovsky M, Minger SL, Moore HD, Mummery CL, Nagy A, Nakatsuji N, O'Brien CM, Oh SKW, Olsson C, Otonkoski T, Park K-Y, Passier R, Patel H, Patel M, Pedersen R, Pera MF, Piekarczyk MS, Pera RAR, Reubinoff BE, Robins AJ, Rossant J, Rugg-Gunn P, Schulz TC, Semb H, Sherrer ES, Siemen H, Stacey GN, Stojkovic M, Suemori H, Szatkiewicz J, Turetsky T, Tuuri T, van den Brink S, Vintersten K, Vuoristo S, Ward D, Weaver TA, Young LA, Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25(7):803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Assou S, Le Carrour T, Tondeur S, Strom S, Gabelle A, Marty S, Nadal L, Pantesco V, Reme T, Hugnot J-P, Gasca S, Hovatta O, Hamamah S, Klein B, De Vos J. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25(4):961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver RT. Germ cell cancer. Curr Opin Oncol. 1999;11(3):236–241. doi: 10.1097/00001622-199905000-00017. [DOI] [PubMed] [Google Scholar]
- Chiou S-H, Wang M-L, Chou Y-T, Chen C-J, Hong C-F, Hsieh W-J, Chang H-T, Chen Y-S, Lin T-W, Hsu H-S, Wu C-W. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer research. 2010;70(24):10433–10444. doi: 10.1158/0008-5472.CAN-10-2638. [DOI] [PubMed] [Google Scholar]
- Garcia P, Clarke M, Vegiopoulos A, Berlanga O, Camelo A, Lorvellec M, Frampton J. Reduced c-Myb activity compromises HSCs and leads to a myeloproliferation with a novel stem cell basis. Embo J. 2009;28(10):1492–1504. doi: 10.1038/emboj.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov KV, Tarasova YS, Tam WL, Riordon DR, Elliott ST, Kania G, Li J, Yamanaka S, Crider DG, Testa G, Li RA, Lim B, Stewart CL, Liu Y, Van Eyk JE, Wersto RP, Wobus AM, Boheler KR. B-MYB is essential for normal cell cycle progression and chromosomal stability of embryonic stem cells. PLoS One. 2008;3(6):e2478. doi: 10.1371/journal.pone.0002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman G, Andersson MK, Andren Y. New tricks from an old oncogene: gene fusion and copy number alterations of MYB in human cancer. Cell cycle (Georgetown, Tex) 2010;9(15):2986–2995. doi: 10.4161/cc.9.15.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8(7):523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- Muller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta. 2000;1470(1):M1–12. doi: 10.1016/s0304-419x(99)00030-x. [DOI] [PubMed] [Google Scholar]
- Olsson AY, Feber A, Edwards S, Te Poele R, Giddings I, Merson S, Cooper CS. Role of E2F3 expression in modulating cellular proliferation rate in human bladder and prostate cancer cells. Oncogene. 2007;26(7):1028–1037. doi: 10.1038/sj.onc.1209854. [DOI] [PubMed] [Google Scholar]
- Robson EJD, He S-J, Eccles MR. A PANorama of PAX genes in cancer and development. Nat Rev Cancer. 2006;6(1):52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- Bokobza SM, Ye L, Kynaston HG, Jiang WG. Growth and differentiation factor-9 promotes adhesive and motile capacity of prostate cancer cells by up-regulating FAK and Paxillin via Smad dependent pathway. Oncol Rep. 2010;24(6):1653–1659. doi: 10.3892/or_00001030. [DOI] [PubMed] [Google Scholar]
- Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends in biochemical sciences. 2008;33(3):122–131. doi: 10.1016/j.tibs.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Conserved POU-binding site linked to SP1-binding site within FZD5 promoter: Transcriptional mechanisms of FZD5 in undifferentiated human ES cells, fetal liver/spleen, adult colon, pancreatic islet, and diffuse-type gastric cancer. International journal of oncology. 2007;30(3):751–755. [PubMed] [Google Scholar]
- Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-L, Chang DC, Ying S-Y, Leu D, Wu DTS. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010;70(22):9473–9482. doi: 10.1158/0008-5472.CAN-10-2746. [DOI] [PubMed] [Google Scholar]
- Lin S-L, Chang DC, Chang-Lin S, Lin C-H, Wu DTS, Chen DT, Ying S-Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. Rna. 2008;14(10):2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS, Kim JS, Cho WJ, Lee MR, Steiner R, Gompers A, Ling D, Zhang J, Strom P, Behlke M, Moon S-H, Salvaterra PM, Jove R, Kim K-S. miR-302b maintains "stemness" of human embryonal carcinoma cells by post-transcriptional regulation of Cyclin D2 expression. Biochemical and biophysical research communications. 2008;377(2):434–440. doi: 10.1016/j.bbrc.2008.09.159. [DOI] [PubMed] [Google Scholar]
- Murray MJ, Saini HK, van Dongen S, Palmer RD, Muralidhar B, Pett MR, Piipari M, Thornton CM, Nicholson JC, Enright AJ, Coleman N. The two most common histological subtypes of malignant germ cell tumour are distinguished by global microRNA profiles, associated with differential transcription factor expression. Mol Cancer. 2010;9:290. doi: 10.1186/1476-4598-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M-R, Lee Y, Kim JY, Kim S-K, Moon S-H, Lee JY, Cha K-Y, Chung HM, Yoon HS, Moon SY, Kim VN, Kim K-S. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270(2):488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Love B, Goff LA, Jornsten R, Graichen R, Hart RP, Chesnut JD. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16(6):1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RAR, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. Embo J. 2011;30(4):756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, Ruzzo WL, Ware C, Radich JP, Gentleman R, Ruohola-Baker H, Tewari M. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26(10):2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Pure E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Wang F, Xue X, Wei J, An Y, Yao J, Cai H, Wu J, Dai C, Qian Z, Xu Z, Miao Y. hsa-miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br J Cancer. 2010;103(4):567–574. doi: 10.1038/sj.bjc.6605724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Zhang Y, Hu N, Shan C, Zhang S, Zhang W, Zhang X, Ye L. miRNA-520b and miR-520e sensitize breast cancer cells to complement attack via directly targeting 3'UTR of CD46. Cancer Biol Ther. 2010;10(3):232–241. doi: 10.4161/cbt.10.3.12277. [DOI] [PubMed] [Google Scholar]
- Hu N, Zhang J, Cui W, Kong G, Zhang S, Yue L, Bai X, Zhang Z, Zhang W, Zhang X, Ye L. miR-520b Regulates Migration of Breast Cancer Cells by Targeting Hepatitis B X-interacting Protein and Interleukin-8. J Biol Chem. 2011;286(15):13714–13722. doi: 10.1074/jbc.M110.204131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller F-J, Lu C, Shamir R, Fan J-B, Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26(6):1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu A-L, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome research. 2008;18(4):610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pan Y-Z, Seigel GM, Hu Z-H, Huang M, Yu A-M. Breast cancer resistance protein BCRP/ABCG2 regulatory microRNAs (hsa-miR-328, -519c and -520h) and their differential expression in stem-like ABCG2+ cancer cells. Biochem Pharmacol. 2011;81(6):783–792. doi: 10.1016/j.bcp.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, Cabo Rd, Gorospe M. miR-519 suppresses tumor growth by reducing HuR levels. Cell cycle (Georgetown, Tex) 2010;9(7):1354–1359. doi: 10.4161/cc.9.7.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends in cell biology. 2008;18(10):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Biology of the Mi-2/NuRD Complex in SLAC (Stemness, Longevity/Ageing, and Cancer) Gene Regul Syst Bio. 2011;5:1–26. doi: 10.4137/GRSB.S6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8(9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Wagner W, Bork S, Horn P, Krunic D, Walenda T, Diehlmann A, Benes V, Blake J, Huber F-X, Eckstein V, Boukamp P, Ho AD. Aging and replicative senescence have related effects on human stem and progenitor cells. PLoS One. 2009;4(6):e5846. doi: 10.1371/journal.pone.0005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. 24 hESC-associated gene sets.
Table S2. 189 hESC-associated transcription factors.
Table S3. 114 hESC-associated microRNAs.
Supplementary references. The list of references for the hESC-associated gene expression signatures identified and human tumor gene expression datasets used.
Table S4. 72 differentially expressed gene sets.
Figure S1. Significance of overlap between hESC and tumor pathways by normal vs. tumor class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S2. Significance of overlap between hESC and tumor pathways by good vs. poor prognosis class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S3. Significance of overlap between hESC and tumor pathways by survival analysis. The detailed description of all the datasets is provided in Additional file 6.
Figure S4. Significance of overlap between hESC and tumor TFs by normal vs. tumor class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S5. Significance of overlap between hESC and tumor TFs by good vs. poor prognosis class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S6. Significance of overlap between hESC and tumor TFs by survival analysis. The detailed description of all the datasets is provided in Additional file 6.
Table S5. 102 miRNAs identified at least in eight different groups.
Figure S7. Significance of overlap between hESC and tumor miRNAs by normal vs. tumor class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S8. Significance of overlap between hESC and tumor miRNAs by good vs. poor prognosis class comparison. The detailed description of all the datasets is provided in Additional file 6.
Figure S9. Significance of overlap between hESC and tumor miRNAs by survival analysis. The detailed description of all the datasets is provided in Additional file 6.