Abstract
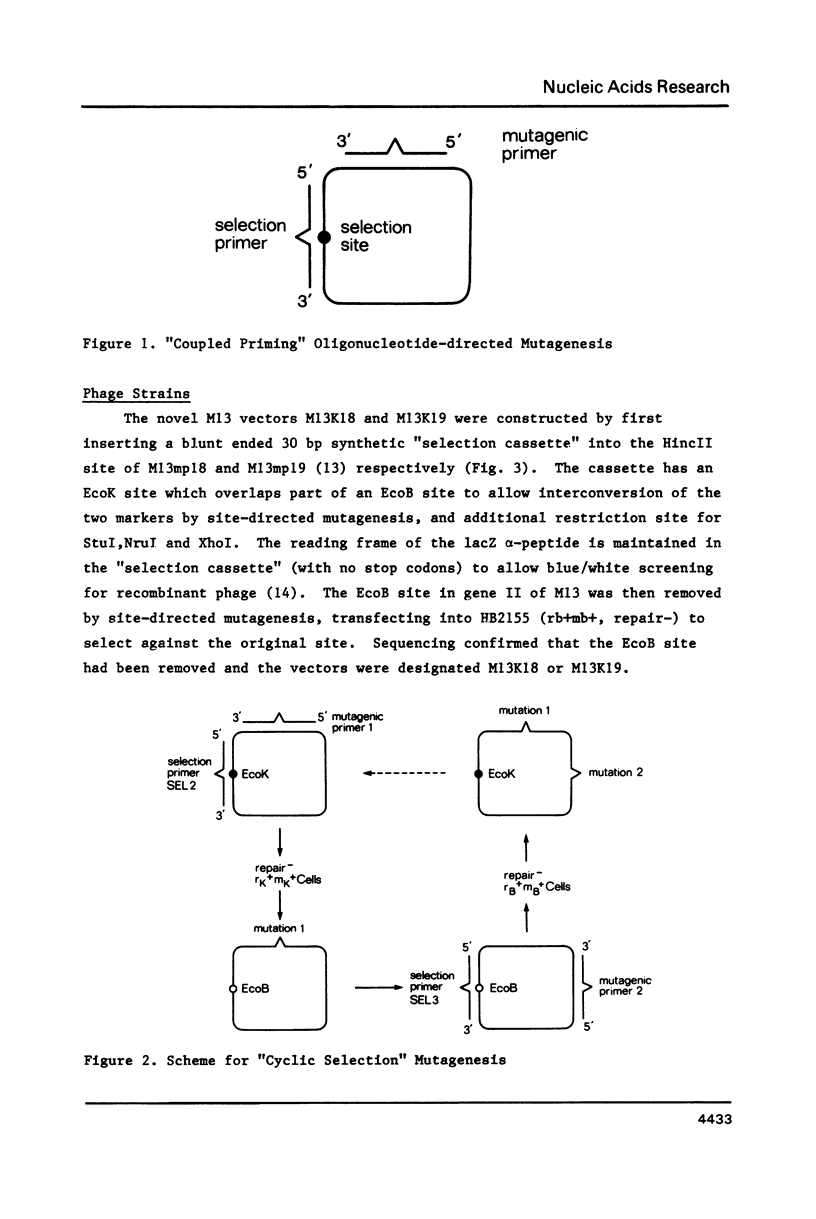
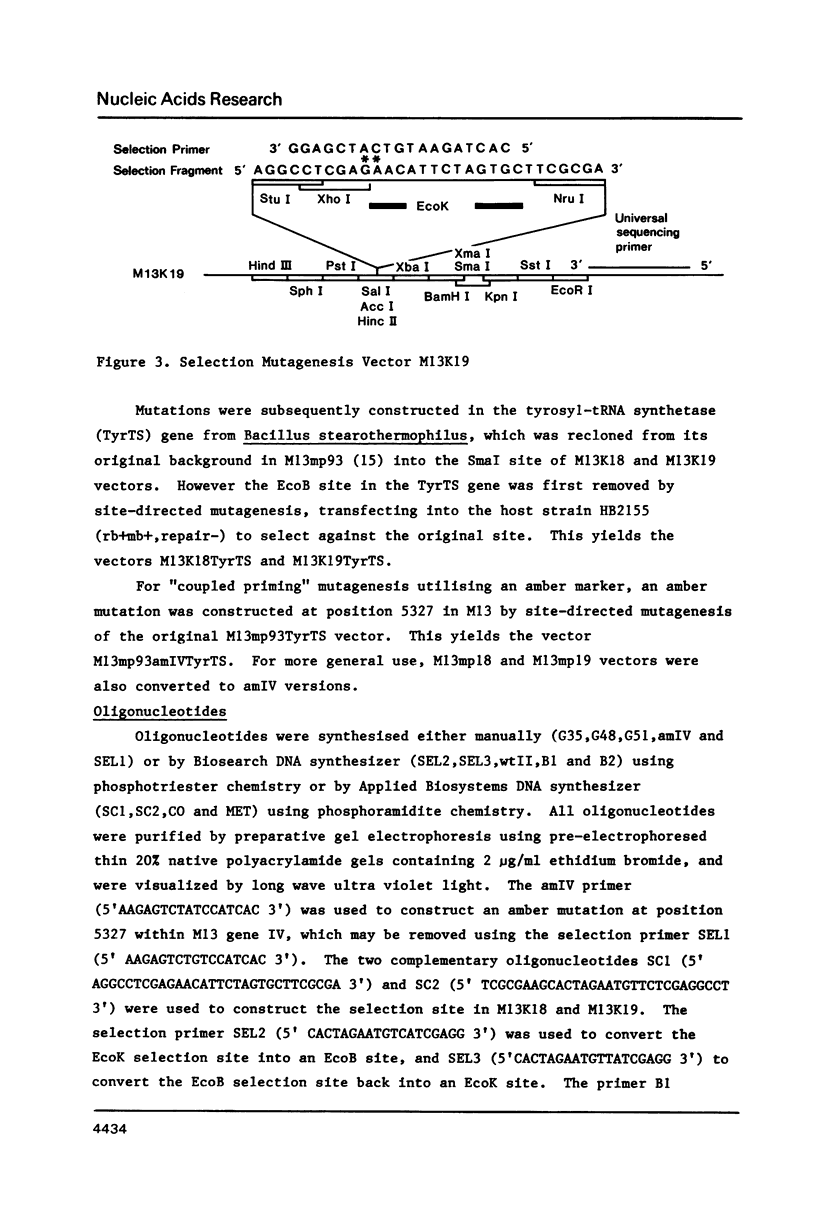
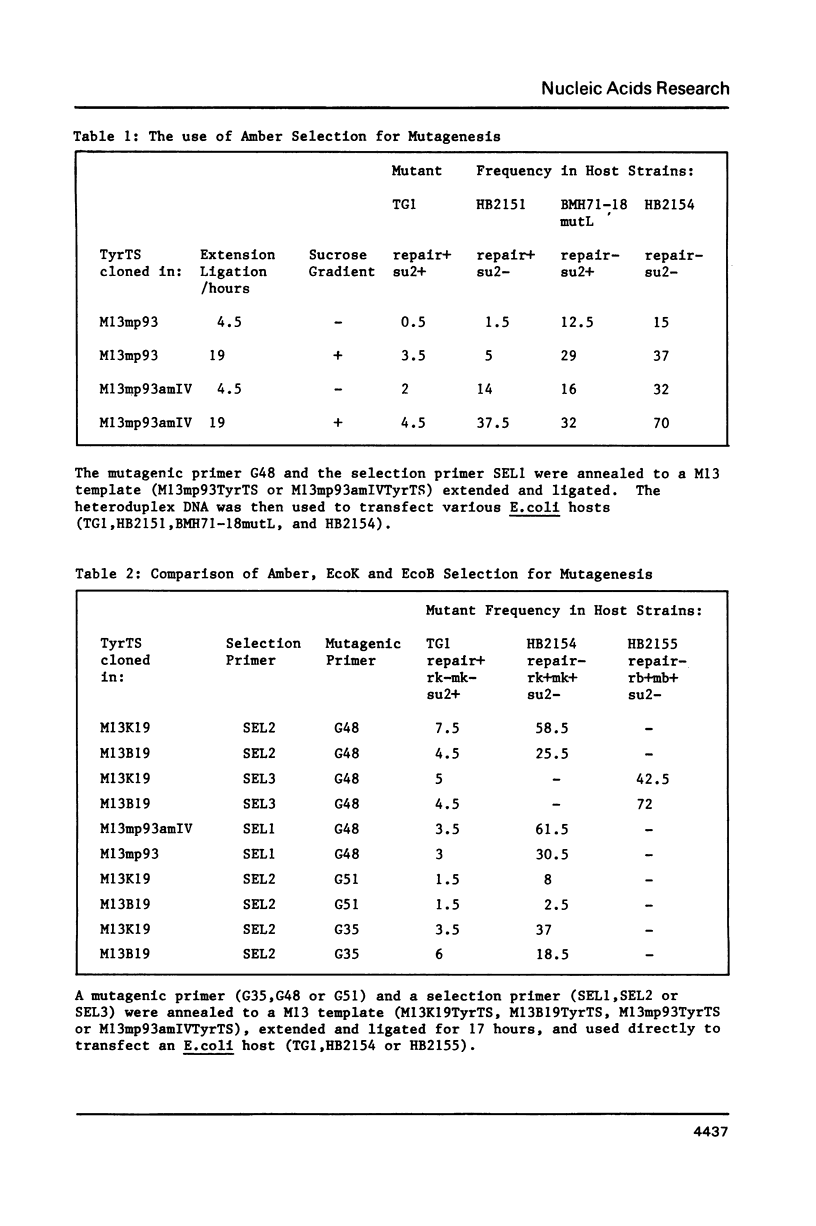
An improved method is described for the construction of mutations in M13 vectors using synthetic oligonucleotides. The DNA is first cloned into a novel M13 vector (based upon M13mp18 or M13mp19), which carries a genetic marker that can be selected against, such as an EcoK or EcoB site, or an amber mutation in an essential phage gene. In this "coupled priming" technique, one primer is used to construct the silent mutation of interest, and a second primer is used to eliminate the selectable marker on the minus strand. After primer extension and ligation, the heteroduplex DNA is transfected into a strain of E. coli which is repair deficient and selects against the plus strand marker. Over 50 mutants have been constructed with this approach, and the yields can be excellent (up to 70%). For the stepwise construction of mutations using separate rounds of mutagenesis, the EcoK and EcoB markers offer a particular advantage over the amber marker. They permit selection in each round, as it is possible to cycle between the two markers. However for construction of multiple mutations over a short region, long synthetic oligonucleotides with multiple mismatches to the template can offer an alternative strategy.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer H. Conjugation in Escherichia coli. J Bacteriol. 1966 May;91(5):1767–1772. doi: 10.1128/jb.91.5.1767-1772.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P. J., Winter G., Wilkinson A. J., Fersht A. R. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell. 1984 Oct;38(3):835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- Enea V., Vovis G. F., Zinder N. D. Genetic studies with heteroduplex DNA of bacteriophage fl. Asymmetric segregation, base correction and implications for the mechanism of genetic recombination. J Mol Biol. 1975 Aug 15;96(3):495–509. doi: 10.1016/0022-2836(75)90175-8. [DOI] [PubMed] [Google Scholar]
- Gronenborn B. Overproduction of phage lambda repressor under control of the lac promotor of Escherichia coli. Mol Gen Genet. 1976 Nov 17;148(3):243–250. doi: 10.1007/BF00332898. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kramer W., Drutsa V., Jansen H. W., Kramer B., Pflugfelder M., Fritz H. J. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984 Dec 21;12(24):9441–9456. doi: 10.1093/nar/12.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer W., Schughart K., Fritz H. J. Directed mutagenesis of DNA cloned in filamentous phage: influence of hemimethylated GATC sites on marker recovery from restriction fragments. Nucleic Acids Res. 1982 Oct 25;10(20):6475–6485. doi: 10.1093/nar/10.20.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Ganem D., Lu P., Schmitz A. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J Mol Biol. 1977 Jan 15;109(2):275–298. doi: 10.1016/s0022-2836(77)80034-x. [DOI] [PubMed] [Google Scholar]
- Modrich P. Structures and mechanisms of DNA restriction and modification enzymes. Q Rev Biophys. 1979 Aug;12(3):315–369. doi: 10.1017/s0033583500005461. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Norris K., Norris F., Christiansen L., Fiil N. Efficient site-directed mutagenesis by simultaneous use of two primers. Nucleic Acids Res. 1983 Aug 11;11(15):5103–5112. doi: 10.1093/nar/11.15.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schold M., Colombero A., Reyes A. A., Wallace R. B. Oligonucleotide-directed mutagenesis using plasmid DNA templates and two primers. DNA. 1984 Dec;3(6):469–477. doi: 10.1089/dna.1.1984.3.469. [DOI] [PubMed] [Google Scholar]
- Wilkinson A. J., Fersht A. R., Blow D. M., Carter P., Winter G. A large increase in enzyme-substrate affinity by protein engineering. Nature. 1984 Jan 12;307(5947):187–188. doi: 10.1038/307187a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson A. J., Fersht A. R., Blow D. M., Winter G. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthetase cysteine-35 to glycine-35 mutation. Biochemistry. 1983 Jul 19;22(15):3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- Winter G., Fersht A. R., Wilkinson A. J., Zoller M., Smith M. Redesigning enzyme structure by site-directed mutagenesis: tyrosyl tRNA synthetase and ATP binding. Nature. 1982 Oct 21;299(5885):756–758. doi: 10.1038/299756a0. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S., Ratti G. The structure of two subgenomic RNAs from human influenza virus A/PR/8/34. Nucleic Acids Res. 1981 Dec 21;9(24):6907–6915. doi: 10.1093/nar/9.24.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Annu Rev Biochem. 1981;50:285–319. doi: 10.1146/annurev.bi.50.070181.001441. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]



