Abstract
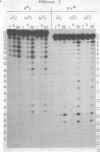
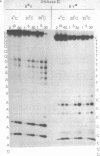
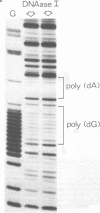
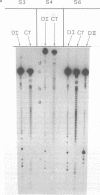
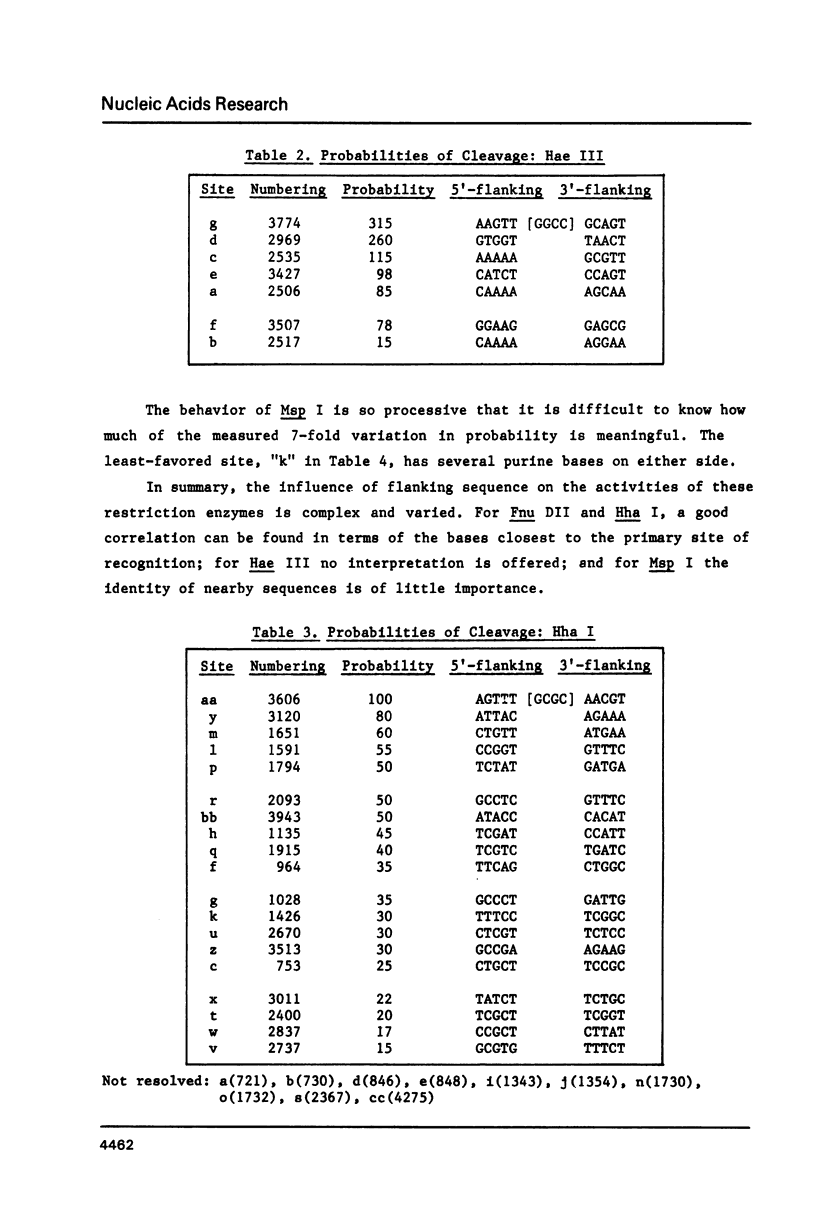
When a protein binds to DNA, the affinity of this protein for its primary site of interaction may be influenced by the nature of flanking sequences. This is thought to be a consequence of local cooperativity in the DNA molecule, where the conformation at one point along the helix can influence the conformation at another, and thereby modulate the free energy of protein-DNA recognition. In order to learn more about this process, we have carried out experiments of two sorts. First, we have constructed sequences of the type (dA)11 (dG)8, where the conformational preferences of the DNA molecule switch from one extreme to another over just a single base pair, and subjected them to digestion by DNAase I and DNAase II. This is to learn whether the structure changes abruptly at the junction point, or more gradually with an influence extending into residues on either side. Secondly, we have subjected long plasmid DNA to digestion by restriction enzymes Fnu DII, Hae III, Hha I and Msp I, to look for correlations between cutting rate and the identity of nucleotides on either side of the restriction site. The influence of flanking sequence on nuclease digestion specificities is clearly evident in both kinds of experiment, but the rules governing this seem complex and not easily formulated. The best that can be done at present is to divide the problem into two parts, "analogue" and "digital", representing sugar-phosphate and base components of recognition.
Full text
PDF



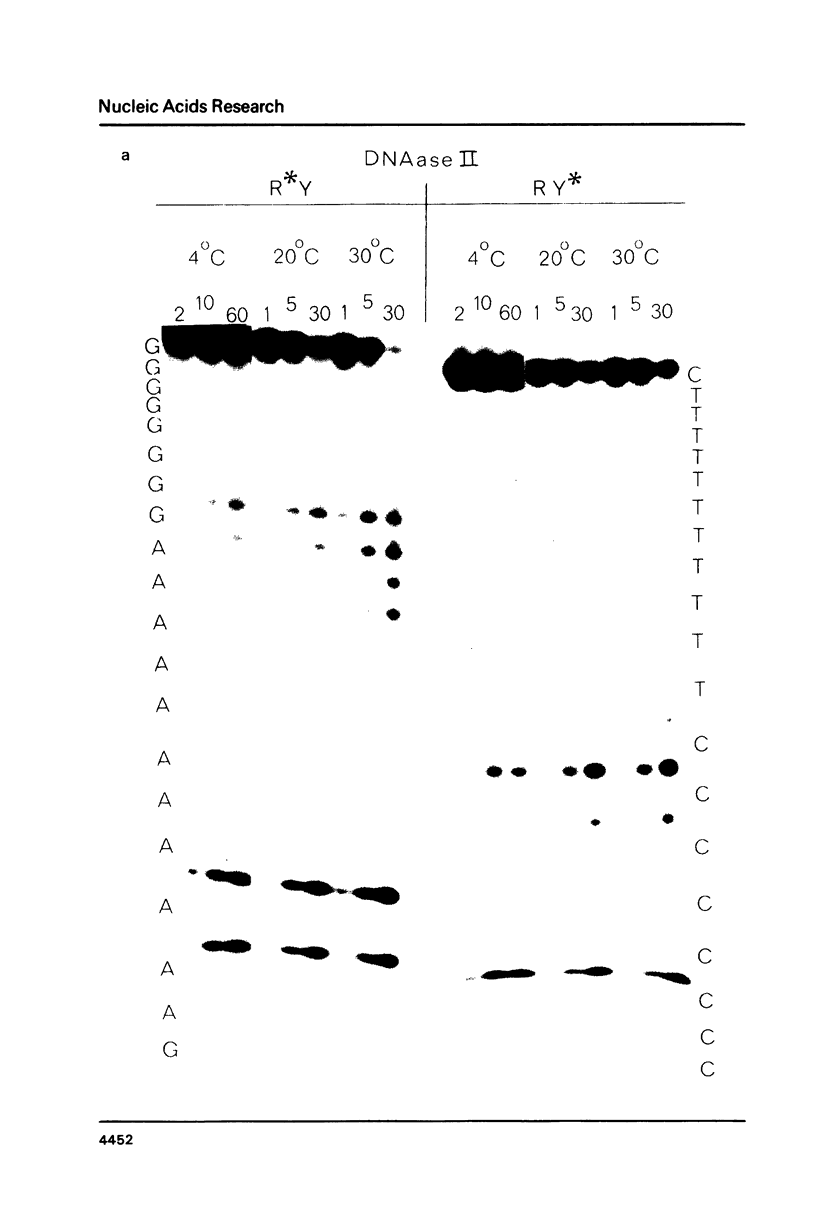
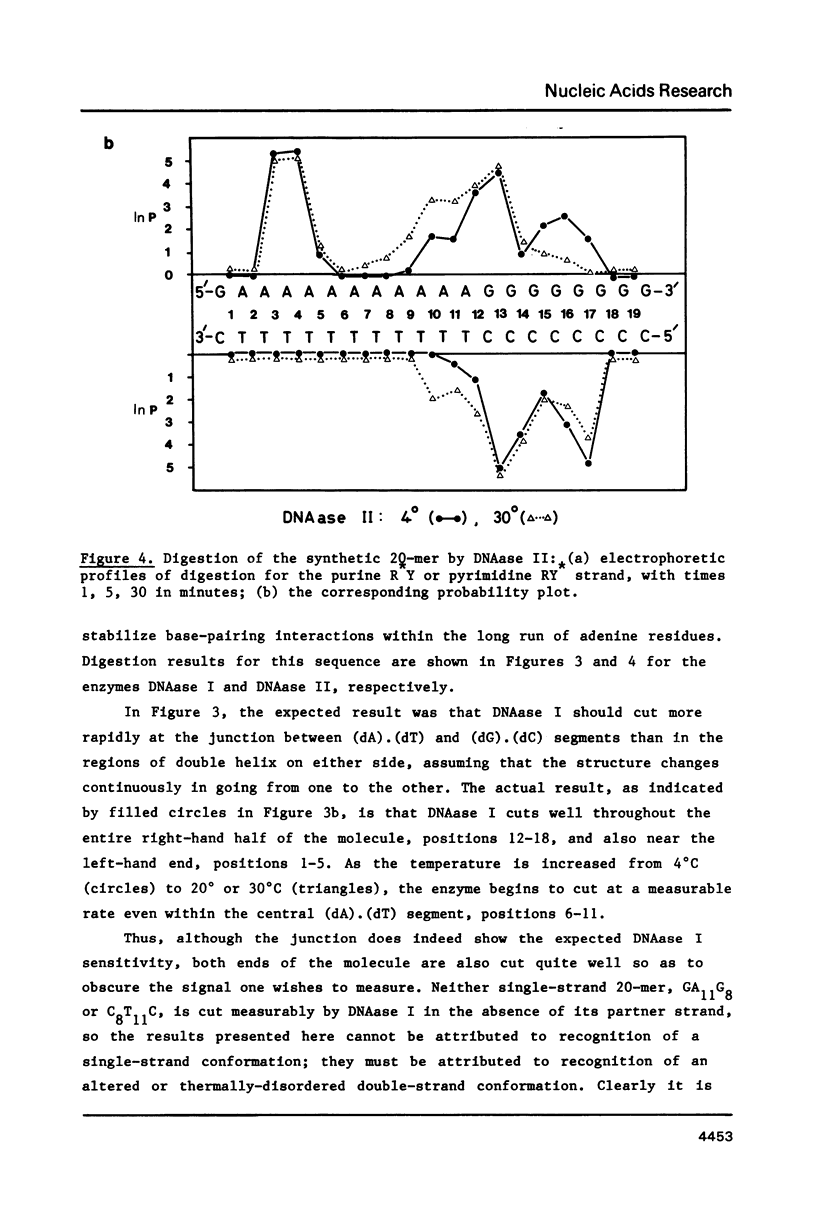
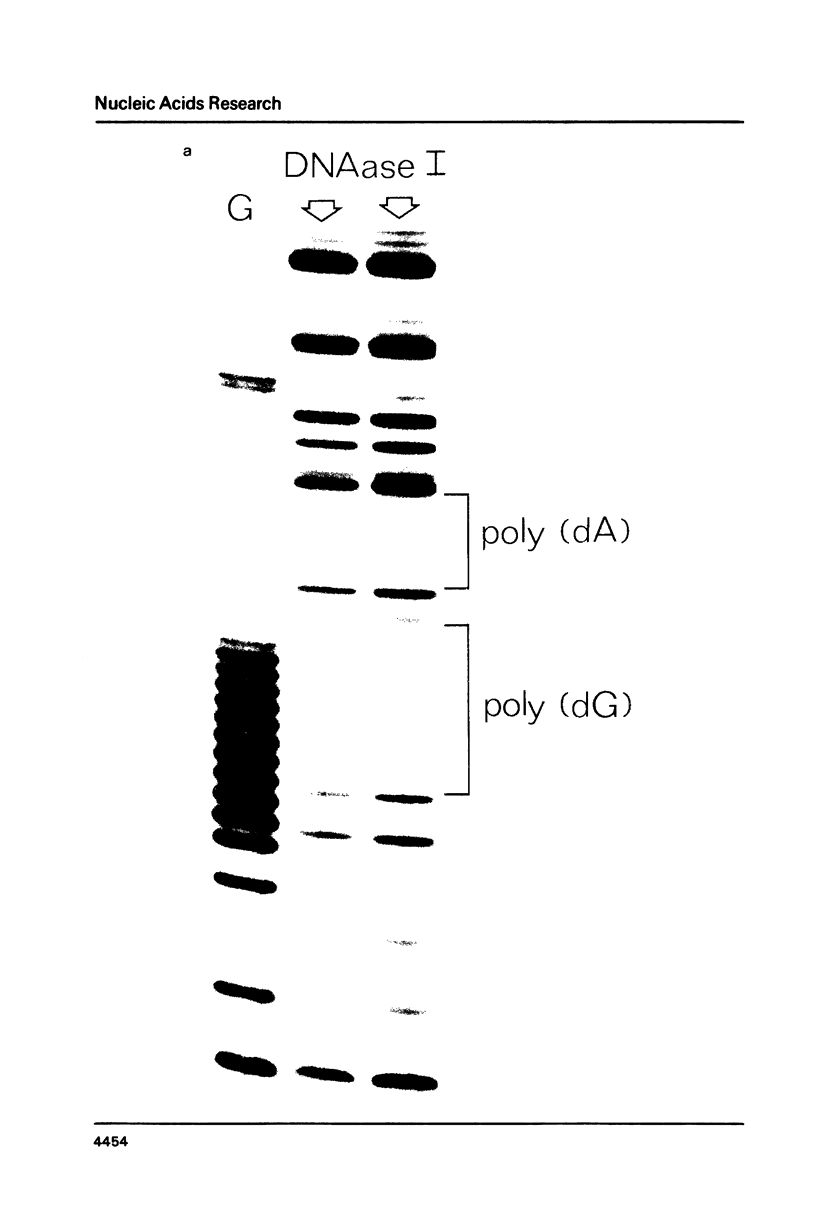
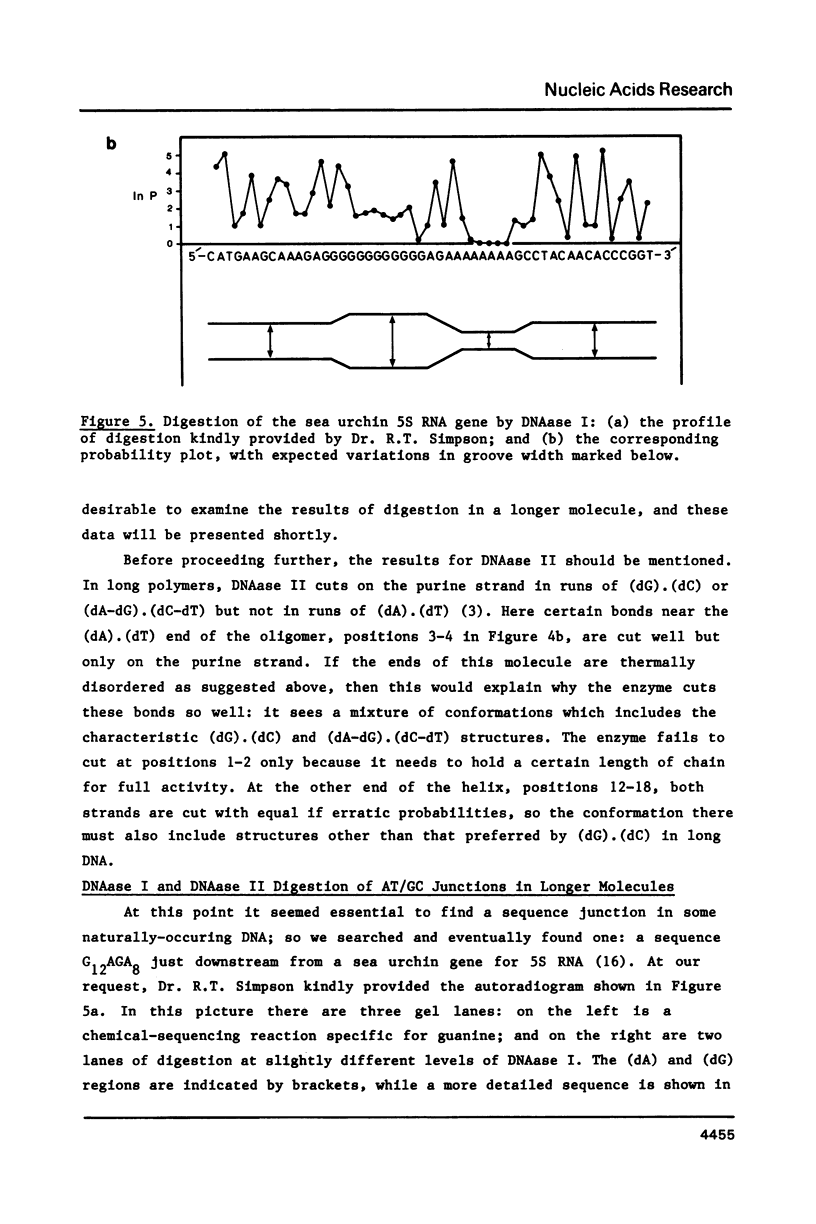







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alves J., Pingoud A., Haupt W., Langowski J., Peters F., Maass G., Wolff C. The influence of sequences adjacent to the recognition site on the cleavage of oligodeoxynucleotides by the EcoRI endonuclease. Eur J Biochem. 1984 Apr 2;140(1):83–92. doi: 10.1111/j.1432-1033.1984.tb08069.x. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Bauer W. R. Site-dependent cleavage of pBR322 DNA by restriction endonuclease HinfI. Nucleic Acids Res. 1983 Jun 25;11(12):4109–4126. doi: 10.1093/nar/11.12.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram S. Secondary structure of DNA depends on base composition. Nat New Biol. 1971 Aug 11;232(2):174–176. doi: 10.1038/newbio232174a0. [DOI] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. A base-centred explanation of the B-to-A transition in DNA. J Mol Biol. 1984 Sep 25;178(3):773–782. doi: 10.1016/0022-2836(84)90251-1. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R., Conner B. N., Kopka M. L., Pjura P. E. Helix geometry and hydration in A-DNA, B-DNA, and Z-DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):13–24. doi: 10.1101/sqb.1983.047.01.004. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Weeks J. R., Travers A. A. Negative supercoiling induces spontaneous unwinding of a bacterial promoter. EMBO J. 1985 Apr;4(4):1025–1032. doi: 10.1002/j.1460-2075.1985.tb03734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Klug A., Jack A., Viswamitra M. A., Kennard O., Shakked Z., Steitz T. A. A hypothesis on a specific sequence-dependent conformation of DNA and its relation to the binding of the lac-repressor protein. J Mol Biol. 1979 Jul 15;131(4):669–680. doi: 10.1016/0022-2836(79)90196-7. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Travers A. A. Requirement for an upstream element for optimal transcription of a bacterial tRNA gene. Nature. 1983 Sep 15;305(5931):248–250. doi: 10.1038/305248a0. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Mellema J. R., Haasnoot C. A., van der Marel G. A., Wille G., van Boeckel C. A., van Boom J. H., Altona C. Proton NMR studies on the covalently linked RNA-DNA hybrid r(GCG)d(TATACGC). Assignment of proton resonances by application of the nuclear Overhauser effect. Nucleic Acids Res. 1983 Aug 25;11(16):5717–5738. doi: 10.1093/nar/11.16.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Pardi A., Itakura K. DNA conformation, dynamics, and interactions in solution. Science. 1982 May 7;216(4546):581–590. doi: 10.1126/science.6280281. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D., Oefner C., Kabsch W. Three-dimensional structure of bovine pancreatic DNase I at 2.5 A resolution. EMBO J. 1984 Oct;3(10):2423–2430. doi: 10.1002/j.1460-2075.1984.tb02149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Ughetto G., Quigley G. J., Hakoshima T., van der Marel G. A., van Boom J. H., Rich A. The molecular structure of a DNA-triostin A complex. Science. 1984 Sep 14;225(4667):1115–1121. doi: 10.1126/science.6474168. [DOI] [PubMed] [Google Scholar]