Abstract
In the current work, we present a hydrogen-bond analysis of 2,673 ligand-receptor complexes that suggests the total number of hydrogen bonds formed between a ligand and its protein receptor is a poor predictor of ligand potency; furthermore, even that poor prediction does not suggest a statistically significant correlation between hydrogen-bond formation and potency. While we are not the first to suggest that hydrogen bonds on average do not generally contribute to ligand binding affinities, this additional evidence is nevertheless interesting. The primary role of hydrogen bonds may instead be to ensure specificity, to correctly position the ligand within the active site, and to hold the protein active site in a ligand-friendly conformation.
We also present a new computer program called HBonanza (hydrogen-bond analyzer) that aids the analysis and visualization of hydrogen-bond networks. HBonanza, which can be used to analyze single structures or the many structures of a molecular dynamics trajectory, is open source and python implemented, making it easily editable, customizable, and platform independent. Unlike many other freely available hydrogen-bond analysis tools, HBonanza provides not only a text-based table describing the hydrogen-bond network, but also a Tcl script to facilitate visualization in VMD, a popular molecular visualization program. Visualization in other programs is also possible. A copy of HBonanza can be obtained free of charge from http://www.nbcr.net/hbonanza.
Keywords: hydrogen bond, computational chemistry, molecular dynamics simulations, molecular recognition, computer-aided drug design
Introduction
Hydrogen bonds, first described in 1912 (1), involve two components: a hydrogen-bond donor, comprised of an electronegative atom covalently bound to a hydrogen-atom partner, and a hydrogen-bond acceptor, also an electronegative heteroatom. The donor heavy atom attracts the electrons normally associated with its hydrogen-atom partner, imparting to that partner a positive partial charge. This positive partial charge can interact with a lone pair of electrons on the hydrogen-bond acceptor, forming a bond that is part electrostatic and part chemical (2). When binding a ligand to its receptor, a hydrogen bond is traditionally thought to contribute between 0.5 and 4.7 kcal/mol to the binding energy (3).
Despite this traditional wisdom, some have speculated that the hydrogen-bond contribution to protein-ligand binding energies is generally overstated, certain convincing exceptions aside (4, 5). Ligand hydrogen-bond donors and acceptors are energetically favorable both in solution, where they form hydrogen bonds with the surrounding water molecules, and when bound to an amenable active site, where they form hydrogen bonds with receptor residues. Thus, ligand binding simply substitutes hydrogen bonds with water for those with the receptor. There is arguably little net gain in energy; binding is merely an exchange process (3).
In the current work, we present an analysis of 2,673 protein-ligand complexes of known binding affinity that suggests hydrogen bonds do not generally contribute to ligand binding energies. Their primary role in ligand binding may instead be to ensure specificity; to correctly position the ligand within the active site, thus facilitating catalysis; and to hold the protein active site in a conformation amenable to ligand binding.
One way to study these important roles is through structural/computational biology. A common task is to perform a hydrogen-bond analysis of an entire molecular-dynamics (MD) trajectory, often consisting of thousands of individual protein conformations. Such an analysis can give a sense of the persistence of selected hydrogen bonds across the many MD frames, providing insight into their relative importance. In our experience, however, performing hydrogen-bond analyses of molecular-dynamics trajectories with existing free software (e.g., (6, 7)) can be tedious. Most of the available free programs produce large text-based charts; the important hydrogen-bond interactions can be easily buried in the data.
In response to this challenge, we created HBonanza (hydrogen-bond analyzer), an open-source, python-implemented computer program that, given a protein structure, can greatly facilitate the identification and analysis of hydrogen-bond networks. HBonanza can be used to analyze single protein structures or entire molecular-dynamics trajectories. While all protein/ligand hydrogen bonds can certainly be identified, the program also facilitates the identification of only those hydrogen-bond networks connected to a selected residue or residues of interest (e.g., the ligands). Importantly, HBonanza outputs a Tcl file that creates an intuitive visualization when loaded into VMD (8), a popular molecular visualization program. Visualization using other programs is also possible.
HBonanza has been tested on Ubuntu 10.04.1 LTS, Mac OS X 10.6.6, and Windows XP using Python versions 2.6.5, 2.6.1, and 2.7.1, respectively. A copy can be obtained free of charge from http://www.nbcr.net/hbonanza.
Materials and Methods
A Database of Experimentally Characterized Ligand-Receptor Complexes
The database of ligand-receptor complexes with experimentally measured binding affinities used in the current work has been described previously (9, 10). In brief, structures with Kd values listed in the PDBbind-CN (11, 12) and MOAD (13) databases were downloaded from the Protein Data Bank (14). Hydrogen atoms were added to all ligands and receptors using the Schrodinger Maestro (Schrodinger) and AutoDockTools 1.5.1 (15) programs. An in-house script was used to optimize the geometry of the hydrogen bonds between ligand and receptor atoms, and the computer program BINANA (9) was used to identify the number of hydrogen bonds formed between the receptor and ligand for each complex. Default BINANA hydrogen-bond parameters were used.
A Molecular Dynamics Simulation of RNA Editing Ligase 1
A molecular model of Trypanosoma brucei RNA editing ligase 1 bound to V2, a recently discovered low-micromolar inhibitor (16), was generated from the 1XDN structure (17) deposited in the Protein Data Bank (14). The ligand, prepared using Schrodinger Maestro (Schrodinger) and AutoDockTools (15), was docked into the 1XDN active site sans the co-crystallized ATP ligand using AutoDock Vina (18). The docked pose with the lowest predicted binding energy was accepted.
Hydrogen atoms were added to the protein receptor using the PDB2PQR server (19, 20), and the receptor was parameterized according to the AMBER-99SB force field (21). Ligand partial charges were assigned using the Gaussian computer program (22) and the RESP module of the Amber molecular-dynamics package (6). The molecular electrostatic potential was calculated at the HF/6–31G* level. Ligand atom types were assigned according to the GAFF force field (23). GAFF does not have parameters for the angle between atom types nh-c3-ca; the angle parameters associated with nh-c3-c3 were used instead.
A box of TIP3P water molecules (24) was built around the protein/ligand model, extending 10 Å beyond the model in all directions, using the xleap module of the Amber molecular-dynamics package. Enough Na+ cations were added to bring the system to electrical neutrality; additional Na+ and Cl− ions were added to approximate 20 mM NaCl solution.
NAMD 2.7b1 (25) was used to minimize the system in four stages. First, the hydrogen atoms of the system were minimized for 5,000 steps. Second, the hydrogen atoms, water molecules, and ions were minimized for 5,000 steps. Third, the hydrogen atoms, water molecules, ions, and protein side chains were minimized for 10,000 steps. Finally, all the atoms of the system were minimized for an additional 25,000 steps. The system was next equilibrated by gradually heating in four stages, beginning at 50°K and ending at 310°K. The simulation was run at each temperature for 250,000 steps with a time step of 1.0 fs. Bonds to hydrogen atoms were held rigid, and Langevin dynamics were performed to maintain constant pressure and temperature.
Following minimization and equilibration, the isothermal-isobaric simulation was continued for an additional 10,000,000 steps using a 2.0 fs time step. The system was saved every 5,000 steps, yielding 2,000 distinct protein conformations that were used for subsequent analysis.
Using HBonanza to Analyze the Molecular Dynamics Trajectory
Default HBonanza values were used for trajectory analysis. Hydrogen bonds were considered to be those that satisfied the following criteria: 1) The heavy-atom participants in the candidate bond were oxygen, nitrogen, fluorine, or sulfur atoms. 2) These heavy atoms were no farther than 3.5 Å apart. 3) The angle between the hydrogen atom, the donor heavy atom, and the acceptor heavy atom was less than 30 degrees. 4) The hydrogen bond as defined above was present in at least 75% of the frames of the trajectory.
Results and Discussion
The purpose of the current work is two fold. First, we will present a small analysis of 2,673 protein-ligand complexes with known binding affinities in order to elucidate the general contribution of hydrogen bonds to ligand binding energies. Second, we will describe in detail a novel computer program called HBonanza (hydrogen-bond analyzer) that has been developed to facilitate the hydrogen-bond analyses of single structures as well as molecular-dynamics trajectories.
The Hydrogen-Bond Contribution to Ligand Binding Energies
We recently described a large structural database of 2,673 ligand-receptor complexes with known binding affinities (9, 10). As the affinities and structures of these complexes have been well characterized experimentally, it is possible to compare their measured Kd values to various binding characteristics. As an example, we used a computer program called BINANA to see how the number of ligand-receptor hydrophobic contacts varied with ligand potency (9).
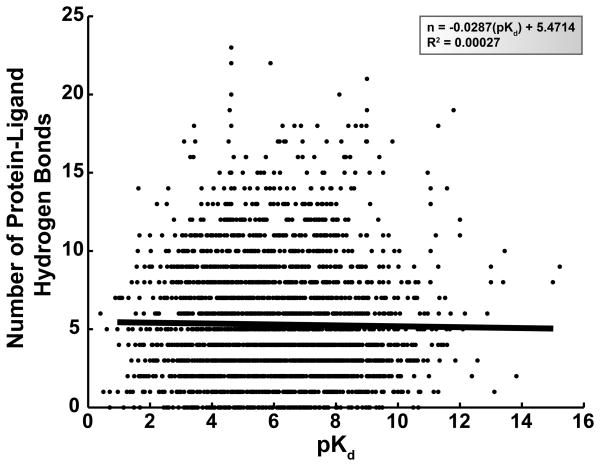
We here present a similar analysis of ligand-protein hydrogen bonds. First, the experimentally measured pKd value of each of the 2,673 complexes was plotted against the BINANA-determined number of ligand-protein hydrogen bonds (Figure 1). In order to determine if these two variables were correlated, linear regression was used to identify a single line that best fit the data. The R2 value of the regression was 2.7 × 10−4, suggesting that the number of ligand-protein hydrogen bonds is a poor predictor of ligand potency if a linear relationship is assumed.
Figure 1.
The experimentally measured pKd values of 2,673 ligand-protein complexes plotted against the corresponding number of ligand-protein hydrogen bonds. A linear regression line is shown in black; the equation of this line and the R2 value of the fit are given in the legend.
The slope, F statistic, and number of degrees of freedom for the regression were −2.9 × 10−2, 0.72, and 2,671, respectively. If there were no linear correlation between ligand potency and the number of ligand-protein hydrogen bonds, one would expect a regression with a slope of 0, a model with only 1 degree of freedom. We accepted as the null hypothesis that the regression model generated from the 2,673 data points does not differ significantly from the linear model of slope 0. This null hypothesis was tested using an F probability distribution; the corresponding p-value was 0.40, suggesting the null hypothesis should not be rejected. Thus, not only is the ligand-protein hydrogen-bond count a poor predictor of ligand potency (based on the R2 value of the linear regression), but even that poor prediction does not suggest a statistically significant correlation between these two variables. We are not the first to suggest that hydrogen bonds do not generally contribute to binding energies, but this additional evidence is nevertheless interesting.
Some may wonder if our database of ligand-protein complexes is biased by the presence of drug-like molecules. Among the ligand-receptor complexes with high potency, are we capturing the characteristics of good drugs rather than the general characteristics of good binders? After all, many drugs conform to Lipinski’s rule (26), which includes constraints on the number of ligand hydrogen-bond donors and acceptors. Additionally, drugs may also have reduced numbers of hydrogen-bond-forming hydroxyl and amino groups in order to avoid phase-II conjugation and to improve cell-membrane and blood-brain-barrier permeability.
In order to address this concern, we searched DrugBank (27) for each of the ligands in our database and determined that only 5.4% (144 ligands) are currently approved drugs. A t-test revealed that the approved drugs are significantly more likely (p = 3.5 × 10−3) to form fewer hydrogen bonds with their respective receptors than ligands that are not approved drugs, but the difference is not great, averaging 4.6 ± 2.9 (standard deviation) vs. 5.3 ± 3.8. When the pKd values of the approved drugs are compared to the pKd values of the remaining ligands in the database, the two groups do not differ significantly (p = 0.46), suggesting that the approved drugs do not tend to be more or less potent than the remaining ligands.
HBonanza: A Novel Computer Program for Characterizing Hydrogen Bonds
In the analysis above, the computer program BINANA (9) was used to identify ligand-receptor hydrogen bonds. BINANA is well suited for counting the number of ligand-receptor hydrogen bonds in a single structure and producing a visualization of those bonds in VMD (8), but it is not well suited for identifying and tallying the hydrogen bonds across an entire molecular-dynamics trajectory, which contains multiple frames with subtly different hydrogen-bond networks. In order to analyze and visualize the hydrogen-bond networks of both trajectory and single-structure PDB files, we have created a new computer program called HBonanza.
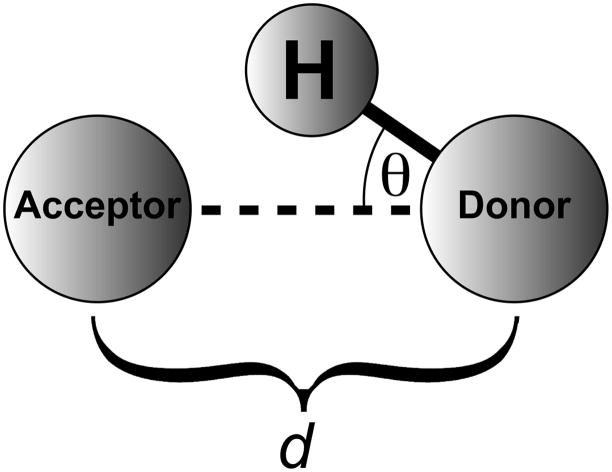
HBonanza considers a number of factors when identifying hydrogen bonds. First, the heavy-atom participants in the potential hydrogen bond must be oxygen, nitrogen, fluorine, or sulfur atoms. Second, the distance between these heavy atoms (d) must be no greater than a user-defined cutoff distance, 3.5 Å by default (Figure 2). Third, the angle between the hydrogen atom, the donor heavy atom, and the acceptor heavy atom (θ) must be less than a user-specified angle, 30 degrees by default (Figure 2). Finally, if an entire MD trajectory is analyzed, the hydrogen bonds as defined above must be well represented among the many frames of the simulation. If a hydrogen bond appears only infrequently, it is less likely to play an important role in ligand binding or protein structure. The required frequency cutoff is user defined; by default, it is set to 75%.
Figure 2.
A schematic of a hydrogen bond. For the purposes of the current work, d is defined as the distance between the donor and acceptor heavy atoms, and θ is defined as the angle formed by the hydrogen atom, the donor heavy atom, and the acceptor heavy atom.
Though it is certainly possible to analyze all of the receptor/ligand hydrogen bonds of a given trajectory, HBonanza may be most useful when one wishes to examine the specific hydrogen-bond network relevant to a residue or residues of interest (i.e., a “seed” residue, often the ligand). HBonanza identifies relevant networks recursively. First, all residues that are connected to the seed residue(s) via hydrogen bonds are identified. Next, all residues that are connected to those that are connected to the seed residues are identified, then all residues that are connected to those, etc., thereby delineating the entire hydrogen-bond network that supports the residue(s) of interest.
Perhaps most helpfully, HBonanza not only prints out a text-based chart describing these hydrogen bonds but also builds a visualizing Tcl script that can be loaded directly into VMD (8), a popular molecular visualization program. The identified hydrogen bonds are colored according to their frequency, allowing the easy identification of the most important bonds (see, for example, Figure 3). For those who do not use VMD, the generated Tcl script contains helpful comments that should make visualization in other software packages easy as well.
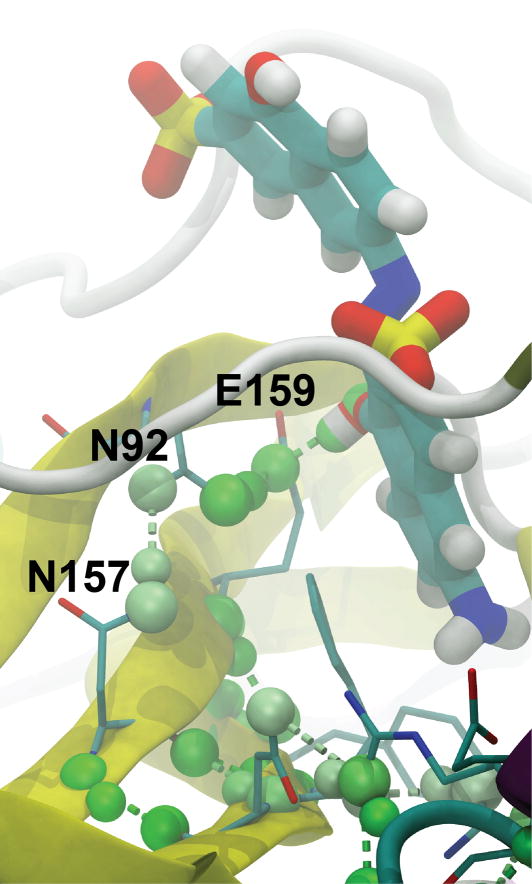
Figure 3.
Predicted hydrogen-bond interactions with V2, a recently identified low-micromolar inhibitor of TbREL1. The visualization shown is representative of the default HBonanza-generated VMD visualization script, rendered with Tachyon. The ligand is shown in thick licorice, protein residues thought to participate in hydrogen-bond networks involving the ligand are shown in thin licorice, and the entire protein is shown in cartoon, colored according to secondary structure. Hydrogen bonds are colored in shades of green according to the frequency with which they appear in the many frames of the MD trajectory. The coloring ranges from white, those bonds present in 75% of the frames, to green, those bonds present in 100% of the frames.
An Example: The Hydrogen-Bond Networks of TbREL1
In order to demonstrate the utility of HBonanza, we analyzed the hydrogen-bond networks of RNA editing ligase 1 from Trypanosoma brucei (TbREL1), a recently discovered therapeutic target useful in the fight against African sleeping sickness. First, a 20-ns molecular dynamics simulation was performed of the protein with a bound, recently discovered inhibitor (V2) (16) whose predicted binding pose was determined using docking. The hydrogen-bond networks that interfaced with the ligand were subsequently identified using HBonanza. The HBonanza output file, visualized in VMD and rendered using Tachyon, is shown in Figure 3.
The analysis revealed that only one hydrogen bond forms between the protein and ligand in more than 75% of the frames of the trajectory. This bond connects a key ligand hydroxyl group to the E159 side-chain carboxylate, which in turn is held in place by a hydrogen bond with the N92 side-chain amine, which is in turn stabilized by a hydrogen bond with the N157 side-chain amine, which may ultimately tie the region of the active-site that interacts with the ligand hydroxyl to the stability of the underlying beta sheet. Of course it is possible to take this kind of analysis too far; residues that are separated from the ligand by many degrees are not likely to be critical to binding. Nevertheless, HBonanza analyses offer interesting insights into the role distant residues play in stabilizing other residues of special interest.
Conclusion
The current work is two fold. First, we presented an analysis of 2,673 ligand-receptor complexes with known binding affinities. We found that the total number of ligand-receptor hydrogen bonds is a poor predictor of ligand potency; furthermore, even that poor prediction does not suggest a statistically significant correlation between these two variables. While we are not the first to suggest that hydrogen bonds do not generally contribute to ligand binding energies, the additional evidence presented in the current work is nevertheless interesting.
Second, we also presented a new computer program called HBonanza (hydrogen-bond analyzer) that, given either a single structure or an entire molecular dynamics trajectory in PDB format, facilitates the identification, analysis, and visualization of hydrogen-bond networks. HBonanza is open source and python implemented, making it easily editable, customizable, and platform independent. Unlike many other freely available hydrogen-bond analysis tools, HBonanza generates not only a text-based table describing the hydrogen-bond network, but also a Tcl script to facilitate visualization in VMD (8), a popular molecular visualization program. Visualization in other programs is also possible. A copy of HBonanza can be obtained free of charge from http://www.nbcr.net/hbonanza.
Acknowledgments
This work was carried out with funding from NIH GM31749, NSF MCB-0506593, and MCA93S013 to JAM. The funding sources had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. Additional support from the Howard Hughes Medical Institute, the National Center for Supercomputing Applications, the San Diego Supercomputer Center, the W.M. Keck Foundation, the National Biomedical Computational Resource, and the Center for Theoretical Biological Physics is gratefully acknowledged. We would also like to thank Aaron Friedman for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moore TS, Winmill TF. The state of amines in aqueous solution. Journal of the Chemical Society, Transactions. 1912;101:1635. [Google Scholar]
- 2.Arunan E, Desiraju GR, Klein RA, Sadlej J, Scheiner S, Alkorta I, et al. Defining the hydrogen bond: an account. IUPAC; 2011. Defining the hydrogen bond: an account. [Google Scholar]
- 3.Davis AM, Teague SJ. Hydrogen Bonding, Hydrophobic Interactions, and Failure of the Rigid Receptor Hypothesis. Angewandte Chemie International Edition. 1999;38:736. doi: 10.1002/(SICI)1521-3773(19990315)38:6<736::AID-ANIE736>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 4.DeChancie J, Houk KN. The origins of femtomolar protein-ligand binding: hydrogen-bond cooperativity and desolvation energetics in the biotin-(strept)avidin binding site. J Am Chem Soc. 2007;129:5419–29. doi: 10.1021/ja066950n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong KA, Tidor B, Cheng AC. Optimal charges in lead progression: a structure-based neuraminidase case study. J Med Chem. 2006;49:2470–7. doi: 10.1021/jm051105l. [DOI] [PubMed] [Google Scholar]
- 6.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 8.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 9.Durrant JD, McCammon JA. BINANA: A novel algorithm for ligand-binding characterization. J Mol Graph Model. 2011;29:888–93. doi: 10.1016/j.jmgm.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durrant JD, McCammon JA. NNScore: A Neural-Network-Based Scoring Function for the Characterization of Protein-Ligand Complexes. J Chem Inf Model. 2010;50:1865–71. doi: 10.1021/ci100244v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Fang X, Lu Y, Wang S. The PDBbind database: collection of binding affinities for protein-ligand complexes with known three-dimensional structures. J Med Chem. 2004;47:2977–80. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Fang X, Lu Y, Yang CY, Wang S. The PDBbind database: methodologies and updates. J Med Chem. 2005;48:4111–9. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 13.Hu L, Benson ML, Smith RD, Lerner MG, Carlson HA. Binding MOAD (Mother Of All Databases) Proteins. 2005;60:333–40. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucl Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 16.Durrant JD, Hall L, Swift RV, Landon M, Schnaufer A, Amaro RE. Novel Naphthalene-Based Inhibitors of Trypanosoma brucei RNA Editing Ligase 1. PLoS Negl Trop Dis. 2010;4:e803. doi: 10.1371/journal.pntd.0000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J Mol Biol. 2004;343:601–13. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 18.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W5. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W7. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–25. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 03. Wallingford CT: Gaussian, Inc; 2004. Gaussian 03. [Google Scholar]
- 23.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–74. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926. [Google Scholar]
- 25.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. Journal of computational chemistry. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 27.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]