Abstract
RNA internal loops often display a variety of conformations in solution. Herein, we visualize conformational heterogeneity in the context of the 5′CUG/3′GUC repeat motif present in the RNA that causes myotonic dystrophy type 1 (DM1). Specifically, two crystal structures are disclosed of a model DM1 triplet repeating construct, 5′r(UUGGGC(CUG)3GUCC)2, refined to 2.20 Å and 1.52 Å resolution. Here, differences in orientation of the 5′ dangling UU end between the two structures induce changes in the backbone groove width, which reveals that non-canonical 1×1 nucleotide UU internal loops can display an ensemble of pairing conformations. In the 2.20 Å structure, CUGa, the 5′UU forms one hydrogen-bonded pairs with a 5′UU of a neighboring helix in the unit cell to form a pseudo-infinite helix. The central 1×1 nucleotide UU internal loop has no hydrogen bonds, while the terminal 1×1 nucleotide UU internal loops each form a one hydrogen-bonded pair. In the 1.52 Å structure, CUGb, the 5′ UU dangling end is tucked into the major groove of the duplex. While the canonical paired bases show no change in base pairing, in CUGb the terminal 1×1 nucleotide UU internal loops form now two hydrogen-bonded pairs. Thus, the shift in major groove induced by the 5′UU dangling end alters non-canonical base patterns. Collectively, these structures indicate that 1×1 nucleotide UU internal loops in DM1 may sample multiple conformations in vivo. This observation has implications for the recognition of this RNA, and other repeating transcripts, by protein and small molecule ligands.
RNA is an important target for small molecules;(1) however, most RNA targets have not been exploited as such because little is known about the RNA motifs that specifically recognize small molecules and the small molecules that specifically bind RNA. One notable class of RNAs that could be targeted by small molecules is triplet and tetranucleotide repeating transcripts.(2) As of 2011, a variety of untreatable diseases are caused by expanded repeating transcripts, including myotonic dystrophy types 1 and 2 (DM1 and DM2, respectively), spinocerebellar ataxia type 3 (SCA3), Fragile X syndrome, and Huntington’s disease (HD).(3)
In contrast to most diseases caused by repeating transcripts, DM1 and DM2 repeating RNAs are not translated into protein. DM1 is caused by an expansion of a CUG repeat in the 3′ untranslated region (UTR) of the dystrophia myotonica protein kinase (DMPK) mRNA (4–6) while DM2 is caused by an expansion of a CCUG repeat in intron 1 of the zinc finger 9 protein (ZNF9) pre-mRNA.(7) A model for how these RNAs contribute to DM1 and DM2 has been developed and is centered on an RNA gain-of-function that occurs upon expansion. Long, toxic repeats fold into hairpins and bind the RNA splicing regulator muscleblind-like protein 1 (MBNL1). Sequestration of MBNL1 by the repeating RNAs causes splicing defects in a subset of pre-mRNAs including the insulin receptor and the muscle main chloride ion channel.(4–6)
Analysis of this disease model provides a strategy for treating DM1 or DM2: a small molecule binds to long CUG repeats and either displaces MBNL1 or inhibits its binding, thereby restoring normal function. Several groups have explored such a strategy. Small molecules have been developed to target DM1 repeats including compounds identified through screening (8) and an acridine-triazole conjugate.(9) Notably, a designer modular assembly strategy, which could be generally applied to all repeating transcripts, provided potent nanomolar in vitro inhibitors of the DM1 RNA-MBNL1 complex. (10–13) Morpholino oligonucleotides (14) and pentamidine (15) correct splicing defects in a DM1 mouse model.
Previously, structural studies have been completed on model RNA systems containing CUG repeats.(16, 17) In these structures, the 1×1 nucleotide UU internal loops adopt either a zero or a one hydrogen-bonded pairing structure. A refined NMR structure and molecular dynamics simulation of 5′r(CCGCUGCGG)2 showed that the 1×1 nucleotide UU internal loop prefers a one hydrogen-bonded structure, but it is dynamic and can interconvert between zero, one, and two hydrogen-bonded pairs without breaking the loop’s closing base pairs.(18)
In this study, two crystal structures of a self-complementary duplex with three copies of the DM1 5′CUG/3′GUC motif are disclosed at 2.20 Å and 1.52 Å resolution. The structures have several notable differences from the structures previously reported. For example, the UU pairs adopt different conformations including pairing geometries that are consistent with zero, one, and two hydrogen-bonded pairs depending upon their position in the helix. The structure of the external 1×1 nucleotide UU loops are different in the two structures due to differences in the structures of the 5′ UU dangling ends. For example, a 1×1 nucleotide internal loop with two hydrogen bonds is observed when the dangling end is tucked into the groove while a one hydrogen-bonded pair is observed when the dangling ends form a pseudo-infinite helix. Evidently, the structure of the dangling end allows for conformational selection of different pairings in the 1×1 nucleotide UU internal loops in the crystal structure. However, in both structures the central 1×1 nucleotide UU internal loop adopts a zero hydrogen-bonded conformation. Collectively, the available information on CUG repeats structures indicate that the 1×1 nucleotide UU internal loops could sample multiple conformations in vivo, which has implications for the recognition of this RNA by protein and small molecule ligands.
MATERIALS AND METHODS
Purification of RNA
Deprotected and desalted RNA was purchased from Integrated DNA Technologies, Inc. The RNA was dissolved in water and purified by HPLC on a Waters HPLC instrument with an attached UV-Vis detector that monitored absorbance at 220 and 254 nm. An XTerra Prep MS C18 column (7.8 ×150 mm, 5 μm) was used. A linear gradient was applied (100% to 0% 10 mM triethylammonium acetate, pH 7.0 in acetonitrile over 55 min) with a flow rate of 2 mL/min; tR = 25 min. Fractions containing RNA were lyophilized, dissolved in DEPC-treated water, and desalted by using a Sephadex PD-10 pre-packed size exclusion column. Fractions containing RNA were combined and lyophilized. The RNA sample was re-dissolved in DEPC-treated water, and the concentration was determined by its absorbance at 260 nm at 95 °C. Molar extinction coefficients were determined by using the Hyther server (Nicolas Peyret and John SantaLucia Jr., Wayne State University, Detroit, MI),(19, 20) which uses parameters based on molar absorptivity of RNA nearest neighbors.(21)
Crystallization of CUG Oligonucleotides
A 1.2 mM solution of RNA duplex (Figure 1a) was prepared in DEPC-treated water. The RNA was folded by heating at 60 °C for 5 min and slowly cooling to room temperature by placing the sample on the bench top. The sitting drop vapor diffusion method and a Qiagen Nucleix Suite kit were used to screen conditions that provided high quality crystals. Samples were placed at 18 °C. The CUGa crystals that diffracted to 2.20 Å resolution were produced from the reservoir solution containing 10 mM magnesium sulfate, 50 mM sodium cacodylate, pH 6.0 and 1.8 M lithium sulfate whereas the CUGb crystals that diffracted to 1.52 Å resolution were produced from the reservoir solution containing 15 mM magnesium acetate, 50 mM sodium cacodylate, pH 6.0 and 1.7 M ammonium sulfate. Both crystals appeared within 2–3 days.
Figure 1.
The secondary structure, refined structure, and crystal packing of the RNA construct. A, the secondary structure of the oligonucleotide r(CUG) duplex model that crystalizes. B and C, the central 5′CUG/3′GUC of the RNA including the electron density map at 1.0σ for CUGa and CUGb respectively. D and E, overlay of the backbone of CUGa (blue) and CUGb (orange) onto the backbone of a model RNA duplex in which the 1×1 nucleotide UU internal loops are replaced with AU pairs (gray). F and G, a view of the stacking of pseudo-infinite or stacked helices on each other in the crystal packing for CUGa and CUGb, respectively. H and I, a view of the stacking of helices next to each other for CUGa and CUGb, respectively.
Data Collection, Structure Determination and Refinement
Crystals used for data collection were flash frozen by immersion in liquid nitrogen. The complete diffraction data sets were collected at the beamline 9-1 at SSRL or the beamline LS-CAT (21-ID) at the Advanced Photo Source (Argonne National Laboratory) under cryostream at 100K using ADSC CCD detectors. Data were processed and scaled using HKL2000.(22) The structural solutions of both crystal forms were determined by molecular replacement using PHASER (23), a component of the PHENIX suite (24), using a search model of a 17 base pair standard A-form RNA generated with Coot. The atomic models were refined with PHENIX suite. The statistics for data collection and processing, and refinements are shown in Supporting Information Table 1. The structures were deposited in the Protein Data Bank (CUGa: 3SZX; CUGb: 3SYW).
Calculation of Electrostatic Potentials
The electrostatic potentials of the CUGa and CUGb structures were calculated from the corresponding PDB files; electrostatic potentials for fully paired RNAs containing AU and GC base pairs were calculated from duplexes that were rebuilt using Amber topology parameters. Hydrogen atoms were added to the RNAs and positioned based on a previously described algorithm.(24) After construction, hydrogen atoms were checked for steric conflicts. Atom partial charges and atomic radii were assigned based on Amber99 force-field using the program AMBER.(25) The RNA molecule was treated as a low dielectric medium within the volume enclosed by its solvent-accessible surface (probe radius = 1.4 Å). A dielectric constant of 2 was used to account for electronic polarizability effects. The surrounding solvent was treated as a continuum with a dielectric constant of 80. To account for ion size on the RNA molecule surface, a 2.0 Å ion-exclusion radius was added. Ten grid points per square angstrom were used to construct molecular surfaces.
All electrostatic calculations were completed at 298 K. In order to solve the numerical equation for calculating the electrostatic surface potential, a focusing method was used.(26) The equation was first solved using a coarse grid, which was then refined to provide a more accurate, finer grid using the Dirichlet boundary condition.(27) Electrostatic potentials shown in depict the solvent excluded molecular surfaces. As RNA molecules are highly negatively charged molecules, most of the molecular surface has a negative electrostatic potential with patches of positive charge.
Calculation of structural parameters
Helical parameters, groove widths and torsion angles were calculated using the program 3DNA.(25) To avoid computational artifacts arising from non-canonical base pairing, sequence independent measurements were made using a vector connecting the C1′ atoms.
RESULTS AND DISCUSSION
The three-dimensional refined structures of an RNA construct with three copies of the 5′CUG/3′GUC motif found in DM1 RNA
The secondary structure of the RNA construct that was crystallized is shown in Figure 1A. Duplex regions adjacent to the 5′CUG/3′GUC repeats were added to stabilize the ends of the duplex and have also been shown to non-covalently bind heavy atoms that can be used for phasing.(26) In the case of the structures presented here, models were able to be built that fit the diffraction data (Figure 1B–I) without having to collect data on heavy atom derivatives. The 5′ UU dangling end was used because it enables crystallization by forming pseudo-infinite helices with neighboring RNAs in a unit cell (Figures 1F–I).(27, 28) The highest quality crystals appeared in 2–3 days, diffracting to 2.20 Å and 1.52 Å (CUGa and CUGb, respectively).
Both of the duplexes crystallize in double stranded helical structures that form a paired region that flanks three 1×1 nucleotide UU internal loops. The secondary structure is similar for both crystal forms, and both crystals display extended helical structures in the packing (Figure 1F–I). The similarity of the structure to canonically paired, A-form RNA is illustrated by an overlay of the backbone from the CUGa structure on a duplex in which the 1×1 nucleotide UU loops were replaced with AU pairs (Figure 1D and E). Globally, these structures have very similar shapes.
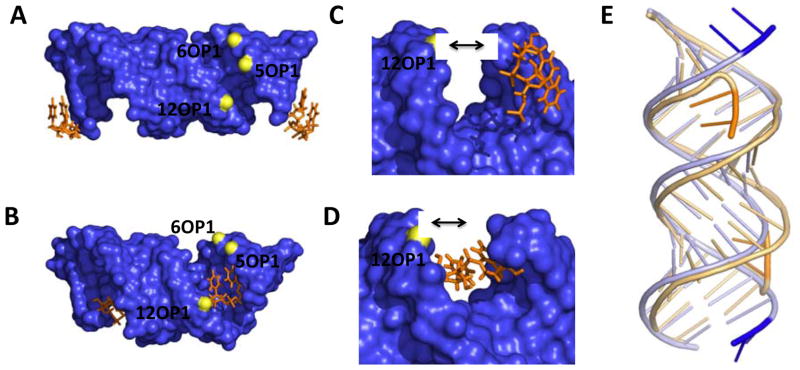
An overlay of CUGa and CUGb and the effect of the orientation of the 5′UU end is shown in Figure 2. The two structures have significant differences in the width of the grooves due to very different orientations of the 5′UU dangling ends (Figure 2A–D). In CUGa, each U in the 5′UU dangling ends forms a one hydrogen-bonded pair with a 5′UU dangling end of a neighboring duplex in the unit cell to form a pseudo-infinite helix as expected (Figure 1F and 2A and C).(27, 28) In CUGb, the 5′UU dangling end curves into the major groove of the RNA duplex (Figures 1G and 2B and D); this orientation is driven by an intramolecular hydrogen bond formed between N3 of U2 and a non-bridging oxygen atom on O2P of G5 (Supporting Information)
Figure 2.
The orientation of the 5′ dangling UU ends in the crystal structures and their effect on groove width. A and B, views down the major and minor groove of CUGa and CUGb, respectively. C and D, close up views of of the 5′UU dangling ends in CUGa and CUGb, respectively. E, an overlay of the backbone of CUGa (blue) onto the backbone of CUGb (orange). The black arrows are used to illustrate the relative difference in the size of the groove.
Comparison of the orientation of the dangling ends in the two structures shows that there is an increase in the width of the major groove when the 5′UU is tucked into it (Figure 2A–D). The distance between the phosphate atoms in P6 and P12 on either end of the groove increases from 12.6 to 17.3 Å, while the distance between phosphate atoms in P5 to P12 increases from 9.4 to 14.5 Å (Figure 2C and D). Such difference in the width can affect solvent and ionic interactions that may not be visible in each of these structures at the reported resolution. These effects could drive the differences in the orientation of the UU loops.
This shift in the backbone grooves is also associated with contraction of the C1′-C1′ distances for CUGb. For CUGa (Figure 3A), distances range from 10.0 to 10.5 Å, which is similar to the C1′-C1′ distance of ~10.5 Å observed in standard A-form RNA helical geometry. For CUGb, the C1′-C1′ distance for the two hydrogen-bonded UU pairs is 8.8 Å, indicating local contraction (Figure 3B), induced by the widening of the adjacent major groove by the dangling UU pair.
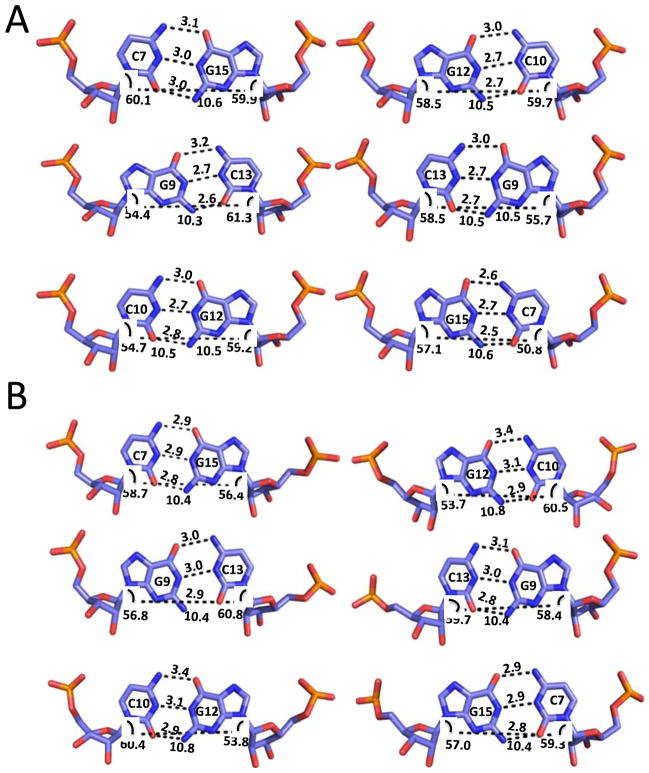
Figure 3.
The three dimensional structures of the 1×1 nucleotide UU internal loops; A and B are the density maps at 1σ and structures of the 1×1 nucleotide UU internal loops in CUGa and CUGb, respectively. Different types of 1×1 nucleotide UU Internal loops are observed in both structures. The dashed lines indicate the lengths of either the C1′-C1′ or hydrogen bond distance and the number above these lines are the measured distance
Structures of the 1×1 nucleotide UU internal loops
Our trapping of the 5′CUG/3′GUC motif into different conformers reveals that 1×1 nucleotide UU internal loops display an ensemble of base pairings between the two structures. Interestingly, there are three different pairing geometries for the 1×1 nucleotide UU internal loops in CUGa (Figure 3A). The two UU pairs in 5′CUG/3′GUC motifs at the ends of the duplex have geometries consistent with a one and a zero hydrogen-bonded structure (Figure 3A). In the one hydrogen-bonded geometry, the hydrogen bond is between the U8N3-U14O4 and has a distance of 3.0 Å. The zero hydrogen-bonded geometry on the other end has a distance of 3.6 Å (between U14N3-U8O4), which is outside of the standard hydrogen bond distance of 3.2 Å. In both of these structures, the geometry is similar in that the U8O4 is inclined towards the U14N3 atom; however, differences in the degree of the base’s incline determines whether a hydrogen bond forms. In contrast, the central 1×1 nucleotide UU internal loop in CUGa has no hydrogen bonds and the carbonyl oxygen atoms are not inclined towards each other.
In CUGb (Figure 3B), the pairings of the 1×1 nucleotide UU internal loops are similar and different than those in CUGa. For example, the central 1×1 nucleotide UU internal loop has no hydrogen bonds and has a very similar orientation as the central 1×1 nucleotide UU internal loop in CUGa. In contrast, the two terminal 1×1 nucleotide UU internal loops have two hydrogen bonds between the U bases (U8N3 with U14O2 and U8O4 with U14N3) and an additional hydrogen bond between the U14O4’s and a water molecule; in CUGa, these pairs had zero and one hydrogen bonds (Figure 3A).
Canonical base pairs and stacking of 1×1 nucleotide UU Internal loops on closing base pairs
The structures of the G-C base pairs that close each of the 1×1 nucleotide UU internal loops have a standard Watson-Crick geometry (Figure 4A). In addition, the terminal base paired regions at the end of the duplex have hydrogen bonding and stacking interactions that are standard for canonically paired duplexes.
Figure 4.
The three dimensional structures of the 1×1 nucleotide UU internal loop closing base pairs in CUGa (A) and CUGb (B). Each of the loop closing pairs has a geometry consistent with that of Watson-Crick GC base pairs.
Figure 5 depicts the stacking interactions for each 1×1 nucleotide UU internal loop on the loop’s closing GC pairs and representative stacking plots for a GC step in the duplex. There is little difference in the stacking of the internal loop U’s on adjacent G-C pairs, for the different types of loops (zero, one, or two hydrogen-bonded), although there is more overlap between the region around U8N3 and the exocyclic amine of a stacked guanine in the two hydrogen-bonded structure. The 3′ closing G-C pair of one 5′CUG/3′GUC motif stacks well on the 5′ closing C-G pair of the next motif (Figure 5). Evidently, differences in stacking of the 1×1 nucleotide UU internal loops does not account for the observed differences in geometry.
Figure 5.
Stacking of the 1×1 nucleotide UU internal loops on the loop closing base pairs. A and B are the structures for CUGa and CUGb, respectively. For each type of 1×1 nucleotide UU internal loop, there is little stacking on the closing pairs. However, the 3′ closing GC pairs of one 5′CUG/3′GUC motif stack well on the 5′ closing GC pairs of the next 5′CUG/3′GUC motif.
Electrostatics of the duplexes in comparison to canonically paired RNAs
Figure 6 provides a comparison between the electrostatic charge distribution of CUGa and CUGb and fully paired RNA duplexes (the 1×1 nucleotide UU internal loops are replaced with either A-U or G-C base pairs). There is an alternating pattern of positive and negative charge distribution in the minor groove in all structures. The density of the partial positive charge distribution is higher in CUGa and CUGb structures compared to the paired structures; however, CUGb has the largest density of partial positive charge.
Figure 6.
Comparison of the electrostatic charge distributions for CUGa and CUGb to canonically paired duplexes. A and B, electrostatic charge distributions of refined CUGa and CUGb, respectively. Panels C and D, electrostatic charge distributions of a structure in which the 1×1 nucleotide UU internal loops in the CUG construct were replaced with AU pairs and GC pairs, respectively.
Comparison of CUGa to CUGb
Globally, the two structures reported herein are similar. However, two significant differences are the orientation of the 5′ double dangling UU end and the orientation of the external 1×1 nucleotide UU internal loops (Figures 1, 2, and 3 and Supporting Information). That is, the orientation of the 5′UU ends is associated with different structures of the internal loops. If the 5′UU dangling ends are in the major groove of the flanking duplexes then the external 1×1 nucleotide UU internal loops form two hydrogen-bonded pairs. If however, the 5′UU dangling ends form base pairs with the adjacent helix, the 1×1 nucleotide UU internal loops have a zero or one hydrogen-bonded geometries. Thus, the shift in width of the backbone allows for conformational selection of different types of 1×1 nucleotide UU pairs in the crystal structure, even though canonical base pairing and stacking are not disrupted (Figures 4 and 5). Conformational selection has been used by small molecules to select different orientations of non-canonically paired or unpaired RNA loops (29–31). In those cases, small molecules make direct contacts to loop nucleotides.
Comparison to other structures of CUG systems
Two crystal structures of RNAs containing the DM1 motif have been previously reported. In the structure of r((CUG)6)2, the majority of the 1×1 nucleotide UU internal loops (5 of 6) have a zero hydrogen bonded conformation, similar to the central 1×1 nucleotide UU loops in both CUGa and CUGb.(16) There is evidence of a minor population of a one hydrogen-bonded conformation in r((CUG)6)2 (1 of 6) that is similar to the structure observed for a terminal UU pair in a 5′CUG/3′GUC motif in CUGa.(16).
In the previously reported structure of r(G(CUG)2C)2, each UU pair forms a one hydrogen bonded conformation that is considered a UU “stretched wobble” pair.(17) In this “stretched” U-U pair, two water molecules were found to interact with the UU pairs by forming contacts to O2 and O4 carbonyl atoms in the major and the minor grooves. The pair has an orientation similar to the one hydrogen-bonded UU pair that was found in CUGa; however, we could not observe specifically bound water molecules, which may be due to the lower resolution of this structure (2.20 Å versus 1.23 Å). A single bound water molecule was observed, however, to two of the three UU pairs in the CUGb structure. The bound water molecules in CUGb interact with the same functional groups in the major groove as was observed in r(G(CUG)2 C)2.
A NMR spectroscopy and molecular dynamics study of an RNA duplex with one copy of 5′CUG/3′GUC or 5′r(CCGCUGCGG)2 has also been disclosed.(18) These studies showed that the lowest energy pairing for the 5′CUG/3′GUC motif is a single hydrogen bonded structure that is similar to the UU pair found in CUGa. However, the 1×1 nucleotide UU internal loop in this structure is dynamic and samples between zero, one, and two hydrogen-bonded structures without breaking the loop closing base pairs.
Comparisons of each of these structural studies with the CUGa and CUGb structures reported herein support a model in which the 1×1 nucleotide UU internal loops in the expanded CUG repeats sample multiple conformations. Collectively, each structure may provide unique snapshots into the ensemble of structures that DM1 repeats sample that could be exploited as binding sites for small molecules. Furthermore, the structures of the central 1×1 nucleotide UU internal loops in both CUGa and CUGb as well as the majority of 1×1 nucleotide internal loops in the structure of r((CUG)6)2 have a zero hydrogen bonded conformation. Since each of the internal 1×1 nucleotide UU internal loops are surrounded by 5′CUG/3′GUC motifs on each slide as would be observed in CUG repeats in vivo, zero hydrogen bonded conformation may be the most populated one in vivo.
Considerations for high affinity recognition of DM1, CUG repeating transcripts
The structures of the CUG repeats could allow for the development of selective and non-toxic small molecules targeting DM1 RNA repeats. In the different hydrogen bonded structures, hydrogen bond acceptors and donors are presented to the major groove that could bind to a small molecule. In the zero, one, and two hydrogen-bonded 1×1 nucleotide UU Internal loop structures, a carbonyl oxygen is available that could serve as a hydrogen bond acceptor for a small molecule. Since the electrostatic potential of the CUG repeats is different than that of base paired RNA, these differences could also be exploited for selective small molecule binding.
Interestingly, several types of small molecules have been found to bind CUG repeats. In general, these compounds are rich in hydrogen bond donors that could form contacts with the available hydrogen bond-accepting carbonyl groups discussed above. For example, Hoechst 33258(11), kanamycin derivatives (10, 12, 13), and pentamidine (15) are all positively charged ligands that could form direct contacts to carbonyl atoms in the CUG repeats. Interestingly, Hoechst 33258 has been shown to interact with DNA by forming contacts between its benzimidazole side chains and the O2 atoms of T in AT rich regions.(32)
Another interesting series of compounds that has been shown to bind CUG repeats are triazine-acridine conjugates.(9) Based on the crystal structures of the central 1×1 nucleotide UU internal loops in CUGa and CUGb, one could envision the triazine moiety easily forming a full complement of three hydrogen bonds to the 1×1 nucleotide UU internal loop with a zero hydrogen-bonded structure. Such interactions could also form with the one and the two hydrogen-bonded UU pairs; however, it would be at the expense of breaking the hydrogen bonds between the U’s, which are each worth about 1 kcal/mol.(33)
Considerations for specific targeting of DM1, CUG repeating transcripts
The most significant barrier for specifically targeting CUG repeats that cause DM1 is the presence of 1×1 nucleotide UU internal loops in other, highly abundant RNA transcripts. Notable is the presence of a 1×1 nucleotide UU internal loop in the Homo sapiens rRNA A-site. Interestingly, analysis of structural data on this UU pair shows that it exists in multiple conformations including one and two hydrogen-bonded pairs. In structures of isolated cytoplasmic and mitochondrial A-sites, (34, 35); Lynch, 2001 #1237) the UU loop has a one or a two hydrogen-bonded conformation. The H. sapiens A-site is an accessible target for small molecules; aminoglycoside antibiotics bind the human A-site and stimulate read-through translation.(36) In a crystal structure of the entire yeast ribosome in the ratcheted state, the 1×1 nucleotide UU internal loop forms a one-hydrogen bonded pair (37) (see Supporting Information). Additionally, many cellular tRNAs also contain 1×1 nucleotide UU internal loops that could bind small molecules that are intended to bind DM1 CUG repeats (Supporting Information).
Selective binding of the CUG hairpin by small molecules is compounded by the relative expression levels of RNA in cells.(38–40) Highly abundant rRNAs and tRNAs comprise ≈ 80% and ≈ 15%, respectively, of total cellular RNA whereas total mRNA constitutes only ≈ 5%. Therefore, specific targeting of a single mRNA is a considerable challenge since the levels of a single mRNA or other low abundance RNA is ≪1% of the total RNA content of cells.
Despite these challenges, strategies are being developed to design specific ligands that target the DM1 RNA, which has regularly repeating copies of the 5′CUG/3′GUC motif. By modularly assembling ligands that bind multiple 5′GUC/3′GUC motifs simultaneously, compounds can be designed that target the DM1 RNA with affinities and specificities that exceed that of MBNL1 by orders of magnitude.(10–13) Assembled compounds also have improved potencies for inhibition of the DM1 RNA-MBNL1 interaction by sequestering larger amounts of surface area on the target RNA than a lower molecular weight compound.(10–13) When using modularly assembled structures, however, a careful balance between potency and size must be considered, as a higher molecular weight compound is likely to be more potent but less cell permeable.
Supplementary Material
ABBREVIATIONS
- DEPC
diethyl pyrocarbonate
- DM1
myotonic dystrophy type 1
- DM2
myotonic dystrophy type 2
- DMPK
dystrophia myotonica protein kinase
- HD
Huntington’s disease
- HPLC
high performance liquid chromatography
- MBNL1
muscleblind-like 1 protein
- MD
molecular dynamics
- mRNA
messenger RNA
- PDB
protein data bank
- rRNA
ribosomal RNA
- SCA3
spinocerebellar ataxia type 3
- tRNA
transfer RNA
- UTR
untranslated region
- ZNF9
zinc finger 9 protein
Footnotes
These structures have been deposited in the PDB (CUGa: 3SZX; CUGb: 3SYW). This work was funded by the National Institutes of Health (3R01GM079235-02S1 and 1R01GM079235-01A2 to MDD) and by The Scripps Research Institute. MDD is a Camille & Henry Dreyfus New Faculty Awardee, a Camille & Henry Dreyfus Teacher-Scholar, and a Research Corporation Cottrell Scholar.
Supporting information. Supporting information is available and describes the crystal parameters and Table of analysis of the structures. This material is available free of charge at www.pubs.acs.org
References
- 1.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 2.Caskey CT, Pizzuti A, Fu YH, Fenwick RG, Jr, Nelson DL, Kuhl DP. Triplet repeat mutations in human disease. Science. 1992;256:784–789. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- 3.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 4.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 5.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 6.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 7.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 8.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: discovery of lead compounds targeting myotonic dystrophy (DM1) J Am Chem Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc Natl Acad Sci U S A. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MM, Childs-Disney JL, Pushechnikov A, French JM, Sobczak K, Thornton CA, Disney MD. Controlling the specificity of modularly assembled small molecules for RNA via ligand module spacing: targeting the RNAs that cause myotonic muscular dystrophy. J Am Chem Soc. 2009;131:17464–17472. doi: 10.1021/ja906877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational Design of Ligands Targeting Triplet Repeating Transcripts That Cause RNA Dominant Disease: Application to Myotonic Muscular Dystrophy Type 1 and Spinocerebellar Ataxia Type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MM, Pushechnikov A, Disney MD. Rational and Modular Design of Potent Ligands Targeting the RNA that Causes Myotonic Dystrophy 2. ACS Chem Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Disney MD, Lee MM, Pushechnikov A, Childs-Disney JL. The Role of Flexibility in the Rational Design of Modularly Assembled Ligands Targeting the RNAs that Cause the Myotonic Dystrophies. Chembiochem. 2010 doi: 10.1002/cbic.200900716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc Natl Acad Sci U S A. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Structural insights into CUG repeats containing the ‘stretched U-U wobble’: implications for myotonic dystrophy. Nucleic Acids Res. 2009;37:4149–4156. doi: 10.1093/nar/gkp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkesh R, Disney MD, Fountain M. NMR Spectroscopy and Molecular Dynamics Simulation of r(CCGCUGCGG)2 Reveal a Dynamic UU Internal Loop Found in Myotonic Dystrophy Type 1. Biochemistry. 2011 doi: 10.1021/bi101896j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A. A, C. C, G. G, and T. T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 20.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci U S A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keel AY, Rambo RP, Batey RT, Kieft JS. A general strategy to solve the phase problem in RNA crystallography. Structure. 2007;15:761–772. doi: 10.1016/j.str.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicens Q, Westhof E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- 28.Wahl MC, Rao ST, Sundaralingam M. The structure of r(UUCGCG) has a 5′-UU-overhang exhibiting Hoogsteen-like trans U. U base pairs. Nat Struct Biol. 1996;3:24–31. doi: 10.1038/nsb0196-24. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Kadakkuzha BM, Zhao L, Fan M, Qi X, Xia T. Dynamic ensemble view of the conformational landscape of HIV-1 TAR RNA and allosteric recognition. Biochemistry. 2011;50:5042–5057. doi: 10.1021/bi200495d. [DOI] [PubMed] [Google Scholar]
- 30.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM. RNA Dynamics by Design: Biasing Ensembles Towards the Ligand-Bound State. Angew Chem Int Ed Engl. 2010 doi: 10.1002/anie.201000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng MK, Usman N, Frederick CA, Wang AH. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG) Nucleic Acids Res. 1988;16:2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SantaLucia J, Jr, Kierzek R, Turner DH. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992;256:217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- 34.Kondo J, Urzhumtsev A, Westhof E. Two conformational states in the crystal structure of the Homo sapiens cytoplasmic ribosomal decoding A site. Nucleic Acids Res. 2006;34:676–685. doi: 10.1093/nar/gkj467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo J, Westhof E. The bacterial and mitochondrial ribosomal A-site molecular switches possess different conformational substates. Nucleic Acids Res. 2008;36:2654–2666. doi: 10.1093/nar/gkn112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo J, Hainrichson M, Nudelman I, Shallom-Shezifi D, Barbieri CM, Pilch DS, Westhof E, Baasov T. Differential selectivity of natural and synthetic aminoglycosides towards the eukaryotic and prokaryotic decoding A sites. Chembiochem. 2007;8:1700–1709. doi: 10.1002/cbic.200700271. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 38.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6. W. H. Freeman and Company; New York: 2007. [Google Scholar]
- 39.Johnson LF, Abelson HT, Penman S, Green H. The relative amounts of the cytoplasmic RNA species in normal, transformed and senescent cultured cell lines. J Cell Physiol. 1977;90:465–470. doi: 10.1002/jcp.1040900310. [DOI] [PubMed] [Google Scholar]
- 40.Johnson LF, Williams JG, Abelson HT, Green H, Penman S. Changes in RNA in relation to growth of the fibroblast. III. Posttranscriptional regulation of mRNA formation in resting and growing cells. Cell. 1975;4:69–75. doi: 10.1016/0092-8674(75)90135-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.