Summary
Several recent studies showed that mouse and human fibroblasts can be directly reprogrammed to induced neuronal (iN) cells, bypassing a pluripotent intermediate state. However, fibroblasts represent heterogeneous mesenchymal progenitor cells that potentially contain neural crest lineages, and the cell of origin remained undefined. This raises the fundamental question whether lineage reprogramming is possible between cell types derived from different germ layers. Here, we demonstrate that terminally differentiated hepatocytes can be directly converted into functional iN cells. Importantly, single cell and genome-wide expression analyses showed that fibroblast- and hepatocyte-derived iN cells not only induced a neuronal transcriptional program, but also silenced their donor transcriptome. The remaining donor signature decreased over time and could not support functional hepatocyte properties. Thus, the reprogramming factors lead to a binary lineage switch decision rather than an induction of hybrid phenotypes but iN cells retain a small but detectable epigenetic memory of their donor cells.
Introduction
Following our initial report that mouse fibroblasts can be converted into functional neuronal cells (iN cells) by the ectopic expression of the three transcription factors Ascl1, Brn2 and Myt1l several additional groups and we have recently shown the induction of neuronal traits also in human fibroblasts based on the same factors (Ambasudhan et al., 2011; Caiazzo et al., 2011; Pang et al., 2011; Pfisterer et al., 2011; Qiang et al., 2011; Son et al., 2011; Vierbuchen et al., 2010; Yoo et al., 2011). A fundamental question not addressed in these studies is whether terminally differentiated cells as well as cells definitely derived from a non-ectodermal lineage can be converted into neurons. Primary fibroblast cultures are inherently heterogeneous in respect of both cell type and maturation stage and can contain neural crest cell derivatives Thus, the identity and the maturation stage of the cells that gave rise to iN cells remained undefined. Given these technical limitations we sought to determine whether better defined and more homogenous cell types can be converted into neuronal cells.
Hepatocytes are considered relatively homogeneous and account for 78% of the liver mass (Zhao and Duncan, 2005) (Braeuning et al., 2006). Here we show that terminally differentiated mouse hepatocytes can be converted into functional iN cells. This is the first proof that an endodermal cell can be converted into an ectodermal cell and that a definitely non-ectodermal (and non-neural crest) cell can be converted into a functional neuronal cell. A well-characterized and specific Albumin-Cre transgenic mouse line combined with a robust fluorescence-based genetic lineage tracing system allowed us to not only unequivocally demonstrate that albumin-expressing hepatocytes were the origin of converted neuronal cells, but also to specifically characterize the hepatocyte-derived (Hep-) iN cells and the reprogramming process. Our data show that both MEF-iN and Hep-iN cells had very efficiently downregulated the MEF and hepatocyte-specific transcriptional network. This result leads to the surprising conclusion that the same neuronal transcription factors can induce the downregulation of two very different transcriptional programs. Similar to induced pluripotent (iPS) cells we observed a small degree of epigenetic memory, which diminished over time (Bar-Nur et al., 2011; Kim et al., 2010; Ohi et al., 2011; Polo et al., 2010). We conclude that iN cells are truly converted cells and not simple hybrid phenotypes between neurons and donor cell types.
Results
Induction of neuronal cells from liver cells
To test whether cells derived from liver can be induced to become neuronal cells, we established primary liver cultures from postnatal days (P) 2-5 wild type and TauEGFP knock-in mice (Tucker et al., 2001; Wernig et al., 2002). Four days after isolation, the majority of cells showed a typical epithelial morphology and expressed albumin, α-fetoprotein and α-anti-trypsin (Figures 1A,J and S1A). One week post explantation, a typical culture was composed of 60% albumin-positive hepatocytes, 16% myeloid cells, 2% Kupffer cells and 2% endothelial cells (Figure S1B,D). Absence of the neuronal or neural progenitor cell markers Sox2, Brn2, MAP2, and NeuN in the culture was confirmed by immunofluorescence (data not shown). The rare (~1/5000) Tuj1-positive cells had a flat morphology, not resembling neuronal cells (Figure S1C). TauEGFP-positive cells were not detectable in these cultures as evaluated by flow cytometry or fluorescence microscopy.
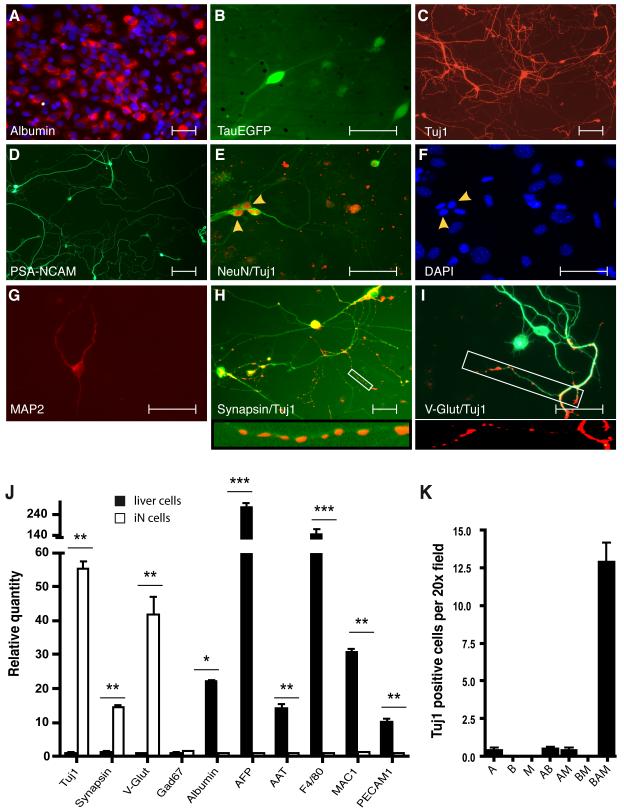
Figure 1. Induction of neuronal cells from liver cells.
(A) Liver cells expressed albumin after isolation (red). (B) TauEGFP-positive cells with neuronal morphology (green, live image) and (C) Tuj1-positive (red) cells 13 and 22 days after dox induction, respectively. (D) PSA-NCAM (green) (E, F) NeuN (red) Tuj1 (green) (G) MAP2 and (H) synapsin-positive cells from liver cultures displayed complex neuronal morphologies. (I) A fraction of the Tuj1-positive cells expressed vGLUT1. (J) qPCR analysis of TauEGFP-positive FACS-sorted cells 21 days after dox (white) compared to liver cells 4 days after purification (black) (n=3). For neuronal markers the liver cell values were set to 1; for liver cell markers the TauEGFP-sorted cell values were set to 1. (K) Efficiency of generating liver-iN cells was quantified in 15 randomly picked 20x microscopic fields (n=3). Scale bars: 100μm (C), 50μm (A, B, D, E, H-I).
The primary liver cultures were then replated and infected with doxycycline (dox)-inducible lentiviruses containing the cDNAs of Ascl1 (A), Brn2 (B) and Myt1l (M) in various combinations. Thirteen days after addition of dox, TauEGFP-positive cells with a complex neuronal morphology were readily detected in the wells that received all three factors (BAM) (Figure 1B). No neuronal cells were found in any other combination (Figure 1K). Immunofluorescence confirmed that all TauEGFP-positive cells generated by the BAM factors were also Tuj1-positive (Figure 1C). When analyzed three weeks after infection, the cells expressed the additional pan-neuronal markers PSA-NCAM, NeuN, MAP2, and synapsin (Figure 1D-H). A fraction (35 out of 200 counted Tuj1-positive cells) of the cells could also be labeled with an antibody against vesicular glutamate transporter 1 (vGLUT1) (Figure 1I). In contrast, no GAD67-, TH-, ChAT-, or serotonin-positive cells were detected (0 out of at least 200 counted Tuj1-positive cells). As neuronal subtype-specific markers are expressed predominantly in mature stages of neuronal differentiation while Tuj1 labels already early postmitotic immature neurons we conclude that the majority of mature iN cells are excitatory neurons. Moreover, qRT-PCR analysis showed that TauEGFP-positive iN cells 3 weeks after infection had not only induced neuronal transcripts but efficiently silenced transcripts characteristic of the starting cell population (Figure 1J).
Genetic proof that iN cells can be derived from albumin-expressing hepatocytes
We then employed the Cre-LoxP system to unambiguously identify hepatocytes and their cellular progeny in primary liver cultures. An Albumin-Cre transgenic mouse strain was used that had been characterized extensively and shown to specifically label hepatocytes in both fetal and adult mice (Postic et al., 1999; Weisend et al., 2009). Albumin-Cre mice were crossed with ROSA26-mTmG reporter mice which express membranous tdTomato before and membranous EGFP after Cre-mediated recombination (Figure 2A). As expected, the EGFP fluorescence was confined to epithelial cells in freshly isolated liver cultures from these mice (Figure 2B). These cultures were typically composed of ~80% EGFP-positive and ~20% tdTomato-positive cells. However, this ratio declined to 60% EGFP-positive 40% tdTomato-positive cells after one week in culture, implying that hepatocytes were lost and/or other cells outgrew the hepatocytes. Next, we infected these cultures with the three BAM factors and 13 days after dox induction we detected both red and green fluorescent cells with neuronal morphologies (Figure 2C,E). EGFP-positive cells also expressed the neuronal markers Tuj1 and PSA-NCAM (Figure 2D,F). Similar results were obtained using an independent reporter allele (ROSA26-Bgeo) (Mao et al., 1999) where expression of β-galactosidase is induced after Cre-mediated recombination (Figure 2G,H). These results unequivocally demonstrate that iN cells can be derived from terminally differentiated hepatocytes. We therefore termed these cells hepatocyte (Hep)-iN cells.
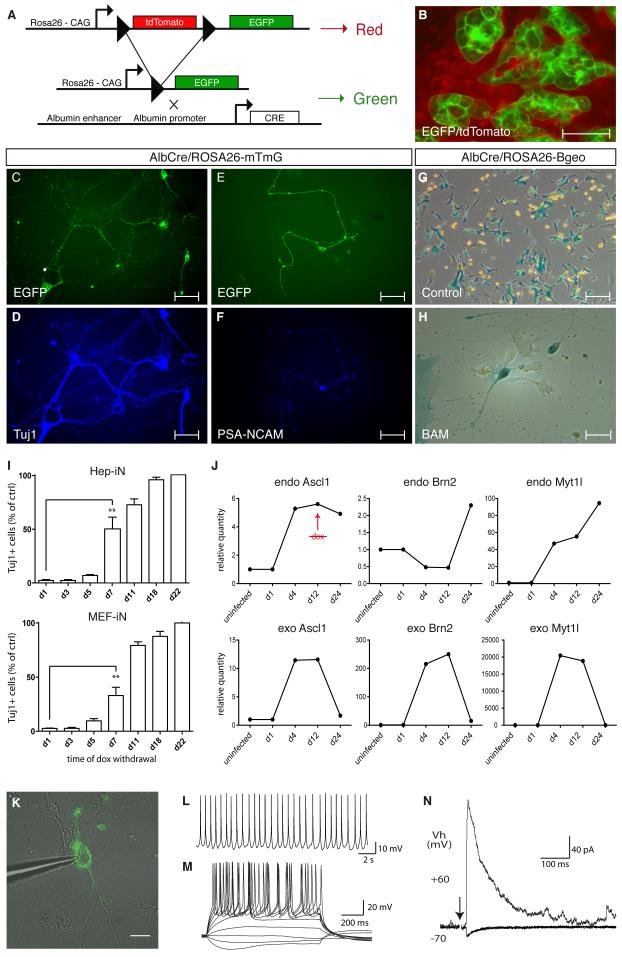
Figure 2. iN cells can be derived from terminally differentiated hepatocytes.
(A) Experimental rationale of the genetic lineage tracing. (B) Albumin-Cre/Rosa26-mTmG liver cultures exhibited EGFP-positive hepatocytes and tdTomato-positive cells 5 days after isolation. (C-F) EGFP-positive Hep-iN cells derived 13 days after dox expressed Tuj1 and PSA-NCAM. (G) Hepatocyte cultures from Albumin-Cre/Rosa26-Bgeo mice showed β-galactosidase activity. (H) After infection both unstained and Xgal-stained neuronal like cells could be identified. (I) Quantification of Hep-iN and MEF-iN cells at day 23 after infection. Dox was removed from culture dishes at time points shown (n=3) (B) qPCR analysis for endogenous mRNA (endo) or viral mRNA (exo). Dox was withdrawn at day 12 (n=2) (K) EGFP-positive cell from Albumin-Cre/Rosa26-mTmG transgenic were characterized by patch clamping. (L) Spontaneous action potentials recorded in Hep-iN cells 30 days after dox induction (n=8). (M) Repetitive action potentials could be induced when increasing amounts of current was injected (n=16). (N) Evoked postsynaptic response recorded from a Hep-iN cell co-cultured with mouse cortical neurons. Arrow indicates time point of stimulation. Scale bars: 20μm (K), 25 μm (H) 50 μm (B, E, F, G), 100 μm (C, D).
Hep-iN cells are independent of transgene expression and have acquired functional properties of mature neurons
To determine whether mature Hep-iN cells require sustained transgene expression in order the maintain their phenotype we removed dox from media at different time points after infection. Surprisingly, as few as 5 days of dox treatment sufficed to generate Hep-iN cells, which were present until at least day 22 after addition of dox. Similar results were obtained with MEF-iN cells (Figure 2I). The longer the transgenes were expressed the more iN cells were generated and efficiencies appeared to plateau at around 11 days of dox treatment. While the 3 exogenous factors were strictly dox-dependent the endogenous genes were induced during the reprogramming process (Figure 2J). To investigate whether Hep-iN cells also possessed functional properties of neurons and whether these properties were stable without transgene expression, we performed patch-clamp recordings with cells that were treated with dox for 12 days and cultured for an additional 18 days in dox-free media. Hep-iN cells were identified as EGFP-positive neuronal cells when derived from Albumin-Cre/ROSA26-mTmG mice. We also recorded from Hep-iN cells identified as EGFP/tdTomato-double positive cells when derived from Albumin-Cre/ROSA26-tdTomato/TauEGFP mice (described below, Figure 2K). The average resting membrane potential of the Hep-iN cells was −50.1±2 mV (n=16). Moreover, spontaneous action potentials were detected in half of the cells (n=8) (Figure 2L). All analyzed Hep-iN cells generated action potentials when depolarized by current injections (Figure 2M) and showed fast inactivation sodium current and outward potassium currents (Figure S1G, H). When Hep-iN cells were FACS-sorted 7 days after dox and cultured together with mouse cortical neuronal cultures for another 4 weeks, postsynaptic responses could be evoked by extracellular stimulation of surrounding neurons (Figure 2N). At holding potentials of −70 mV a small inward current was detected, presumably mediated by AMPA receptors and/or GABAA receptors. At +60 mV a large outward current was evoked, presumably mediated by NMDA and/or GABAA receptors.
Reprogramming efficiencies and kinetics are similar between fibroblasts and hepatocytes
To gain insight into the process of iN cell reprogramming, we first evaluated the cell division frequency after induction of the BAM transgenes in liver cultures by a 5-bromodeoxyuridine (BrdU) incorporation assay. When BrdU was present from the day of infection (i.e. one day before dox) throughout the time of iN cell generation, only 12% of the Tuj1-positive cells at day 13 incorporated BrdU. When BrdU treatment was begun on the day of transgene induction (dox addition) only 1% of the Tuj1-positive cells were BrdU-positive (Figure 3A,B). Thus, the vast majority of hepatocytes were reprogrammed to iN cells without mitosis.
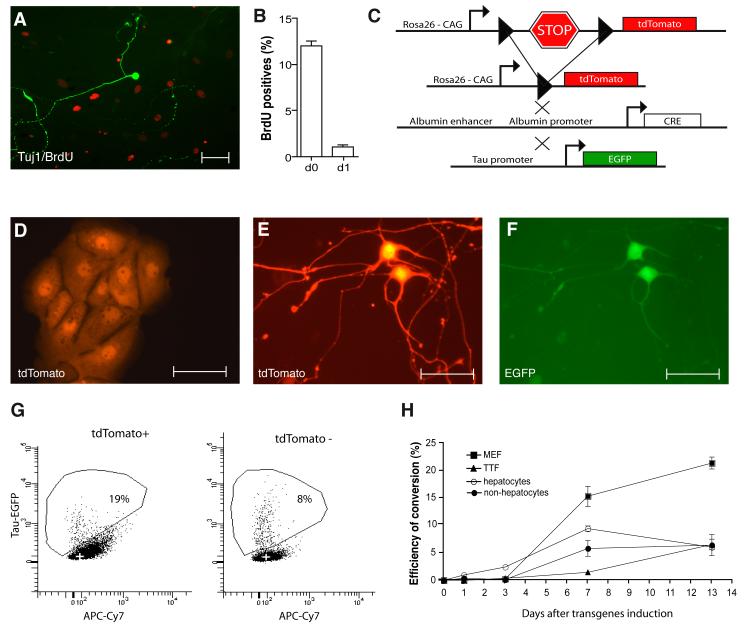
Figure 3. Efficiencies and timing of Hep-iN cell generation.
(A) Most Tuj1-positive cells (green) were BrdU negative (red). (B) Quantification of BrdU/Tuj1 double-positive liver-iN cells after BrdU treatment from day 0–13 or day 1–13 after dox induction (n=3). (C) Lineage tracing outcome in Albumin-Cre/Rosa26-tdTomato/TauEGFP mice. (D) Freshly isolated hepatocytes (E,F) Hep-iN cells are double positive. (G) Representative FACS plots showing iN cells (EGFP) from hepatocytes (tdTomato positive) and non-hepatocyte (tdTomato negative) 13 days after dox. (H) Efficiency of conversion to iN cells in mouse embryonic fibroblasts (MEF ■), postnatal tail tip fibroblasts (TTF ▲), hepatocytes (□), non-hepatocyte liver cells (●) expressed as percentage of infected cells that successfully activate the TauEGFP reporter. Scale bars: 50μm (A, D, E, F).
To address the reprogramming kinetics we generated triple transgenic mice containing the TauEGFP allele together with Albumin-Cre and a ROSA26-tdTomato reporter. In this lineage tracing setting albumin-positive hepatocytes and their progeny constitutively express tdTomato while non-hepatocyte-derived cells remain without fluorescent label (Figure 3C and S2A). We established primary hepatocyte cultures from these mice and as expected 13 days after transduction with the BAM factors Tau-EGFP/ tdTomato-double positive Hep-iN cells appeared (Figure 3D-F). Surprisingly, as early as one day after transgene expression, some infected hepatocytes expressed TauEGFP (Figure 3G-H). Over time the generation of EGFP-positive cells steadily increased with similar kinetics for hepatocytes and fibroblasts. On day 13, the conversion efficiencies relative to number of plated cells of hepatocytes were similar to postnatal fibroblasts (ca. 6%) but lower than embryonic fibroblasts (ca. 20%) (Figure 3H and supplementary methods). When cultured in keratinocyte serum free media (KSFM), a media reported to prevent de-differentiation of cultured hepatocytes (Li et al., 2007) we observed conversion efficiencies similar to our regular hepatocyte growth media (Figure S1E,F).
Finally, we asked whether iN cell reprogramming could be extended to more mature hepatocytes. Following infection with the BAM viruses we could generate iN cells from 1 year old TauEGFP or mice Albumin-Cre/ROSA26-mTomato/mGFP reporter mice (Figure S2C). Correcting for an assumed infection rate of 30%, we estimated a conversion efficiency of 2.7±1.4% (Figure S2D,E).
MEF-iN and Hep-iN cells show global transcriptional remodeling
In order to characterize iN cell formation on the molecular level we determined the gene expression profiles of FACS-purified iN cells 13 and 22 days after dox from hepatocytes, MEFs and TTFs using Illumina’s MouseRef-8 v2.0 Expression BeadChip microarrays (Figure 4A). In addition we profiled the starting populations of Albumin-Cre/Rosa26-tdTomato-positive hepatocytes (FACS-sorted) and MEFs as well as primary neonatal cortical neurons (CN) and neurons derived from fetal (E13.5) forebrain neural progenitor cells (NPCs) 7 and 13 days after differentiation, sorted for TauEGFP expression.
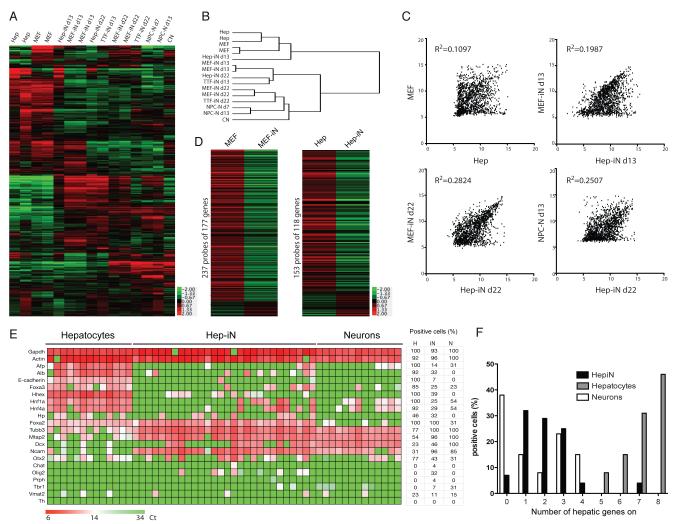
Figure 4. Global transcriptional remodeling during lineage conversion.
(A) Heatmap of microarray data illustrating all differentially expressed genes among hepatocytes, d13 and d22 Hep-iN cells, MEF-iN and TTF-iN cells, d7 and d13 NPC-derived neurons and cortical neurons (12,275 genes). Expression levels are shown as mean centered log2 values. Red indicates up-regulated genes whereas green indicates down-regulated genes. The scale extends from 1.988- to 15.691-fold over mean (−2 to +2 in log2 space) as is indicated on the bottom. (B) Hierarchical clustering among all the samples analyzed. (C) Pearson’s distribution and correlation analysis of MEF vs. Hepatocytes, MEF-iN vs. Hep-iN at different stages of conversion and Hep-iN d22 vs. NPC-N d13. Shown are 1522 probes that are at least 4-fold differentially expressed. (D) Heatmaps showing the MEF and liver signature genes in donor and respective day 22 iN cells. (E) Single-cell gene expression profiling. Rows represent the evaluated genes and columns represent individual cells. Heat map represents the threshold Ct values. Table indicates percentage of positive cells in the three populations (H= hepatocytes, iN= Hep-iN cells, N= cortical neurons). (F) Distribution of hepatocytes, neurons, and Hep-iN cells as function of how many hepatic genes are expressed per single cell.
We first considered only those genes that were differentially expressed (genes with expression changes of at least 3-fold) between hepatocytes, d22 Hep-iN cells, and NPC-derived neurons (13 days after differentiation). Unsupervised clustering identified 3 major clusters of genes and revealed that the vast majority of transcriptional changes in d22 Hep-iN cells approached the levels of primary neurons (Figure S3A). The largest cluster (cluster B) contained mostly genes with higher expression levels in Hep-iN cells and neurons than hepatocytes. Accordingly, in this cluster the top 8 most significantly enriched gene ontology (GO) terms are associated with neuronal function and development (Figure S3A). The second largest cluster (cluster C) consisted of mostly those genes downregulated in iN cells and primary neurons (Figure S3A). Within this cluster many GO terms typical of liver function such as coagulation, wound healing, and inflammatory response were among the most significantly enriched. The analysis also revealed a cluster of genes that were low in hepatocytes and Hep-iN cells but high in neurons (cluster A). This may indicate a group of genes that failed to be induced in iN cells. Indeed, GO terms associated with more mature neuronal function (regulation of membrane and action potential in neuron) were significantly enriched in this cluster (Figure S3A). However, even more GO terms reflecting glial function were similarly enriched (axon and neuron ensheathment, myelination, lipid biosynthesis, and regulation of action potential). Thus, (i) contaminating glial cells in the NPC-derived cultures may have contributed to many genes in this cluster and (ii) d22 Hep-iN cells may represent a less differentiated state than the primary neurons.
We then performed unsupervised hierarchical clustering of all samples based on 12,275 genes (Figure 4A,B). Most iN cell samples clustered together with the primary neuron samples indicating that their overall transcriptome is more similar to neurons than to their starting cell types. Surprisingly, NPC-derived neurons were more similar to two iN cell populations (d22 MEF-iN and d22 TTF-iN) than to neonatal cortical neurons. Thus, the transcriptional variability between 2 different primary neuronal populations was greater than between iN cells and a specific population of primary neurons. The various iN cell samples fell into 3 groups: (a) the d22 fibroblast-iN cells, which are most closely approaching primary neurons, (b) d22 Hep-iN cells and d13 fibroblast-iN cells, which are still closer to primary neurons than to fibroblasts or hepatocytes and (c) d13 Hep-iN cells, which are more similar to hepatocytes than primary neurons. This suggests that hepatocytes are harder to reprogram and take longer to induce a complete neuronal program than fibroblasts. The corresponding heatmap showing expression changes of all the 12,275 genes illustrates the genome-wide remodeling of iN cells towards primary neurons (Figure 4A). In addition, Pearson correlation analysis of genes differentially expressed across all samples by at least 4-fold revealed that d13 MEF-iN and Hep-iN cells are much better correlated (R2=0.1987) than MEFs and hepatocytes (R2=0.1097) (Figure 4C). Of note, the increased overall correlation is caused by a subset of genes being almost perfectly correlated while the remaining genes appear uncorrelated and higher expressed in Hep-iN cells (Figure 4C). This may suggest that at 13 days the reprogramming factors have induced a portion of the transcriptional program that is similar between Hep- and MEF-iN cells. Our preliminary analysis of expression data directly in response to the BAM factors suggest that the correlated genes are not predominantly direct target genes indicating that the transcription factors induce this pattern through secondary changes (not shown). This is in agreement with the finding that the two different iN cell states become much more similar to each other after 22 days (R2=0.2824) and the distinct group of uncorrelated genes decreases (Figure 4C compare d13 with d22 MEF and Hep-iN cell plots). Conversely, d22 Hep-iN cells and hepatocytes appear quite unrelated (R2=0.02104) while many genes are well correlated between Hep-iN and NPC-derived neurons (R2=0.2507) (Figure 4C and S3B). Thus, based on these data the two iN cell samples are more similar to each other than the donor cell types and iN cells are gradually and increasingly approximating the state of primary neurons.
The BAM factors induce silencing of both MEF and liver-specific transcriptional programs
A key question in lineage reprogramming is whether the induced cell types may represent hybrid phenotypes composed of similarly dominant donor and target cell features or whether reprogrammed cells have efficiently extinguished the donor cell-specific identity. To test the hypothesis whether the BAM pool of transcription factors was capable of silencing the two donor program we first identified a MEF- and liver-specific expression signature by comparing publicly available microarray data from 20 different tissues (Figure S3C). We then evaluated the expression levels of these genes in iN cells and their donor cells. Strikingly, for both MEF- and Hep-iN cells those donor-specific programs were extensively downregulated. The MEF signature contained 221 probes and 209 (95%) and 201 (91%) were downregulated at least 2-fold in MEF-iN cells at days 13 and 22, respectively. Similarly, the liver-specific signature was composed of 149 probes and 113 (76%) were downregulated at least 2-fold in Hep-iN at day 13 and 126 (85%) at day 22 (Figure 4D). To quantify the extent of silencing we compared expression levels of genes from the liver signature in Hep-iN and neurons. Strikingly, as many as 45% of these liver genes could be considered “turned off” (i.e. showed expression levels lower or up to maximal 2-fold higher than in neurons) (Figure S3D). Furthermore, we found that Hep-iN cells have completely lost hepatocyte-specific functional properties such as albumin secretion and urea production (Figure S2F-G).
We then asked what the extent of reprogramming in Hep-iN cells is on the single cell level. Twenty-eight single Hep-iN cells 32 days after dox treatment from two independently infected cultures, 13 primary Tau-EGFP-positive cortical neurons cultured for 5 days and 13 Albumin-Cre/Rosa26-tdTomato-positive hepatocytes cultured for 6 days were picked and analyzed using Fluidigm dynamic RT–PCR arrays. Figure 4E shows that robust expression of pan-neuronal markers was found in 27/28 Hep-iN cells (β-III-tubulin, Map2, Ncam). Surprisingly, many primary neurons expressed some of the 8 analyzed liver signature genes, illustrating the transcriptional noise of assumed cell type-specific genes. Similar to neurons, Hep-iN cells were randomly positive for one or more liver markers while hepatocytes expressed most of those genes (Figure 4E). When plotting the cells based on how many liver genes were expressed, we found essentially no overlap between hepatocytes and neurons or Hep-iN cells (Figure 4F). On the other hand, the distribution of neurons and Hep-iN cells are overlapping but distinct. Thus, while some Hep-iN cells appear to be indistinguishable from primary neurons there is a trend that Hep-iN cells express slightly more liver genes than neurons. This finding shows that Hep-iN cells do not represent hybrid phenotypes of neurons and donor cell types but possess an epigenetic memory of their cells of origin. The lack of detectable hepatic functional properties suggests that this epigenetic memory has little if any functional consequence.
Discussion
In this report we show that albumin-expressing hepatocytes can be converted into functional neuronal cells. Our results unequivocally prove that terminally differentiated somatic cells can be directly converted into a distantly related somatic cell type. In contrast to previous studies with fibroblasts we can now formally rule out the possibility that a specific subpopulation potentially enriched in stem or progenitors are the origin of iN cells. Also, it is the first demonstration that cells from definitive endoderm can be directly converted into functional ectodermal cells. Hepatocytes have been reprogrammed to other functional but closely related lineages (Horb et al., 2003; Sapir et al., 2005; Zhou et al., 2008). Together with the finding that iN cells can be generated also from non-hepatocyte liver cells as well as connective tissue fibroblasts this raises the possibility that any cell type that can be cultured in vitro may be able to be converted into iN cells using the same or similar reprogramming factors.
The fact that the vast majority of hepatocytes are reprogrammed in the absence of cell division argues that the genome-wide transcriptional remodeling is an active process and does not require DNA synthesis. Several aspects were similar between the Hep- and fibroblast-iN cells such as the expression of glutamatergic markers, reprogramming efficiencies, and timing of the induction of neuronal reporter genes. However, gene profiling revealed that Hep-iN cells required more time to induce a transcriptional neuronal program than fibroblast-iN cells. These findings suggest that besides the key role of the BAM transcription factors the epigenetic state of the donor cells does have an important influence on the reprogramming process and outcome. It therefore may be possible to identify a more optimal donor cell type for iN cell generation than fibroblasts or hepatocytes and some donor cell types may be more prone to generate one particular neuronal subtype than others.
Finally, we addressed the question whether iN cells derived from hepatocytes or fibroblasts had not only induced neuronal features but also silenced their donor cell type-specific transcriptional network. We made the remarkable observation, that the exact same three transcription factors can efficiently downregulate both a fibroblast- and a liver-specific gene expression program through direct and/or indirect events and hepatic functions were extinguished. This result was unexpected given that those transcriptional regulators act in well-defined progenitor cell contexts during development and one would not necessarily have expected a mutually exclusive mechanism for cell-type specific gene expression programs. One possibility is that the BAM factors target and inhibit a large number of key lineage-determining factors representing many non-neuronal cell fates. Alternatively, the mutual lineage switch could be caused by a more general mechanism such as competition of the lineage-determining factors with a finite amount of ubiquitously required co-factors which would lead to an obligatory extinction of any other developmental lineage once differentiating cells have committed to one lineage. The ubiquitously expressed E-proteins could represent such critical co-factors as they are known to heterodimerize with several different lineage-specific bHLH transcription factors (Massari and Murre, 2000). While our findings reassuringly demonstrate that iN cells predominantly display features of only the target cell lineage it raises new questions about the molecular mechanisms of cell fate decisions in the embryo and induced by expression of ectopic factors in vitro.
Experimental Procedures
Hepatocyte Culture
Disaggregated mouse liver cells were isolated by an adaptation of the two-step collagenase perfusion technique. Liver was extirpated 2 to 5 days after birth, incised, washed, minced and digested in Kreb’s Ringer Buffer 0.15mM CalCl2, 0.54 mg/ml of collagenase type I (Sigma C0130) 40min at 37 °C. Cells were centrifuged at 100g for 3 minutes and plated on Collagen coated plates in hepatocytes plating media. After 4 hours media was changed to hepatocyte culturing media.
Immunofluorescence, RT–PCR, and Flow Cytometry
Neuronal cells were defined as cells that stained positive for Tuj1 and had a process at least three times longer than the cell body. Immunofluorecence stainings were performed as previously described(Vierbuchen et al., 2010). EGFP and tdTomato-expressing cells were analyzed and sorted on a FACS Aria II and flow cytometry data were analyzed using FACS Diva Software (Becton Dickinson).
Gene expression analysis
Total RNA was isolated using the Qiagen RNAeasy kit according to the manufacturer’s instruction (Qiagen). 200 nanograms of total RNA were reverse transcribed using SuperScript® First-Strand Synthesis System (Invitrogen). Real-time polymerase chain reaction (PCR) was performed using the 7900HT Real-Time PCR System (Applied Biosystems) using SYBR Green I dye. Primers used are reported in Table S1. Expression profiling was performed using Illumina’s MouseRef-8 v2.0 Expression BeadChip. Data were deposited under the GEO accession number: GSE30102.
Electrophysiology
Cells were analyzed at indicated time points after dox induction. Action potentials were recorded with current-clamp whole-cell configuration (Maximov et al., 2007; Vierbuchen et al., 2010).
Statistical methods
Results are presented as mean ±SD. The Student’s t test was used to estimate statistical significance. * represents P<0.05, ** P<0.01 and *** P<0.001.
Supplementary Material
Highlights.
Direct lineage reprogramming is possible across different defined germ layers
Terminally differentiated endodermal cells can be directly converted to neurons
Reprogramming factors induce silencing of the donor-specific transcriptional program
Induced neuronal cells retain a small but detectable epigenetic memory
Acknowledgments
We would like to thank T. Vierbuchen for providing MEF, TTF and NPC cultures and critical comments on the manuscript, A. Lanctot for help with mouse genotyping, P. Lovelace for support with FACS, and C. Kuo, I. Graef for mice. M.W. is a NYSCF-Robertson Investigator. In addition, this work was supported by the Dean’s Postdoctoral Fellowship at the Stanford School of Medicine (S.M.), NARSAD Young Investigator Award (Z.P.P.), the Stinehart-Reed Foundation, (M.W.) the Ellison Medical Foundation (M.W.), the Howard Hughes Medical Institute (H.Y.C., T.C.S.), and the NIH grants RC4NS073015-01 (M.W., H.Y.C.) and R01MH092931-01 (M.W., T.C.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct Reprogramming of Adult Human Fibroblasts to Functional Neurons under Defined Conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet Beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Braeuning A, Ittrich C, Kohle C, Hailfinger S, Bonin M, Buchmann A, Schwarz M. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006;273:5051–5061. doi: 10.1111/j.1742-4658.2006.05503.x. [DOI] [PubMed] [Google Scholar]
- Caiazzo M, Dell’anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011 doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Horb ME, Shen CN, Tosh D, Slack JM. Experimental conversion of liver to pancreas. Curr Biol. 2003;13:105–115. doi: 10.1016/s0960-9822(02)01434-3. [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WC, Ralphs KL, Slack JM, Tosh D. Keratinocyte serum-free medium maintains long-term liver gene expression and function in cultured rat hepatocytes by preventing the loss of liver-enriched transcription factors. Int J Biochem Cell Biol. 2007;39:541–554. doi: 10.1016/j.biocel.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci U S A. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov A, Pang ZP, Tervo DG, Sudhof TC. Monitoring synaptic transmission in primary neuronal cultures using local extracellular stimulation. J Neurosci Methods. 2007;161:75–87. doi: 10.1016/j.jneumeth.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011 doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, et al. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sapir T, Shternhall K, Meivar-Levy I, Blumenfeld T, Cohen H, Skutelsky E, Eventov-Friedman S, Barshack I, Goldberg I, Pri-Chen S, et al. Cell-replacement therapy for diabetes: Generating functional insulin-producing tissue from adult human liver cells. Proc Natl Acad Sci U S A. 2005;102:7964–7969. doi: 10.1073/pnas.0405277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KL, Meyer M, Barde YA. Neurotrophins are required for nerve growth during development. Nat Neurosci. 2001;4:29–37. doi: 10.1038/82868. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisend CM, Kundert JA, Suvorova ES, Prigge JR, Schmidt EE. Cre activity in fetal albCre mouse hepatocytes: Utility for developmental studies. Genesis. 2009;47:789–792. doi: 10.1002/dvg.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Tucker KL, Gornik V, Schneiders A, Buschwald R, Wiestler OD, Barde YA, Brustle O. Tau EGFP embryonic stem cells: an efficient tool for neuronal lineage selection and transplantation. J Neurosci Res. 2002;69:918–924. doi: 10.1002/jnr.10395. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011 doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Duncan SA. Embryonic development of the liver. Hepatology. 2005;41:956–967. doi: 10.1002/hep.20691. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.