Abstract
The glutathione transferase GSTP is overexpressed in many human cancers and chemotherapy-resistant cancer cells, where there is evidence that GSTP may have additional functions beyond its known catalytic role. Based on evidence that Gstp-deficient mice have a comparatively higher susceptibility to skin carcinogenesis, we investigated whether this phenotype reflected an alteration in carcinogen detoxification or not. For this study, Gstp−/− mice were interbred with Tg.AC mice which harbor initiating H-ras mutations in the skin. Gstp−/−/Tg.AC mice exposed to the pro-inflammatory phorbol ester TPA exhibited higher tumor incidence and multiplicity with a significant thickening of skin after treatment illustrating hyperproliferative growth. Unexpectedly, we observed no difference in cellular proliferation or apoptosis or in markers of oxidative stress, although higher levels of the inflammatory marker nitrotyrosine were found in Gstp−/−/Tg.AC mice. Instead, gene set enrichment analysis of microarray expression data obtained from skin revealed a more pro-apoptotic and pro-inflammatory environment shortly after TPA treatment. Within 4 weeks of TPA treatment, Gstp−/−/Tg.AC mice displayed altered lipid/sterol metabolism and Wnt signalling along with aberrant processes of cytoskeletal control and epidermal morphogenesis at both early and late times. In extending the evidence that GSTP has a vital role in normal homeostatic control and cancer prevention, they also strongly encourage the emerging concept that GSTP is a major determinant of the pro-inflammatory character of the tumor microenvironment.
This study demonstrates that the glutathione transferase GstP plays a major role in carcinogenesis distinct from its role in detoxification, and provides evidence that the enzyme is a key determinant of the pro-inflammatory tumour environment.
Keywords: skin, cancer, promotion, glutathione, mouse
Introduction
Glutathione S-transferases (GST) are a multi-gene family of dimeric enzymes playing an important role in chemical detoxification due to their capacity to catalyse the addition of reduced glutathione to reactive electrophiles [1]. GSTP has received particular attention because of its association with carcinogenesis, drug resistance and chemical toxicity [2-4]. In order to define the in vivo functions of this protein, we have generated Gstp-null mice; these mice develop normally, are fertile and show no obvious abnormalities [5]. When Gstp-null mice are submitted to a two-stage skin tumorigenesis protocol, involving topical application of the polycyclic aromatic hydrocarbon (PAH) and tumour initiator 7,12-dimethylbenz[a]anthracene (DMBA), followed by the tumour promoting agent 12-0-tetradecanoylphorbol-13-acetate (TPA), there was a significant increase in the number of papillomas in null animals, demonstrating that GSTP is an important determinant in PAH-induced skin cancer susceptibility [5]. Similarly, increased adenoma formation in the lungs of Gstp-null mice was observed following the administration of benzo[a]pyrene, 3-methylcholanthene (3-MC) and urethane. In these studies, no increase in pulmonary DNA adducts was found in 3-MC treated tissue from Gstp-null mice relative to wild-type controls, suggesting GSTP may be acting in a manner distinct from its role as a detoxification enzyme [6]. In this regard, we have also recently reported a marked increase in colon adenoma formation, when APCMin mice are on a GstP null background [7]. GSTP has now been linked to a wide range of cellular functions, including modulation of JNK and TRAF2 signalling [8, 9], glutathionylation [10] and in inflammation [7]. Also, mice nulled at the Gstp locus exhibit significant changes in mRNA expression profiles in liver, lung and colon [6, 7], and the fact that the GSTP gene becomes hypermethylated, and as a consequence inactivated, in certain human cancers all point to novel functions of this protein [11, 12].
To further explore the role(s) of Gstp in carcinogenesis, we have crossed the Gstp-null mouse with the Tg.AC line which is predisposed to the development of skin cancer. The Tg.AC mouse contains the oncogenic v-Ha-ras transgene (mutated at codons 12 and 59) [13] which when treated topically with variety of tumour promoters, develop multiple papillomas [14]. Since the tumorigenic response observed in Tg.AC mice occurs independently of the initiation step, the Tg.AC mouse has been characterised as a ‘genetically initiated’ model for mouse skin tumorigenesis, allowing us to determine whether GSTP is involved in the initiation or promotion steps, or both.
Methods
Reagents
All chemicals were of the highest grade available and were purchased from Sigma (Poole, UK) or Fisher Scientific Ltd. (Loughborough, UK).
Animals
All experiments were undertaken in accordance with the Animals (Scientific Procedures) Act 1986 and approved by the Animal Ethics Committees of the University of Dundee and Cancer Research UK. Gstp1/p2 null and wild-type mouse lines, on a 129 × MF1 background, were generated and maintained by random inter-crossing as previously reported [5]. Tg.AC mice, on a FVB background, were purchased from Taconic, and were crossed with Gstp-null or Gstp-wt mice to generate Gstp+/+/Tg.AC or Gstp−/−/Tg.AC mice.
Genotyping
Gstp genotyping was carried out as previously described [7]. Tg.AC genotype was determined by Southern blotting (Taconic); only those mice with the Tg.AC responder genotype were used.
Chemical carcinogenesis protocol
TPA (6 μg) was dissolved in acetone (200 μl) and applied twice weekly to the shaved backs of 6- to 9-week-old mice. Matched cohorts of mice were treated with acetone alone, or left untreated. All mice were monitored for papilloma growth twice weekly. The date of first papilloma incidence was recorded; papillomas that grew to ≥1mm were counted. All animals entered into the study were included in the final analysis.
Histology
Tumors were fixed in PBS-formalin (10%), transferred to 80% ethanol and processed to wax for sectioning. Tissue sections were stained with haematoxylin and eosin (H & E) and examined by a pathologist blinded to sample identity.
Microarray Analysis
Mice were sacrificed by a rising concentration of CO2 and skin immediately removed for preparation of RNA using TRIzol (Invitrogen) and an RNeasy Mini Kit (Qiagen). RNA was pooled from two animals of each genotype, and subsequent hybridisations were carried out in triplicate. A260/280 ratio of total RNA was typically ≥ 1.9. RNA quality was assessed on an Agilent 2100 Bioanalyzer.
Total RNA (1μg) was labelled with Cyanine 3 (Cy3)-CTP (Agilent One-Colour Microarray-Based Gene Expression Analysis protocol, v5.0.1) using the Low Input RNA Fluorescent Linear Amplification Kit (Agilent). Agilent 4×44K Whole Mouse Genome Oligo Microarray slides were hybridized, washed and scanned at 5μM resolution on an Agilent Microarray Scanner. Scanner images were processed using Agilent Feature Extraction Software v9.1. The microarray scanned image and intensity files were imported into Rosetta Resolver™ gene expression analysis software v6.0.0.0.1. Individual expression profiles from the various genotypes (Gstp+/+/Tg.AC and Gstp−/−/Tg.AC) and treatments (no treatment, acetone and TPA) were pooled in silico by calculating an error weighted mean and compared to build ratios. Data were analyzed using the bioinformatics platform Bioconductor 2.7, running on R 2.12.1. Chip expression data were quantile normalized and a linear model fitted to determine the effects of genotype, time and stimulation using the LIMMA package [51]. Differential genes were selected by applying a 0.05 false discovery rate threshold to p values corrected using the Benjamini and Hocberg method. Differential genes were used for enrichment analysis using Genego’s Metacore pathway tool to identify enriched pathways and biological processes.
Gene set enrichment of Biocarta pathways was determined by using the function “geneSetTest” within the LIMMA package to see if pathways are either up or down regulated using the t-statistics for each factor. Pathways with a false discovery rate of < 0.05 were called enriched. GO annotation enrichments were also determined to see which biological processes and molecular functions were enriched using the same method.
Immunohistochemistry
Sections were de-waxed and rehydrated, and then underwent microwave antigen retrieval using 10mM citrate buffer for 10 min. Immunohistochemistry was carried out using the Dako Envision staining system and an anti-BrdU antibody (Becton Dickinson) or anti-nitrotyrosine antibody (Millipore). Signals were developed by standard techniques.
TUNEL assay
The terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay to determine levels of apoptosis was carried out using the In situ cell death detection kit, POD (Roche). For analysis, the total number of cells and the number of labelled cells for 3 separate fields/slide were counted.
Statistical analysis
Statistical analysis was performed using GraphPad Quickcalcs Online statistical calculator. Significant differences when comparing two groups were determined by unpaired t-test. P values were considered significant if they were less than 0.05.
Results
Papilloma pathology and growth
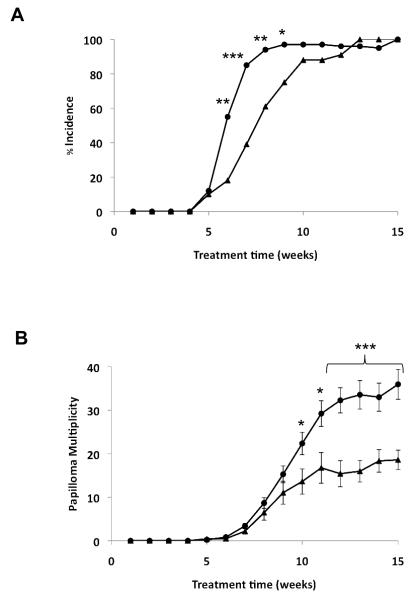
Wild-type mice on a Tg.AC background (Gstp+/+/Tg.AC) and Gstp null mice on a Tg.AC background (Gstp−/−/Tg.AC) were born with normal Mendelian frequency and did not differ in size, weight or growth characteristics. To assess the role of Gstp in the promotion stage of tumorigenesis we treated Gstp+/+/Tg.AC and Gstp−/−/Tg.AC mice with TPA. Both genotypes developed papillomas, which were indistinguishable either in gross anatomical appearance or after histological examination (Fig. 1). The first detectable tumours (>1 mm) on both Gstp+/+/Tg.AC and Gstp−/−/Tg.AC mice were seen at 5 weeks (Fig. 2a). However, by week 6, whereas only 18% of Gstp+/+/Tg.AC mice had papillomas, in 55% of Gstp−/−/Tg.AC mice, tumours were observed. This significantly accelerated appearance of papillomas in Gstp−/−/Tg.AC mice continued through weeks 6-9. By week 15, all mice of both genotypes had papillomas. Tumour multiplicity was also significantly higher in Gstp−/−/Tg.AC animals, this difference reaching statistical significance after 9 weeks of treatment (Fig. 2b). These results demonstrated that loss of Gstp markedly increased both the incidence and multiplicity of skin papillomas in Gstp−/−/Tg.AC mice.
Figure 1. Gross histology of skin papillomas did not differ differ between Gstp+/+/Tg.AC and Gstp−/−/Tg.AC mice.
Haematoxylin and Eosin stained sections of papillomas from (a) Gstp+/+/Tg.AC and (b) Gstp−/−/Tg.AC genotypes. Photomicrographs are representative of each genotype.
Figure 2. Increased number and rate of papilloma formation in Gstp−/−/Tg.AC mice.
(a) Papilloma incidence (number of mice with at least one papilloma) and (b) multiplicity (mean number of papillomas per mouse) was determined twice-weekly over a 15-week period in Gstp−/−/Tg.AC (circles) and Gstp+/+/Tg.AC mice (triangles). n=29 wt, 33 null at experimental initiation, (* p<0.05, ** p<0.01, *** p<0.001). Data are mean + standard deviation.
Effect of Gstp-null genotype on skin thickness, cell proliferation, apoptosis and oxidative stress
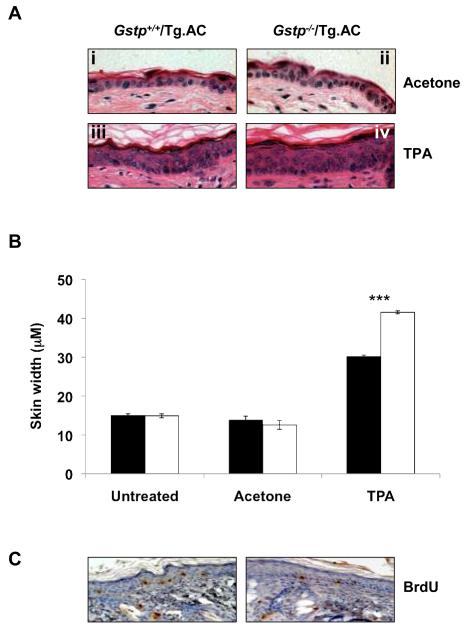
In order to evaluate the effects of the Gstp-null genotype, we looked for changes in skin morphology and epidermal proliferation rates following TPA exposure. Following 6 weeks treatment with TPA, a marked increase in the thickness of the epidermis of both genotypes was observed, consistent with the known ability of TPA to induce cellular proliferation (Fig. 3a). This increase was much more marked (28% higher) in Gstp−/−/Tg.AC mice relative to Gstp+/+/Tg.AC animals (Fig 3a, 3b). These effects were not observed in acetone-treated controls. Investigation of increased cellular proliferation rates using in vivo BrdU labelling experiments showed that there was no difference in epidermal cellular proliferation rate between mouse lines (Fig 3c). Although the percentage of cells in apoptosis appeared to be slightly higher in Gstp+/+/Tg.AC mice than in null animals (69% ± 5 vs. 58% ± 3 respectively), this difference was not statistically significant. Western blotting of skin tissue from Gstp+/+/Tg.AC and Gstp−/−/Tg.AC mice was carried out for markers of oxidative stress (Supplementary Figure 1); haem oxygenase-1 (HO-1) expression was marginally induced, and NADH quinone oxidoreductase (NQO1) suppressed, 6 hours after TPA treatment in Gstp+/+/Tg.AC mice, but both returned to untreated levels at 4 weeks. GSTP behaved in a similar manner to NQO1. Similar changes were observed in Gstp−/−/Tg.AC mice for NQO1 and HO-1, and the alterations in protein expression were confirmed by RT-PCR, along with Nrf2, a transcription factor considered as a master regulator of the antioxidant response. (Supplementary Figure 2).
Figure 3. Increased skin thickness, but no change in BrdU labelling, in Gstp−/−/Tg.AC mice treated with TPA for 6 weeks.
(a) Increase in skin thickness relative to acetone controls in both Gstp+/+/Tg.AC (compare i and iii) and Gstp−/−/Tg.AC mice (compare ii and iv) following TPA treatment, whilst comparison of iii and iv clearly shows the increased thickness of the epidermal layer in Gstp−/−/Tg.AC mice relative to Gstp+/+/Tg.AC mice. *** p<0.001; 6 fields/slide, 5 measurements/field; n=3, (b) Skin thickness in Gstp+/+/Tg.AC (black bars) and Gstp−/−/Tg.AC (white bars) mice following TPA treatment, (c) No difference in BrdU-labelled cells between Gstp+/+/Tg.AC mice and Gstp−/−/Tg.AC mice. Sections shown are representative of each genotype (3 fields/slide, n= 3 for each genotype).
Gene expression changes in the skin of Gstp-null and Gstp-WT mice
Despite no histological differences being evident between untreated wild-type and Gstp-null mouse skin (Fig. 1), the expression of 169 genes was increased, and 175 genes reduced, by a factor of 2-fold or greater (Supplementary Table 1). Ten of the 30 genes increased to the greatest extent were unannotated; of the remainder, the most induced was armadillo repeat-containing 10 (Armc10) (~12-fold), followed by murinoglobulin 1 (Mug1) and mitogen-activated protein kinase kinase kinase kinase 2 (Map4k2) (both ~11-fold). Among the genes whose expression was most reduced approximately one-third were unannotated; the most suppressed gene (~10-fold) was Fin15, (fibroblast growth factor inducible 15). Further analysis using Metacore pathway software failed to find enrichment of pathways in either set of genes, although within the set of induced genes a number of biological processes, rather than pathways, were significantly enriched in Gstp-null mouse skin, including cell inflammation and cell adhesion (data not shown).
Gene expression changes in the skin of Gstp−/−/Tg.AC mice following TPA treatment
To evaluate the effects of TPA on gene expression in the skin in the presence or absence of Gstp, we carried out gene expression profiling following both short- (6 hours) and long-term (4 weeks) exposure of Gstp−/−/Tg.AC and Gstp+/+/Tg.AC mice to TPA.
Genes whose expression was significantly changed in Gstp−/−/Tg.AC and Gstp+/+/Tg.AC mice after 6h of TPA treatment were compared to vehicle (acetone)-treated animals. The expression of more than 3900 genes was changed in both groups. At 6h, within the top 50 induced or suppressed genes, there was a high degree of similarity, forty-five genes being found in both genotypes (Supplementary Table 2). However, some variation in the extent of induction was observed; expression of 27 genes induced in the skin of both Gstp−/−/Tg.AC and Gstp+/+/Tg.AC mice differed in fold-change by a factor of three or more, 16 to a greater extent in Gstp−/−/Tg.AC mice - Chi3l3, Trpm2, AK048833, Nrg1, Prg4, Clec4d, AK031717, Ccl22, Klk9, Tnc,, Slc26a4, Ccl3, Ccl17, Serpinb1c, IL19, Il1rl1 - several of which are associated with inflammatory response. Eleven genes were induced to a greater extent in the skin of Gstp+/+/Tg.AC mice - Zdhhc21, CCl24, Ier2, Tnip3, AK078295, Lce3f, RSG2, Mmp10, A330043J11Rik, Gfpt1, Mcpt8.
The expression of many common genes was also reduced, with the majority of the most suppressed genes present in both genotypes (Supplementary Table 2). Similar to the induced genes, the fold-change varied between the genotypes. The expression of thirteen genes was decreased in both genotypes with fold-change ratios differing by a factor of three or more, two to a greater extent in Gstp−/−/Tg.AC (Pla2g2d, Pamr1) and eleven in Gstp+/+/Tg.AC mice (Adh7, Eml16, Edar, 3830408C21Rik, Cttnbp2, Cldn8, Dlg2, 2130005G13Rik, Pramef12, AK089297, BC034902). A single gene - Bclp2 - was induced in Gstp+/+/Tg.AC mice (3.2-fold) and suppressed in Gstp−/−/Tg.AC mice (2.6-fold).
In addition to the similarities, there were many significant differences between the effects of TPA on gene expression between Gstp−/−/Tg.AC and Gstp+/+/Tg.AC mice. After 6h of TPA treatment a total of 1750 genes were altered uniquely in Gstp+/+/Tg.AC mice relative to acetone-treated controls and TPA-treated Gstp−/−/Tg.AC animals, ie changes in the Gstp+/+/Tg.AC were observed which did not occur in the Gstp−/−/Tg.AC mice. The expression of 840 genes was reduced and 910 genes increased by more than 2-fold. The top 50 genes in each category are shown in Table 1. Similarly,, the expression of 2170 genes were uniquely altered in Gstp−/−/Tg.AC mice 6h after TPA treatment; of these, 1058 were expressed at higher levels and 1112 at lower levels by more than 2-fold; the top 50 genes in each category are shown in Table 2.
Table 1.
mRNA expression profiles in skin of Gstp+/+/Tg.AC (WT) uniquely changed 6h after TPA treatment, relative to untreated animals. Microarray analysis was carried out as described in Methods section. Data are expressed as fold-changes between untreated and TPA treatment after subtraction of effects of acetone vehicle on gene expression.
| Symbol | Fold-change |
|---|---|
| Ccl7 | 25.1 |
| A530023O14Rik | 17.6 |
| Pcdh21 | 15.5 |
| Msx3 | 10.6 |
| Mmp12 | 9.0 |
| Dsc2 | 8.2 |
| AK087584 | 8.1 |
| Ccl24 | 8.0 |
| Ifi205 | 8.0 |
| 9130204L05Rik | 7.6 |
| Rprm | 7.5 |
| F830004M19Rik | 7.3 |
| 4930519N16Rik | 7.2 |
| Msx3 | 7.2 |
| 4732457N14 | 7.1 |
| Ltf | 7.1 |
| Rprm | 6.7 |
| Chrna4 | 6.7 |
| Bhlhb5 | 6.4 |
| Alkbh1 | 6.4 |
| Itpkc | 6.2 |
| Cd9 | 6.2 |
| Elf5 | 6.0 |
| 2810403A07Rik | 5.9 |
| Gpr26 | 5.9 |
| 1700095B10Rik | 5.8 |
| NAP093165-001 | 5.7 |
| Rgs4 | 5.6 |
| Bhlhb5 | 5.6 |
| Lsg1 | 5.5 |
| NAP027952-1 | 5.5 |
| Grid2ip | 5.4 |
| Gm1967 | 5.3 |
| Rgs4 | 5.1 |
| Rgs4 | 5.1 |
| Hoxd13 | 4.9 |
| ENSMUST0000006029 8 |
4.7 |
| Tnfsf11 | 4.7 |
| Twsg1 | 4.7 |
| Abhd2 | 4.7 |
| LOC638575 | 4.7 |
| C330005M16Rik | 4.6 |
| Klk1b27 | 4.6 |
| C430019N01Rik | 4.6 |
| Mesdc2 | 4.6 |
| AK041408 | 4.6 |
| Fblim1 | 4.6 |
| Dtl | 4.5 |
| Abhd2 | 4.5 |
| Rassf8 | 4.4 |
| AK082583 | −4.8 |
| D030022P07Rik | −4.8 |
| Msh3 | −4.9 |
| NAP062641-1 | −4.9 |
| Atp6v1g3 | −4.9 |
| Pde8b | −4.9 |
| E030025L21Rik | −4.9 |
| AK089839 | −5.0 |
| 37865 | −5.0 |
| Abcg3 | −5.0 |
| Il22ra2 | −5.1 |
| AK078633 | −5.1 |
| D830026I12Rik | −5.1 |
| 4930506C02Rik | −5.1 |
| Rnf43 | −5.1 |
| 9630031F12Rik | −5.1 |
| Wdr76 | −5.2 |
| 5730590G19Rik | −5.2 |
| AK086288 | −5.2 |
| Armc2 | −5.3 |
| Tmc5 | −5.3 |
| B230114P17Rik | −5.4 |
| Tnrc9 | −5.4 |
| Lrrc44 | −5.4 |
| Gm711 | −5.5 |
| AK028935 | −5.5 |
| Abcb1a | −5.6 |
| 8430408G22Rik | −5.6 |
| AI428226 | −5.6 |
| AK045549 | −5.6 |
| AK037956 | −5.7 |
| Dlg7 | −5.8 |
| A230048G03Rik | −5.9 |
| Dlec1 | −5.9 |
| 5031426D15Rik | −6.0 |
| Nnat | −6.2 |
| Zfp30 | −6.2 |
| 9030407P20Rik | −6.3 |
| AK027992 | −6.3 |
| Prom1 | −6.4 |
| AK043317 | −6.4 |
| AK085674 | −6.5 |
| 1700041G16Rik | −6.7 |
| Lama1 | −6.9 |
| 9530077C05Rik | −7.8 |
| NAP028040-1 | −8.5 |
| 5430435G22Rik | −8.8 |
| Ak5 | −9.8 |
| TC1668068 | −11.0 |
| Add2 | −24.6 |
Table 2.
mRNA expression profiles in skin of Gstp−/−/Tg.AC (NULL) uniquely changed 6h after TPA treatment, relative to untreated animals. Microarray analysis was carried out as described in Methods section. Data are expressed as fold-changes between untreated and TPA treatment after subtraction of effects of acetone vehicle on gene expression.
| Symbol | Fold- change |
|---|---|
| Atp11b | 52.8 |
| Ubxd2 | 51.3 |
| 5830417I10Rik | 47.6 |
| Dio3 | 44.9 |
| Uts2 | 44.1 |
| Sult4a1 | 18.1 |
| 5430405N12Rik | 16.1 |
| Il8rb | 15.8 |
| Mmp7 | 15.0 |
| Setd7 | 14.5 |
| 1700102J08Rik | 14.4 |
| 5830417I10Rik | 10.2 |
| E130310K16Rik | 10.0 |
| Serpine1 | 9.1 |
| AK033022 | 8.9 |
| Cdk5r2 | 8.7 |
| 1200011M11Rik | 8.6 |
| C530042K13Rik | 8.5 |
| Cxcl11 | 7.9 |
| Cacna1b | 7.7 |
| D330037H05Rik | 7.6 |
| Olfr218 | 7.3 |
| 2600011E07Rik | 7.2 |
| 4930593A02Rik | 7.0 |
| NAP028432-1 | 7.0 |
| Gpr35 | 7.0 |
| Slc7a11 | 6.8 |
| AK082839 | 6.7 |
| Olfr665 | 6.6 |
| Gpr83 | 6.4 |
| ENSMUST00000094393 | 6.4 |
| Mill1 | 6.4 |
| Ntrk1 | 6.3 |
| Mtrr | 6.2 |
| Il17c | 6.0 |
| Hemt1 | 6.0 |
| Olfr711 | 6.0 |
| 4930555F03Rik | 6.0 |
| A_52_P852662 | 5.9 |
| Itga2 | 5.9 |
| Xlkd1 | 5.9 |
| Prss16 | 5.8 |
| Gnaz | 5.8 |
| B3gnt5 | 5.7 |
| Olfr689 | 5.7 |
| 2810430M08Rik | 5.6 |
| Atp12a | 5.6 |
| Tekt1 | 5.5 |
| Olfr1148 | 5.5 |
| NAP027441-1 | 5.4 |
| Pln | −4.0 |
| 9030617O03Rik | −4.0 |
| H1fx | −4.0 |
| AK036030 | −4.1 |
| Sp5 | −4.1 |
| Serpinb12 | −4.1 |
| Agxt2l1 | −4.1 |
| Pon1 | −4.1 |
| Slc6a7 | −4.1 |
| Dync1i1 | −4.2 |
| AK081832 | −4.2 |
| Rab27a | −4.2 |
| Spic | −4.2 |
| Spic | −4.3 |
| Cyp3a25 | −4.3 |
| Kcnk2 | −4.3 |
| 2810032G03Rik | −4.3 |
| C330050A14Rik | −4.3 |
| LOC620355 | −4.4 |
| AF143539 | −4.4 |
| D230012E17Rik | −4.4 |
| NAP027004-1 | −4.4 |
| Klhl4 | −4.4 |
| Thrsp | −4.5 |
| 2810030E01Rik | −4.5 |
| AK087309 | −4.5 |
| E030026E10Rik | −4.6 |
| 4933400E14Rik | −4.6 |
| BC062109 | −4.6 |
| Tef | −4.8 |
| AK034975 | −5.0 |
| AK076276 | −5.0 |
| Sema3d | −5.1 |
| Gpc2 | −5.1 |
| Cbs | −5.1 |
| Dtl | −5.2 |
| Hoxa9 | −5.3 |
| Kcnj10 | −5.5 |
| Neil3 | −5.7 |
| NAP025218-001 | −5.8 |
| Sfrp2 | −6.1 |
| BB744776 | −6.3 |
| Rab36 | −6.5 |
| A630012P03Rik | −6.5 |
| Car8 | −7.0 |
| Gabrp | −7.6 |
| AK046982 | −8.3 |
| TC1658561 | −8.9 |
| Susd4 | −9.5 |
| Tead2 | −14.8 |
After 4 weeks of TPA treatment 950 genes were altered in both Gstp−/−/Tg.AC and Gstp+/+/Tg.AC mice, compared to control animals; the top 50 induced or repressed genes are shown in Supplementary Table 4. Again, there was a very strong similarity between the Gstp+/+/Tg.AC and Gstp−/−/Tg.AC groups. As with the 6h timepoint, there were, however, differences in the fold-changes in the expression of these genes between the groups. Fifteen genes were increased in the skin of both mouse strains and differed in fold-change ratio by a factor of 3 or more, 14 to a greater extent in Gstp−/−/Tg.AC mice (Krtap 31-1, Stfa2, Krt84, 2310033E01Rik, Spink12, Lrrc15, Lce3f, Oca2, Sprrl1, Sprr2d, Krt16, Scrg1, Uox, Dlx4), and one in Gstp+/+/Tg.AC mice (Prss51). Two genes were suppressed in both genotypes with a fold-change ratio greater than three, one to a greater extent in Gstp−/−/Tg.AC mice (Kel), and one to a greater extent in Gstp+/+/Tg.AC mice (1500015O10Rik). Five genes were induced in Gstp+/+/Tg.AC mice but suppressed in Gstp−/−/Tg.AC mice (Cacnb2, Dmrtc2, Olf346, Mpp1, A630077J23Rik).
In addition to the similarities to the gene expression profiles at 4 weeks, there were also significant differences. In Gstp+/+/Tg.AC mice, the expression of 1868 genes was uniquely altered in their expression as compared to controls and Gstp-null animals; 937 were higher, and 931 lower, by more than 2-fold. The top 50 genes in each category are shown in Table 3. In Gstp−/−/Tg.AC mice, a total of 1772 genes were uniquely altered 4 weeks after TPA treatment, 910 genes being increased and 862 decreased by more than 2-fold. The top 50 genes in each category are shown in Table 4. It is interesting to note that in Gstp−/−/Tg.AC mice, 40 of the top 50 genes which were found to be increased were keratins or keratin-associated proteins (induced 200 - 2000-fold).
Table 3.
mRNA expression profiles in skin of Gstp+/+/Tg.AC (WT) uniquely changed 4 weeks after TPA treatment, relative to untreated animals. Microarray analysis was carried out as described in Methods section. Data are expressed as fold-changes between untreated and TPA treatment after subtraction of effects of acetone vehicle on gene expression.
| Symbol | Fold- change |
|---|---|
| Trim12 | 45.6 |
| Prdm16 | 32.8 |
| Olfr1240 | 9.6 |
| Trim12 | 9.6 |
| Hsd3b2 | 8.5 |
| Pcnx12 | 8.5 |
| AK036971 | 8.0 |
| Olfr739 | 7.9 |
| CA560305 | 7.7 |
| 4930434F21Rik | 7.7 |
| Hadha | 7.4 |
| BB262475 | 7.2 |
| Olfr291 | 7.1 |
| Rdhe2 | 7.0 |
| AK020422 | 6.8 |
| Tpcn2 | 6.6 |
| AK083252 | 6.6 |
| unknown | 6.5 |
| Olfr1337 | 6.5 |
| Rhox5 | 6.4 |
| unknown | 6.3 |
| 1700013E18Rik | 6.2 |
| unknown | 6.2 |
| Gprc2a-rs1 | 6.1 |
| 1700121L16Rik | 6.1 |
| 4932443L11Rik | 6.1 |
| Olfr944 | 6.1 |
| Mbnl1 | 6.0 |
| unknown | 6.0 |
| Aldh1a3 | 5.9 |
| Olfr444 | 5.8 |
| Tdrd9 | 5.8 |
| unknown | 5.8 |
| unknown | 5.7 |
| Tac4 | 5.7 |
| Gpr21 | 5.6 |
| Gin1 | 5.6 |
| Olfr497 | 5.5 |
| Olfr1255 | 5.4 |
| ENSMUST00000062608 | 5.4 |
| AK013499 | 5.4 |
| C730043O17 | 5.4 |
| Mylk3 | 5.4 |
| Rhcg | 5.3 |
| V1ra6 | 5.3 |
| 4921523P09Rik | 5.2 |
| AK029175 | 5.2 |
| 4930564C03Rik | 5.2 |
| Wdr66 | 5.2 |
| Olfr1420 | 5.1 |
| unknown | −5.5 |
| Clvs2 | −5.5 |
| Pparbp | −5.5 |
| Mmp9 | −5.6 |
| Cts7 | −5.6 |
| Kcnq3 | −5.6 |
| unknown | −5.7 |
| ENSMUST00000074654 | −5.7 |
| Pdgfd | −5.7 |
| Klrb1a | −5.7 |
| LOC665043 | −5.8 |
| Pcdhac2 | −5.8 |
| Klra7 | −5.8 |
| Car8 | −5.8 |
| Arhgap26 | −5.9 |
| MGC41410 | −5.9 |
| 2310050B05Rik | −6.0 |
| Klra15 | −6.0 |
| Sox8 | −6.1 |
| unknown | −6.2 |
| AK084908 | −6.3 |
| 9630013D21Rik | −6.3 |
| TC1744477 | −6.6 |
| TC1646808 | −6.6 |
| AK076536 | −6.6 |
| 0610037P05Rik | −6.7 |
| D430006K04 | −6.9 |
| 5730410E19Rik | −7.1 |
| C030002O17Rik | −7.2 |
| Masp1 | −7.2 |
| Fkbp11 | −7.3 |
| LOC620807 | −7.3 |
| Kcnip4 | −7.3 |
| unknown | −7.4 |
| AK083452 | −7.5 |
| Map4k2 | −7.7 |
| Neto1 | −7.8 |
| AK086129 | −7.9 |
| AK045549 | −7.9 |
| Agtr2 | −8.2 |
| Spata17 | −8.9 |
| AK035641 | −9.0 |
| Trim16 | −9.1 |
| Scn2a1 | −9.2 |
| Pgr | −9.7 |
| Tnfsf18 | −11.9 |
| 1110037P09 | −12.7 |
| Klra20 | −12.9 |
| Zfp319 | −20.9 |
| Klra22 | −21.8 |
Table 4.
mRNA expression profiles in skin of Gstp−/−/Tg.AC (NULL) uniquely changed 4 weeks after TPA treatment, relative to untreated animals. Microarray analysis was carried out as described in Methods section. Data are expressed as fold-changes between untreated and TPA treatment after subtraction of effects of acetone vehicle on gene expression.
| Symbol | Fold-change |
|---|---|
| Krtap16-9 | 2036 |
| Krt25 | 1899 |
| Ktrap22-2 | 1808 |
| Krtap8-1 | 1572 |
| Krtap3-1 | 1537 |
| Krtap16-8 | 1330 |
| Krtap14 | 1207 |
| LOC435285 | 1101 |
| Ktrap9-3 | 1048 |
| Ktrap6-1 | 1032 |
| Krt34 | 966 |
| Krt33b | 921 |
| high-glycine/tyrosine protein | 916 |
| Krtap16-5 | 842 |
| Krtap16-1 | 824 |
| Krtap8-1 | 820 |
| Krt26 | 754 |
| Ktrap4-2 | 684 |
| Krtap6-2 | 672 |
| Krtap16-10 | 619 |
| Krtap13-1 | 618 |
| Krtap4-7 | 610 |
| Prr9 | 586 |
| Krt27 | 558 |
| Ktrap26-1 | 547 |
| Krtap3-1 | 522 |
| Krt33a | 520 |
| Krt71 | 506 |
| Ktrap1-5 | 503 |
| 1110025L11Rik | 501 |
| Krtap6-1 | 480 |
| glycine tyrosine-rich hair keratin | 467 |
| Krtap16-4 | 441 |
| LOC432600 | 419 |
| Krtap16-7 | 414 |
| LOC432600 | 413 |
| Ktrap4-13 | 411 |
| 5430421N21Rik | 376 |
| Krt31 | 365 |
| Krt2-25 | 338 |
| A030003K21Rik | 324 |
| Krtap2-4 | 324 |
| Krt35 | 323 |
| 5530401N06Rik | 303 |
| Krt85 | 292 |
| Krtap6-3 | 275 |
| Krtap5-1 | 267 |
| Krtap8-2 | 245 |
| Trichohyalin | 227 |
| Ktrap17-1 | 211 |
| Lin7a | −4.4 |
| Il20ra | −4.4 |
| Folh1 | −4.6 |
| Serpinb7 | −4.6 |
| Fut9 | −4.6 |
| Paqr7 | −4.6 |
| D430015B01Rik | −4.6 |
| Islr2 | −4.7 |
| Hsd17b13 | −4.7 |
| D430041B17Rik | −4.8 |
| Gls | −4.8 |
| Fhl5 | −4.8 |
| E330013P04Rik | −4.8 |
| Gpha2 | −4.8 |
| Stk40 | −4.9 |
| Nalp4b | −5.0 |
| Myh1 | −5.0 |
| E230001N04Rik | −5.0 |
| Tmod2 | −5.0 |
| AK080997 | −5.1 |
| Fkbp5 | −5.1 |
| Pde1c | −5.2 |
| Siglecf | −5.2 |
| Trpc2 | −5.3 |
| Cxcl13 | −5.3 |
| 4930567K12Rik | −5.3 |
| Tef | −5.4 |
| Slc23a1 | −5.5 |
| C730027J19Rik | −5.6 |
| Per1 | −5.8 |
| Defb15 | −6.0 |
| Cyp2g1 | −6.0 |
| Pde1c | −6.0 |
| Il31ra | −6.1 |
| 1110012N22Rik | −6.1 |
| Tsc22d3 | −6.8 |
| Tsc22d3 | −6.8 |
| Fkbp5 | −6.9 |
| 6030422H21Rik | −7.3 |
| Tsc22d3 | −7.4 |
| Cyp2e1 | −7.5 |
| Cyp2g1 | −7.7 |
| Myoz2 | −8.4 |
| Wbp2nl | −8.7 |
| Il22ra2 | −9.7 |
| Cyp2a4 | −9.8 |
| Cyp2a5 | −10.5 |
| Cyp2a4 | −11.7 |
| Il22ra2 | −11.7 |
| Hamp2 | −83.8 |
In order to define the pathways that were uniquely up- or down-regulated in Gstp−/−/Tg.AC mice, and identify those biological processes or molecular functions that were statistically enriched at both time-points, the unfiltered microarray data from Gstp−/−/Tg.AC and Gstp+/+/Tg.AC mice treated with TPA for 6h or 4 weeks were subject to ANOVA and subsequent gene set enrichment analysis (GSEA). There were 101 pathways up-regulated in Gstp−/−/Tg.AC compared to Gstp+/+/Tg.AC mice at 6h after TPA treatment, and the same number up-regulated at the 4 week time-point. Seventeen pathways were uniquely up-regulated at each time-point: at 6h, pathways were associated with apoptosis, cell-mediated immunity, T cell activation and differentiation, inflammation and cytokine production, while at 4 weeks apoptotic pathways were essentially absent although pathways related to cell proliferation and the immune system were still up-regulated in Gstp−/−/Tg.AC mice compared to Gstp+/+/Tg.AC mice (Supplementary Table 3). With regard to pathways uniquely down-regulated in Gstp−/−/Tg.AC mice, there were a smaller number at both 6 hours (8) and 4 weeks (1), and none in common; keratinocyte differentiation was represented at both time-points, while pathways down-regulated at 6h also included those involved in cell cycle, inflammatory response and T cell activation (Supplementary Table 5). Gene ontology gene sets were analysed using GSEA to identify biological processes or molecular functions that were statistically enriched (up or down) in Gstp−/−/Tg.AC compared to Gstp+/+/Tg.AC mice at both time-points (Supplementary Table 8). Interestingly, with regard to up-regulation, there was a good deal of conformity between the two time-points; in both cases there were biological processes and molecular functions associated with the actin cytoskeleton, chromatin remodeling and transcriptional regulation, as well as kinase signaling, blood vessel development and epithelial/endothelial cell proliferation and differentiation. However, whilst at 6 hours there was a predominance of anti-apoptotic biological processes, at the later time-point pro- and anti-apoptotic processes were more balanced. Furthermore, cytokine production and cytokine-mediated signalling observed at 6 hours were absent at 4 weeks, while altered lipid and sterol metabolic processes, and Wnt receptor signalling, present at 4 weeks were not found at the earlier time-point (Supplementary Table 8). For down-regulated GO annotations, there were relatively few, and little or no overlap between time-points: at 6 hours down-regulated molecular functions included aromatase and oxidoreductase activities, while at 4 weeks signal transduction, G protein coupled receptor activity and a number of processes related to pheromones and olfaction were down-regulated (data not shown).
Gene expression changes in papillomas from Gstp−/−/Tg.AC mice
To investigate the molecular mechanism(s) underlying the increased incidence and multiplicity of tumour in Gstp−/−/Tg.AC mice, mRNA profiles for papillomas from each genotype were determined. In Gstp−/−/Tg.AC mice only 87 genes were expressed at higher and 127 genes at lower levels, by a factor of 2-fold or greater relative to Gstp+/+/Tg.AC mice (see Supplementary Table 6). The genes that were expressed at higher levels included the β-galactoside-binding proteins galectin-4 (lgals4, 3.4 fold) and galectin-6 (lgals6, 8.5 fold), and Mug1 (3.3-fold). Genes repressed in Gstp−/−/Tg.AC mice vs. Gstp+/+/Tg.AC mice were chitinase-like proteins Chi3l4 (17-fold down) and Chi3l3 (6-fold down); Clec2g (C-type lectin domain family 2, member g, 3.8-fold down); and the tumour suppressor Dbc1 (Deleted in bladder cancer protein 1, 3.7-fold), [15]. Unbiased analysis of the gene expression profiles in Gstp−/−/Tg.AC mice compared to Gstp+/+/Tg.AC mice failed to identify specific biochemical pathways significantly altered in mice lacking Gstp. However, a number of biological processes were found to be enriched in Gstp−/−/Tg.AC mice, including keratinization, keratinocyte differentiation, epidermal morphogenesis, development and differentiation, as well as several processes related to steroid and lipid metabolism (Supplementary Table 7), all of which could potentially be mechanistically associated with the increased rate of papilloma development observed in the Gstp−/−/Tg.AC mice following TPA treatment (Figure 2)
Discussion
Our previous work with the GstP null mouse and skin carcinogenesis showed that GstP can play an important role in the aetiology of skin and lung tumours [5, 6]. We have also recently reported that by crossing Gstp−/− mice onto APCMin mice enhanced the incidence of adenomas arising in the colon [7]. However, the mechanism(s) by which GstP protected against tumour formation was unclear and did not appear to be solely due to its role as a detoxification enzyme. In this study we have demonstrated that GstP can have a profound effect on skin tumorigenesis and initiated by the presence of the mutant Ras oncogene, i.e. in the absence of a chemical carcinogen. There is now also a growing body of evidence suggesting that Gstp is also an important determinant in inflammatory disease [16-18].
In the experiments described here, there are a number of other ways in which Gstp could influence carcinogenesis. It could alter the signaling pathways induced by the presence of mutant Ras protein, or it could alter the promotional effects of TPA. Alternatively, because TPA and Ras act at least in part through AP1 signalling, Gstp could act through a combination of the two mechanisms. However, It is also important to consider the role of GstP in the detoxification of endogenous toxins as a possible explanation for the increased tumorigenesis noted in the null animals. The generation of endogenous reactive oxygen species (ROS) by NADPH oxidase-1 (Nox1), which catalyses the reduction of molecular oxygen to superoxide, has been recently found to be required for Ras mediated oncogenic transformation [19, 20]. In addition Nox1 is thought to be responsible for the generation of ROS in human keratinocytes following UVA irradiation [21]. Given the well-known activity of GstP in the detoxification of genotoxic lipid peroxidation products such as acrolein [22], it is possible that the increased tumorigenesis noted in GSTP null animals is a direct consequence of their reduced ability to deal with endogenously produced toxic metabolites of oxidative stress. However, expression of oxidative stress markers such as HO-1 and NQO1, and the transcription factor Nrf2, although altered at 6 hours after TPA, were essentially unchanged from control levels at the 4 week time-point (Supplementary Figures 1 & 2), indicating that while oxidative stress may play a role in the early stages of tumorigenesis, it appears to have less influence in the longer term. The GstP null background seems to potentiate the pathways activated by TPA and is manifest in the increased skin thickness observed in the GstP null animals. However, these effects do not seem to be mediated by an increased cell proliferation rate, as measured by BrdU, or by changes in rates of apoptosis; rationalisation of these effects requires further study.
In order to gain further insights into the mechanism of action of Gstp in skin tumorigenesis, we carried out a detailed gene expression profile analysis on mice where Gstp is either present or absent. In normal skin the annotated mRNA most induced in Gstp−/−/Tg.AC mice was Armc10 (~12 fold); this protein has been implicated in cell survival and growth, has been reported to suppress p53 activity and thus apoptosis [23], and is also up-regulated in hepatocellular carcinoma [24]. We also found that Armc10 expression is elevated (~4-fold) in the lungs of Gstp null mice which had a significantly higher incidence of chemically-induced pulmonary adenomas [6]. Also elevated significantly (~11-fold) was Map4k2, a member of the serine/threonine protein kinase family and which can be activated by TNFα, poly(IC), LPS and IL-1, thus mediating an array of inflammatory responses, and couples TNF signalling to the p38 MAPK and SAPK pathways [25, 26]. Deletion of this gene in mice renders them resistant to endotoxin-mediated lethality [27]. Interestingly, serum amyloid a protein 2 (Saa2) was also expressed at significantly higher levels (6.8-fold) in Gstp−/−/Tg.AC mice; Saa2 is a member of a family of highly-conserved acute-phase proteins, secreted during inflammation in response to the cytokines IL-1, IL-6 and TNFα, and which aid in the recruitment of immune cells to inflammatory sites. Saa2 is expressed in a variety of tissues, including skin, primarily in epithelial cells [28]. Similarly to Armc10, another Saa family member, Saa3, was significantly induced (~6-fold) in the lungs of Gstp null mice, relative to WT [6]. Interestingly, skin sections from mice treated with TPA and stained for the inflammatory marker nitrotyrosine, show higher levels of expression in the absence of Gstp (Supplementary Figure 3). The presence of genes associated with inflammation was also observed in the colonic epithelium of Gstp−/−:: APCMin mice [7]. Together, these data add weight to the argument that the absence of Gstp creates an elevated level of basal inflammation in a number of different tissues – skin (this study), lung [6] and colon [7]. However, such an inflammatory environment, while pro-tumorigenic, is clearly not sufficient for tumour development, since Gstp null mice do not develop cancer spontaneously [29].
Many genes induced in the skin by TPA on a Tg.AC background were also induced in the Gstp−/−/Tg.AC background. These data are reassuring and provide confidence about the significance of the differences between the Gstp+/+/Tg.AC and Gstp−/−/Tg.AC genotypes. It is interesting to note that in many cases the genes were induced to a higher level in the Gstp−/− mice compared to mice carrying the Tg.AC alone. Among the most prominent genes induced by TPA administration in both genotypes (but to a significantly greater extent in Gstp−/−/Tg.AC mice) at 6h or 4 weeks were s100A8, Sprr2a, Sprr2d, and Stfa1, Stfa2, Stfa3 (Supplementary Tables 2 & 4), previously described as being present in clusters of differentially-expressed genes from the skin of Tg.AC mice in which papillomagenesis had been induced by abrasion [30].
Strikingly, more than 90% of the top 50 genes encoded for keratins or keratin-associated proteins and were expressed at several orders of magnitude higher in Gstp−/−/Tg.AC mice, compared to wild-type at the 4 week timepoint (Table 4). Interestingly, a recent study by Quigley et al [31] in which the authors mapped genetic loci contributing to skin tumour susceptibility in mice identified a network of 62 genes involved in control of the hair follicle containing 37 keratins or keratin-associated proteins, all of which were found on our list of genes uniquely up-regulated in Gstp−/−/Tg.AC mice 4 weeks after TPA treatment (Table 4). Furthermore, of the remaining genes in this network, all but 3 (Lyg2, Pdzm3, Vsig8) were also up-regulated (3- to 227-fold) only in Gstp−/−/Tg.AC mice 4 weeks after TPA treatment, strongly suggesting that dysregulation of hair follicle function occurs to a considerably greater extent in the absence of GstP and may play a significant role in the accelerated development of papillomas in this genotype. Quigley et al [31], also identified the intestinal stem cell marker Lgr5 (leucine rich repeat containing G protein coupled receptor 5) as the best candidate for a master regulator of the hair follicle network; interestingly, although Lgr5 expression is down-regulated in both Gstp+/+/Tg.AC and Gstp−/−/Tg.AC mice 6h after TPA treatment (12- and 9-fold, respectively), it is up-regulated (3.2-fold) uniquely in Gstp−/−/Tg.AC mice 4 weeks after treatment.
Also of note, one α-defensin (Defa-rs12, down 3.2-fold) and two β-defensin proteins (Defb14, up 4-fold; Defb15, down 6-fold) were altered in their expression in Gstp−/−/Tg.AC mice relative to Gstp+/+/Tg.AC; these antimicrobial peptides are involved in the defence response to bacterial infection [32], and interestingly other proteins from this family (α-defensins) were found be significantly down-regulated in both normal colonic tissue and adenomas from GstP-null mice which also carried the APCMin+ mutation [7]. Interestingly, at this stage rather than key induced pathways, many genes associated with inflammatory responses appeared to be repressed in the Gstp−/−/Tg.AC mice relative to the Gstp+/+/Tg.AC animals.
At both time-points after TPA treatment a large number of genes were either induced or repressed in the Gstp−/−/Tg.AC mouse skin relative to Gstp+/+/Tg.AC Tg.AC mice alone (Supplementary Tables 2 & 4). Although there were a handful of genes found in the top 50 upregulated genes at both timepoints (Sprr2d, Uox, Lcn2, S100A8, S100A9) the transcript profiles of skin at 4 weeks after treatment were markedly different from that at 6 hours; this probably reflects the emerging complexity and crosstalk between many regulatory networks prior to overt tumour formation. However, the experimental design employed in the current study allowed us to carry out GSEA on the unfiltered data generated from the microarray protocols [33, 34]. All microarray data was subjected to ANOVA analysis to examine the effect of time, treatment and genotype on gene expression, and Biocarta pathways were mapped onto the gene expression data (Supplementary Tables 3 & 5). Since we were primarily interested in the role of Gstp in the process of skin tumorigenesis, we focussed on the effect of genotype in the pathway and gene ontology analysis. As shown in Supplementary Table 8, the up-regulated biological processes and molecular functions statistically enriched in Gstp−/−/Tg.AC mice relative to Gstp+/+/Tg.AC were highly similar, with the exception of a more anti-apoptotic and pro-inflammatory environment at the early timepoint, and alterations to lipid and sterol metabolism, and the Wnt receptor signaling pathway, at 4 weeks. The latter is interesting since transient activation of β-catenin signalling has previously been shown to be required for the initiation of hair follicle development [35], but higher and more sustained, or continuous, activation is required to maintain tumours derived from hair follicles such as those generated in the model used in this study [36].
Although the changes observed at 4 weeks could reflect changes in the constitution of the skin, those at 6h must reflect changes in short-term regulatory control and not be related to cell type. It is important to note that the high number of papillomas seen in these studies suggest that the 4 week time-point the entire skin is highly predisposed to tumour formation, reflected in the altered pathways of hair follicle morphogenesis and development, and epidermal morphogenesis observed, and suggesting a more de-differentiated phenotype in the skin of Gstp−/−/Tg.AC mice.
We also carried out analysis of patterns of gene expression in papillomas from Gstp−/−/Tg.AC vs. Gstp+/+/Tg.AC animals. Although there were some differences in patterns of gene expression between the two genotypes, we were unable to identify regulatory networks from which these gene expression changes were derived. Relative to the gene expression differences in normal skin, remarkably few changes were observed in the papillomas of the different genotypes, further strengthening the evidence that Gstp affects tumour promotion rather than tumour genetics. However, some of the differences observed are worthy of note. Galectin-4 and galectin-6 were elevated in Gstp−/−/Tg.AC mice; galectins have been implicated in a number of biological processes, including inflammation, innate and adaptive immunity, and cance - indeed, galectin-4 has been proposed as a marker for breast cancer [37]. Mug1 expression was induced both in papillomas (3.2-fold) and skin (11.3-fold) from Gstp−/−/Tg.AC mice. Mug1 is a novel protease inhibitor and a member of the α2-macroglobulin family whose members act as binding proteins for growth factors and cytokines including TNFα, TGFβ, and interleukins; Mug1 knockout mice are more susceptible to diet-induced acute pancreatitis [38]. The elevated expression of Mug1 in both skin and papillomas from Gstp−/−/Tg.AC mice may reflect the presence of a significantly enhanced inflammatory environment, both basally and following TPA treatment. CHi3l3 and Chi3l4 mRNA were both significantly lower in papillomas from Gstp−/−/Tg.AC mice; chitinase-like proteins have been associated with infection, T cell-mediated inflammation & allergy [39]. Interestingly, Chi3l3 was induced in the skin of both Gstp+/+/Tg.AC and Gstp−/−/Tg.AC mice 6h after TPA treatment (Supplementary Table 2), but to a significantly greater extent in the latter (6-fold vs 84-fold). These data demonstrate that the absence of Gstp can cause marked changes in gene expression profiles in tumours containing dominant oncogenes. Such changes could explain why GSTP is methylated in human tumours of prostate, liver and breast [12, 40, 41].
A number of studies have reported a reduction in papilloma multiplicity following crossing of the Tg.AC mouse with different transgenic mouse strains [42-44], whilst others have shown the transgene to initiate earlier onset of papillomagenesis [45] or an increase in tumour size [46]. However, as reported in this study the effects of the absence of Gstp on the Tg.AC background appear to be particularly marked with regards to both the incidence and multiplicity of papillomas.
Ras-related signaling has been show to be activated by mutation in a significant number of skin cancers in man, for example B-Raf mutations are commonly found in melanoma [47, 48]. The finding that Gstp can alter tumours induced by Ras signaling raises the interesting possibility that it may affect susceptibility to skin cancer in humans. In support of this possibility, polymorphisms in GSTP have been reported to increase susceptibility to basal cell and squamous cell carcinomas [49, 50]. In future work it will be interesting to establish whether GSTP also alters tumour incidence in other models of skin cancer such as the conditional B-Raf mutant mouse and whether tumorigenicity in these models can be inhibited by exogenous agents.
Supplementary Material
Acknowledgements
We are grateful to Catherine Hughes, Susanne van Schelven and Jennifer Kennedy for assistance with mouse work. Dr Shaun Walsh (Pathology, NHS Tayside) is thanked for expert analysis of histology sections and Probir Chakravarty (Cancer Research UK London Research Institute) is thanked for bioinformatic analyses. This work was funded by a Programme Grant awarded to CRW by Cancer Research UK (C4639/A5661).
Abbreviations
- BrdU
bromodeoxyuridine
- DMBA
7,12-dimethylbenz[a]anthracene
- GSEA
gene set enrichment analysis
- GST
glutathione S-transferase
- H & E
haematoxylin and eosin
- HO-1
haem oxygenase-1
- 3-MC
3-methylcholanthene
- PAH
polycyclic aromatic hydrocarbon
- PBS
phosphate-buffered saline
- NQO1
NADH quinone oxidoreductase
- Nrf2
nuclear factor erythroid 2-related factor 2
- Saa
serum amyloid A
- TPA
12-0-tetradecanoylphorbol-13-acetate
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- WT
wild-type
References
- 1.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 2.McIlwain CC, Townsend DM, Tew KD. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene. 2006;25:1639–48. doi: 10.1038/sj.onc.1209373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–75. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wareing CJ, Black SM, Hayes JD, Wolf CR. Increased levels of alpha-class and pi-class glutathione S-transferases in cell lines resistant to 1-chloro-2,4-dinitrobenzene. European Journal of Biochemistry. 1993;217:671–6. doi: 10.1111/j.1432-1033.1993.tb18292.x. [DOI] [PubMed] [Google Scholar]
- 5.Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci U S A. 1998;95:5275–80. doi: 10.1073/pnas.95.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritchie KJ, Henderson CJ, Wang XJ, Vassieva O, Carrie D, Farmer PB, et al. Glutathione transferase pi plays a critical role in the development of lung carcinogenesis following exposure to tobacco-related carcinogens and urethane. Cancer Res. 2007;67:9248–57. doi: 10.1158/0008-5472.CAN-07-1764. [DOI] [PubMed] [Google Scholar]
- 7.Ritchie KJ, Walsh S, Sansom OJ, Henderson CJ, Wolf CR. Markedly enhanced colon tumorigenesis in ApcMin mice lacking glutathione S-transferase Pi. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0911351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, et al. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–34. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Fan Y, Xue B, Luo L, Shen J, Zhang S, et al. Human glutathione S-transferase P1-1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene. 2006;25:5787–800. doi: 10.1038/sj.onc.1209576. [DOI] [PubMed] [Google Scholar]
- 10.Townsend DM, Manevich Y, He L, Hutchens S, Pazoles CJ, Tew KD. Novel role for glutathione S-transferase pi. Regulator of protein S-Glutathionylation following oxidative and nitrosative stress. J Biol Chem. 2009;284:436–45. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao P, Yang X, Xue YW, Zhang XF, Wang Y, Liu WJ, et al. Promoter methylation of glutathione S-transferase pi1 and multidrug resistance gene 1 in bronchioloalveolar carcinoma and its correlation with DNA methyltransferase 1 expression. Cancer. 2009;115:3222–32. doi: 10.1002/cncr.24369. [DOI] [PubMed] [Google Scholar]
- 12.Yang B, House MG, Guo M, Herman JG, Clark DP. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol. 2005;18:412–20. doi: 10.1038/modpathol.3800287. [DOI] [PubMed] [Google Scholar]
- 13.Leder A, Kuo A, Cardiff RD, Sinn E, Leder P. v-Ha-ras transgene abrogates the initiation step in mouse skin tumorigenesis: effects of phorbol esters and retinoic acid. Proc Natl Acad Sci U S A. 1990;87:9178–82. doi: 10.1073/pnas.87.23.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spalding JW, Momma J, Elwell MR, Tennant RW. Chemically induced skin carcinogenesis in a transgenic mouse line (TG.AC) carrying a v-Ha-ras gene. Carcinogenesis. 1993;14:1335–41. doi: 10.1093/carcin/14.7.1335. [DOI] [PubMed] [Google Scholar]
- 15.Nishiyama H, Gill JH, Pitt E, Kennedy W, Knowles MA. Negative regulation of G(1)/S transition by the candidate bladder tumour suppressor gene DBCCR1. Oncogene. 2001;20:2956–64. doi: 10.1038/sj.onc.1204432. [DOI] [PubMed] [Google Scholar]
- 16.Conklin DJ, Haberzettl P, Lesgards JF, Prough RA, Srivastava S, Bhatnagar A. Increased sensitivity of glutathione S-transferase P-null mice to cyclophosphamide-induced urinary bladder toxicity. J Pharmacol Exp Ther. 2009;331:456–69. doi: 10.1124/jpet.109.156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Townsend DM, Tew KD, He L, King JB, Hanigan MH. Role of glutathione S-transferase Pi in cisplatin-induced nephrotoxicity. Biomed Pharmacother. 2009;63:79–85. doi: 10.1016/j.biopha.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J, Wolf CR, Henderson CJ, Cai Y, Board PG, Foster PS, et al. Glutathione transferase P1: an endogenous inhibitor of allergic responses in a mouse model of asthma. Am J Respir Crit Care Med. 2008;178:1202–10. doi: 10.1164/rccm.200801-178OC. [DOI] [PubMed] [Google Scholar]
- 19.Mitsushita J, Lambeth JD, Kamata T. The superoxide-generating oxidase Nox1 is functionally required for Ras oncogene transformation. Cancer research. 2004;64:3580–5. doi: 10.1158/0008-5472.CAN-03-3909. [DOI] [PubMed] [Google Scholar]
- 20.Shinohara M, Shang WH, Kubodera M, Harada S, Mitsushita J, Kato M, et al. Nox1 redox signaling mediates oncogenic Ras-induced disruption of stress fibers and focal adhesions by down-regulating Rho. The Journal of biological chemistry. 2007;282:17640–8. doi: 10.1074/jbc.M609450200. [DOI] [PubMed] [Google Scholar]
- 21.Valencia A, Kochevar IE. Nox1-based NADPH oxidase is the major source of UVA-induced reactive oxygen species in human keratinocytes. The Journal of investigative dermatology. 2008;128:214–22. doi: 10.1038/sj.jid.5700960. [DOI] [PubMed] [Google Scholar]
- 22.Berhane K, Widersten M, Engstrom A, Kozarich JW, Mannervik B. Detoxication of base propenals and other alpha, beta-unsaturated aldehyde products of radical reactions and lipid peroxidation by human glutathione transferases. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1480–4. doi: 10.1073/pnas.91.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Yang G, Huang R, Chen X, Hu G. SVH-B interacts directly with p53 and suppresses the transcriptional activity of p53. FEBS Lett. 2007;581:4943–8. doi: 10.1016/j.febslet.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 24.Huang R, Xing Z, Luan Z, Wu T, Wu X, Hu G. A specific splicing variant of SVH, a novel human armadillo repeat protein, is up-regulated in hepatocellular carcinomas. Cancer Res. 2003;63:3775–82. [PubMed] [Google Scholar]
- 25.Yuasa T, Ohno S, Kehrl JH, Kyriakis JM. Tumor necrosis factor signaling to stress-activated protein kinase (SAPK)/Jun NH2-terminal kinase (JNK) and p38. Germinal center kinase couples TRAF2 to mitogen-activated protein kinase/ERK kinase kinase 1 and SAPK while receptor interacting protein associates with a mitogen-activated protein kinase kinase kinase upstream of MKK6 and p38. J Biol Chem. 1998;273:22681–92. doi: 10.1074/jbc.273.35.22681. [DOI] [PubMed] [Google Scholar]
- 26.Zhong J, Kyriakis JM. Germinal center kinase is required for optimal Jun N-terminal kinase activation by Toll-like receptor agonists and is regulated by the ubiquitin proteasome system and agonist-induced, TRAF6-dependent stabilization. Mol Cell Biol. 2004;24:9165–75. doi: 10.1128/MCB.24.20.9165-9175.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong J, Gavrilescu LC, Molnar A, Murray L, Garafalo S, Kehrl JH, et al. GCK is essential to systemic inflammation and pattern recognition receptor signaling to JNK and p38. Proc Natl Acad Sci U S A. 2009;106:4372–7. doi: 10.1073/pnas.0812642106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Urieli-Shoval S, Cohen P, Eisenberg S, Matzner Y. Widespread expression of serum amyloid A in histologically normal human tissues. Predominant localization to the epithelium. J Histochem Cytochem. 1998;46:1377–84. doi: 10.1177/002215549804601206. [DOI] [PubMed] [Google Scholar]
- 29.Henderson CJ, Wolf CR. Disruption of the glutathione transferase pi class genes. Methods in enzymology. 2005;401:116–35. doi: 10.1016/S0076-6879(05)01007-4. [DOI] [PubMed] [Google Scholar]
- 30.Dang H, Trempus C, Malarkey DE, Wei SJ, Humble M, Morris RJ, et al. Identification of genes and gene ontology processes critical to skin papilloma development in Tg.AC transgenic mice. Mol Carcinog. 2006;45:126–40. doi: 10.1002/mc.20154. [DOI] [PubMed] [Google Scholar]
- 31.Quigley DA, To MD, Perez-Losada J, Pelorosso FG, Mao JH, Nagase H, et al. Genetic architecture of mouse skin inflammation and tumour susceptibility. Nature. 2009;458:505–8. doi: 10.1038/nature07683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, Wang C, Zhou Y, Speed TP. Gene set enrichment analysis made simple. Statistical methods in medical research. 2009;18:565–75. doi: 10.1177/0962280209351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Developmental cell. 2002;2:643–53. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 36.Lo Celso C, Prowse DM, Watt FM. Transient activation of beta-catenin signalling in adult mouse epidermis is sufficient to induce new hair follicles but continuous activation is required to maintain hair follicle tumours. Development. 2004;131:1787–99. doi: 10.1242/dev.01052. [DOI] [PubMed] [Google Scholar]
- 37.Huflejt ME, Leffler H. Galectin-4 in normal tissues and cancer. Glycoconj J. 2004;20:247–55. doi: 10.1023/B:GLYC.0000025819.54723.a0. [DOI] [PubMed] [Google Scholar]
- 38.Umans L, Serneels L, Overbergh L, Stas L, Van Leuven F. alpha2-macroglobulin- and murinoglobulin-1-deficient mice. A mouse model for acute pancreatitis. Am J Pathol. 1999;155:983–93. doi: 10.1016/s0002-9440(10)65198-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutherland TE, Maizels RM, Allen JE. Chitinases and chitinase-like proteins: potential therapeutic targets for the treatment of T-helper type 2 allergies. Clin Exp Allergy. 2009;39:943–55. doi: 10.1111/j.1365-2222.2009.03243.x. [DOI] [PubMed] [Google Scholar]
- 40.Jeronimo C, Varzim G, Henrique R, Oliveira J, Bento MJ, Silva C, et al. I105V polymorphism and promoter methylation of the GSTP1 gene in prostate adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2002;11:445–50. [PubMed] [Google Scholar]
- 41.Lasabova Z, Tilandyova P, Kajo K, Zubor P, Burjanivova T, Danko J, et al. Hypermethylation of the GSTP1 promoter region in breast cancer is associated with prognostic clinicopathological parameters. Neoplasma. 2010;57:35–40. doi: 10.4149/neo_2010_01_035. [DOI] [PubMed] [Google Scholar]
- 42.Lozano J, Xing R, Cai Z, Jensen HL, Trempus C, Mark W, et al. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res. 2003;63:4232–8. [PubMed] [Google Scholar]
- 43.Murphy JE, Morales RE, Scott J, Kupper TS. IL-1 alpha, innate immunity, and skin carcinogenesis: the effect of constitutive expression of IL-1 alpha in epidermis on chemical carcinogenesis. J Immunol. 2003;170:5697–703. doi: 10.4049/jimmunol.170.11.5697. [DOI] [PubMed] [Google Scholar]
- 44.Robles AI, Rodriguez-Puebla ML, Glick AB, Trempus C, Hansen L, Sicinski P, et al. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 1998;12:2469–74. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larcher F, Murillas R, Bolontrade M, Conti CJ, Jorcano JL. VEGF/VPF overexpression in skin of transgenic mice induces angiogenesis, vascular hyperpermeability and accelerated tumor development. Oncogene. 1998;17:303–11. doi: 10.1038/sj.onc.1201928. [DOI] [PubMed] [Google Scholar]
- 46.Tober KL, Cannon RE, Spalding JW, Oberyszyn TM, Parrett ML, Rackoff AI, et al. Comparative expression of novel vascular endothelial growth factor/vascular permeability factor transcripts in skin, papillomas, and carcinomas of v-Ha-ras Tg.AC transgenic mice and FVB/N mice. Biochem Biophys Res Commun. 1998;247:644–53. doi: 10.1006/bbrc.1998.8787. [DOI] [PubMed] [Google Scholar]
- 47.Banerji U, Affolter A, Judson I, Marais R, Workman P. BRAF and NRAS mutations in melanoma: potential relationships to clinical response to HSP90 inhibitors. Mol Cancer Ther. 2008;7:737–9. doi: 10.1158/1535-7163.MCT-08-0145. [DOI] [PubMed] [Google Scholar]
- 48.Packer LM, East P, Reis-Filho JS, Marais R. Identification of direct transcriptional targets of (V600E)BRAF/MEK signalling in melanoma. Pigment Cell Melanoma Res. 2009;22:785–98. doi: 10.1111/j.1755-148X.2009.00618.x. [DOI] [PubMed] [Google Scholar]
- 49.Fryer AA, Ramsay HM, Lovatt TJ, Jones PW, Hawley CM, Nicol DL, et al. Polymorphisms in glutathione S-transferases and non-melanoma skin cancer risk in Australian renal transplant recipients. Carcinogenesis. 2005;26:185–91. doi: 10.1093/carcin/bgh291. [DOI] [PubMed] [Google Scholar]
- 50.Marshall SE, Bordea C, Haldar NA, Mullighan CG, Wojnarowska F, Morris PJ, et al. Glutathione S-transferase polymorphisms and skin cancer after renal transplantation. Kidney Int. 2000;58:2186–93. doi: 10.1111/j.1523-1755.2000.00392.x. [DOI] [PubMed] [Google Scholar]
- 51.Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.