Abstract
Background
Di-(2-ethylhexyl)-phthalate (DEHP) is a commonly used plasticizer in polyvinylchloride (PVC) formulations and a potentially non-genotoxic carcinogen. The aim of this study was to identify genes whose level of expression is altered by DEHP by using a global wide-genome approach in Syrian hamster embryo (SHE) cells, a model similar to human cells regarding their responses to this type of carcinogen. With mRNA Differential Display (DD), we analysed the transcriptional regulation of SHE cells exposed to 0, 12.5, 25 and 50 μM of DEHP for 24 hrs, conditions which induced neoplastic transformation of these cells. A real-time quantitative polymerase chain reaction (qPCR) was used to confirm differential expression of genes identified by DD.
Results
Gene expression profiling showed 178 differentially-expressed fragments corresponding to 122 genes after tblastx comparisons, 79 up-regulated and 43 down-regulated. The genes of interest were involved in many biological pathways, including signal transduction, regulation of the cytoskeleton, xenobiotic metabolism, apoptosis, lipidogenesis, protein conformation, transport and cell cycle. We then focused particularly on genes involved in the regulation of the cytoskeleton, one of the processes occurring during carcinogenesis and in the early steps of neoplastic transformation. Twenty one cytoskeleton-related genes were studied by qPCR. The down-regulated genes were involved in focal adhesion or cell junction. The up-regulated genes were involved in the regulation of the actin cytoskeleton and this would suggest a role of cellular plasticity in the mechanism of chemical carcinogenesis. The gene expression changes identified in the present study were PPAR-independent.
Conclusion
This study identified a set of genes whose expression is altered by DEHP exposure in mammalian embryo cells. This is the first study that elucidates the genomic changes of DEHP involved in the organization of the cytoskeleton. The latter genes may be candidates as biomarkers predictive of early events in the multistep carcinogenic process.
Background
Di-(2-ethylhexyl)-phthalate (DEHP) is a commonly used plasticizer in polyvinylchloride (PVC) formulations which have a number of applications, especially in food packaging, medical devices or cosmetics. Phthalates are not chemically bound to PVC and can migrate from PVC-containing products to the environment, resulting in significant environmental contamination and human exposure [1,2].
DEHP experiments have revealed toxicities including carcinogenesis and endocrine-disrupting effects, but no genotoxicity has been recorded [3]. DEHP is capable of disturbing the reproductive process by mimicking or antagonizing steroid hormone action [4] and its effects on testosterone, luteinizing hormone or estrogen-like activity have been reported [5,6]. DEHP has been shown to decrease free testosterone levels in humans after occupational exposure [7] and thyroid hormone levels in adult men otherwise exposed [8].
DEHP has been classified as a peroxisomal proliferator and as a non-genotoxic carcinogen in animals [9]. Experimental studies using rodents and in vitro assays showed that DEHP and its active metabolite MEHP (mono-(2-ethylhexyl)-phthalate) can interact with nuclear receptors like PPARα [10] or PPARγ [11]. Oxidative stress, as a result of peroxisome proliferation, and DNA damage have been described in the human prostate adenocarcinoma cell line LNCaP [12,13] and the mouse Leydig tumor cell line MA-10 [14] exposed to high concentrations of DEHP (3 mM). Peroxisome proliferation is one of the mechanisms that produce liver tumors in rats or mice, but this mechanism was not judged to be relevant in humans [15]. The liver is not the sole target for DEHP carcinogenicity: testicular tumors [16] and pancreatic acinar adenomas have also been reported [17]. Other studies have pointed out that peroxisome proliferation is not a necessarily pathway in the carcinogenicity of DEHP [18] and more liver tumors occurred in PPARα-null mice than in wild type animals [19]. Transcriptional changes independent of PPARα were also found in rats and mice exposed to DEHP [20]. Several non-PPARα mechanisms were addressed: activation of p38 mitogen-activated protein kinase not involved in peroxisome proliferations [21]; stimulation of growth regulatory pathways, mitogen-activated protein kinase, extracellular signal-regulated kinase and p38 phosphorylation [22]. Other mechanisms related to non-genotoxic carcinogenicity, like inhibition of gap junctional intercellular communication [23] or inhibition of apoptosis, were reported. Apoptosis was shown to be suppressed by DEHP through different pathways. An interference with the cytokine TGF-β1 (transforming growth factor-β1) [24] or with TNF-α (tumor necrosis factor-α) has been described [25]. An increased level of Bcl-2 and negative regulation of c-Myc expression has been related to inhibition of apoptosis in Syrian hamster embryo cells treated with 50 μM of DEHP [26].
Several authors have demonstrated that DEHP and its active metabolite MEHP induce morphological transformation of SHE cells [27-29], indicating the carcinogenic potency of the two chemicals. Although phthalate toxicity has been extensively investigated over the past 10 years, the mechanisms of DEHP carcinogenicity have not been elucidated. It was recently stated by the International Agency for Research on Cancer (IARC) that PPAR-independent mechanisms of DEHP carcinogenesis are necessary to be studied [http://monographs.iarc.fr/ENG/Publications/techrep42/TR42-18.pdf].
The choice of cellular models and methodologies is critical to the study of the phenomenon of carcinogenesis. Syrian hamster embryo cells are a relevant model for mechanistic studies of chemical carcinogenicity. SHE cells, unlike mouse and rat cells, are less responsive to peroxisomal proliferation and, in this respect, more similar to human cells. SHE cells are normal, diploid, genetically stable and primary cells which are metabolically competent for procarcinogen activation. Therefore they are used to study mechanisms of in vitro carcinogenesis [30]. SHE cells are obtained from embryos after removal of the differentiated tissues, and the population is mainly composed of epithelial and fibroblastic cells [31]. SHE cells from colonies having been morphologically transformed after short exposure to chemical carcinogens induced tumours when transplanted back into hamsters [32]. This validated the model and the cell transformation criteria for in vitro carcinogenicity. Recently, the SHE cell transformation assay has been recommended by OECD in 2007 as in vitro method of screening chemical carcinogens on the basis of its performances to detect non-genotoxic as well as genotoxic carcinogens [http://www.oecd.org/dataoecd/56/5/37863750.pdf].
The aim of this work was to use a global transcriptomic approach to understand the molecular mechanisms of cell transformation induced by DEHP in SHE cells. The objectives were to identify changes in gene expression occurring in the early steps of cell transformation as well as pathway disturbances that may trigger a carcinogenic process. A characterization of the genes expressed in SHE cells at DEHP concentrations inducing cell transformation may give information on PPAR-independent mechanisms and alternative pathways of DEHP carcinogenicity. The transcriptomic changes induced by DEHP in SHE cells were analyzed in the first hours of exposure. We focused secondly on changes of cytoskeleton-related genes underlying morphological transformation in SHE cells. Indeed, cell transformation is expressed by the alteration of cell morphology, a disorganized pattern of colony growth and the acquisition of anchorage-independent growth which is predictive of their ability to induce tumors when injected into syngenic animals [33]. Despite the central role of the actin cytoskeleton throughout the life cycle, little is known about the gene expression changes involved in deregulation of its dynamic in the first stages of tumorigenesis. Cytoskeleton defects in relation to cancer have been mostly studied in the late stages of cell invasion and metastasis.
Differential Display was chosen to identify differentially-expressed genes in SHE cells and to explore the entire genome. The mRNA differential display described by Liang and Pardee [34] is a powerful approach for transcriptomic analysis. This methodology has become popular as a tool for non-model organisms because of lack of requirement of previous genomic information about the species of interest. As the genome of hamster is partly characterized so far, Differential Display appeared quite appropriate to study DEHP dose-dependent effects in SHE cells. We applied the current methodology that uses a combination of 3 "anchored" oligo-dT primers (to divide the cDNA population in 3 subsets) and 80 "arbitrary" primers of 13-mers. The 240 primer combination allowed us to obtain a level of 95% gene coverage [35].
DD was applied to cells exposed for 24 hrs to DEHP. Genes corresponding to differentially-expressed fragments were characterized. Differential Display allowed us to screen a set of differentially-expressed fragments in treated cells, among which 122 genes were identified as targeted by a 24 hr-DEHP exposure. These genes were involved in such functions as transcription signalling pathways, cytoskeleton regulation, apoptosis, metabolism. As Differential Display is a semi-quantitative method, the expression changes of the genes we were interested in, were checked by qPCR using hamster specific primers. qPCR was applied to RNAs not only from 24 hr-treated cells, but also from cells treated for 5 hrs in order to study the cell response in the meantime. We particularly focused on changes of cytoskeleton-related genes underlying morphological transformation in SHE cells. The objective was to explain from a mechanistic point of view the gene expression changes after DEHP exposure. To the best of our knowledge, this exercise has never been done previously.
Results
Identification of DEHP-responsive genes using Differential Display
The Differential Display technique was used to identify genes differentially expressed in SHE cells, after 24 hrs of treatment with DEHP. An illustration of differentially-expressed fragments is given in Figure 1 which shows gels obtained after the DD protocol and highlights fragments regulated more than 2-fold by DEHP. Using 3 anchored primers and 80 arbitrary primers, 178 differentially expressed fragments were identified (115 up-regulated and 63 down-regulated). Among these transcripts, 141 (79%) showed homology to known genes in the RefSeq database (mouse or human) of Genbank, while 37 (21%) had no homology or homology to hypothetical proteins. The sequences of the fragments obtained by DD have been deposited in the Genbank dbEST database (LIBEST_027390 Syrian Hamster Embryo cells library). These 141 fragments corresponded to 122 genes that are listed in table 1 with their accession numbers and the tblastx expected (the threshold for tblastx comparison was p ≤ 0.001). These genes were classed according to 8 biological functions with reference to the GO process database. These functions included signal transduction and transcription, cytoskeleton regulation, xenobiotic metabolism, apoptosis, lipidogenesis, protein conformation or transport and cell cycle. The regulation of the cytoskeleton was one of the most impacted pathways. Indeed, 21 genes involved in this function were differentially expressed after DEHP exposure. Ten genes were up-regulated, and 11 were down-regulated.
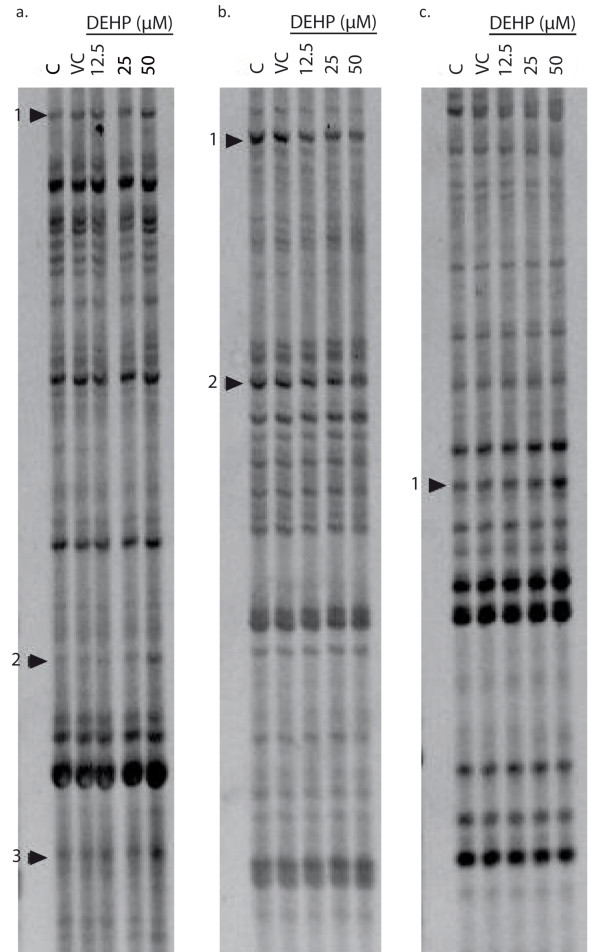
Figure 1.
Representative Differential Display. This figure shows results obtained with control (C), vehicle control (VC) and DEHP-treated (12.5, 25 and 50 μM) SHE cells mRNA, after 24 hrs of exposure. Arrows indicate fragments regulated more than 2-fold and used for the next step of analysis. The DD fragments were generated using the A-anchored primer (H-dT+A) combined with the arbitrary primer H-AP37 (lane a) or H-AP52 (lane b) and the G-anchored primer (H-dt+G) combined with the arbitrary primer H-AP40 (lane c). After characterization by sequencing, we obtained the following genes: a1:Rlp37a/a2:Rlp27a/a3:Col1a1/b1:Cttnbip1/b2: Enah/c1: Coro1C.
Table 1.
List of genes identified by Differential Display after DEHP exposure and classified by major biological function, according to GO process
| Id | Official symbol | Gene Name | Accession Nr | Tblastx expect | Effect of DEHP |
|---|---|---|---|---|---|
| 1-Transcription and Signal Transduction | |||||
| A0601 | Hsp90 | Heat shock protein 90 kDa protein | NM_010478 | 9E-25 | + |
| A0602 | Hsp70 | Heat shock protein 70 kDa protein | NM_005346 | 1E-24 | + |
| A3201 | Cebpd | CCAAT/enhancer binding protein delta | NM_005195 | 9E-10 | - |
| A4102 | Ncbp2 | nuclear cap binding protein subunit 2 | NM_007362 | 6E-21 | + |
| A5101 | Mettl5 | methyltransferase like 5 | NM_029280 | 7E-10 | + |
| A5102 | Ogt | O-linked N-acetylglucosamine transferase | NM_181672 | 5E-28 | + |
|
A6101 A6102 |
Nr1i2 | nuclear receptor subfamily 1I2 | NM_003889 | 1E-158 3E-74 |
+ |
| A6302 | Mapk15 | mitogen-activated protein kinase 15 | NM_177922 | 3E-16 | + |
| A6801 | Rab1b | member of RAS oncogene family | NM_030981 | 1E-22 | ++ |
| A7502 | Mdb1 | methyl-CpG binding domain protein 1 | NM_013594 | 2E-16 | -- |
| C0602 | Gper | G protein-coupled estrogen receptor 1 | NM_001098201 | 6E-68 | + |
| C1001 | Hmbox1 | Homeobox 1 | NM_024567 | 2E-32 | ++ |
| C1002 | Mapk3 | mitogen-activated protein kinase 3 | NM_001109891 | 9E-60 | - |
| C1301 | Usp3 | Ubiquitin peptidase 3 | NM_006537 | 9E-34 | + |
| C1401 | Psmc5 | 26S protease regulatory subunit ATPase 5 | NM_002805 | 2E-23 | - |
| C4601 | Hsph1 | heat shock 105kDa/110kDa protein | NM_006644 | 5E-07 | + |
| C4802 | Irf2 | interferon regulatory factor 2 | NM_008391 | 8E-19 | + |
| C7101 | Lmo4 | LIM domain only 4 | NM_006769 | 7E-09 | + |
| G0301 | Smarcc1 | SWI/SNF related actin dependent regulator of chromatin | NM_003074 | 3E-20 | - |
| G1001 | Chd4 | ATP dependant hélicase 4 | NM_001273 | 1E-45 | - |
| G1504 | Foxp1 | Forkhead box P1 | NM_032682 | 3E-50 | ++ |
| G1701 | Creb3l1 | cAMP responsive element binding protein 3-like 1 | NM_052854 | 8E-19 | + |
| G1902 | Pbrm | Polybromo domain | NM_001081251 | 8E-49 | + |
| G3301 | Rxfp2 | relaxin/insulin-like family peptide receptor 2 | NM_130806 | 1E-24 | + |
| G3501 | Akap5 | A kinase anchor protein 5 | NM_004857 | 1E-05 | + |
|
G3801 G7001 |
Eif1 | eukaryotic translation initiation factor 1 | NM_005801 | 3E-42 1E-15 |
+ |
| G4801 | Spry3 | sprouty homolog 3 | NM_005840 | 0.0002 | + |
| G6001 | Foxa3 | forkhead box A3 | NM_008260 | 9E-17 | - |
| G6201 | Mett5d1 | methyltransferase 5 containing 1 | NM_029790 | 0.0038 | + |
| 2-Regulation of cytoskeleton | |||||
| A0201 | Cttnbp2 | Cortactin binding protein 2 | NM_030249 | 0.0001 | + |
| A1901 | Snx6 | sorting nexin 6 | NM_021249 | 8E-10 | - |
| A2401 | Lrrc8a | leucine rich repeat containing 8A | NM_177725 | 5E-32 | - |
|
A3501 A3502 C3501 G3502 G3503 |
Actb | beta-actin | NM_007393 | 3E-51 1E-106 2E-21 7E-114 3E-48 |
+ |
| A3702 | Col1a1 | collagen, type I, alpha 1 | NM_000088 | 1E-21 | + |
| A3801 | Nrp2 | neuropilin 2 | NM_0010774 | 1E-22 | - |
| A5201 | Ctnnbip1 | catenin beta interacting protein 1 | NM_020248 | 0.0008 | - |
| A5202 | Enah | enabled homolog | NM_008680 | 2E-09 | - |
| A5402 | Kif23 | kinesin family member 23 | NM_024245.4 | 1E-25 | ++ |
| A7501 | Cdh3 | cadherin 3 | NM_007665 | 0.0007 | - |
| C4502 | Nid2 | nidogen 2 | NM_008695 | 0.006 | - |
|
C6603 C6604 C6605 |
Crip1 | cysteine-rich protein 1 | NM_007763 | 1E-31 9E-08 3E-61 |
+ |
| G0601 | Thy1 | Thy-1 cell surface antigen | NM_009382.3 | 0.001 | - |
| G0801 | Calml3 | calmodulin-like 3 | NM_027416.3 | 0.0004 | + |
| G1102 | Flrt2 | fibronectin leucine rich transmembrane protein 2 | NM_201518 | 2E-08 | - |
| G1301 | Has2 | hyaluronan synthase 2 | NM_008216 | 8E-18 | - |
| G1901 | Plekha5 | Pleckstrin homology domain A5 | NM_019012 | 3E-06 | + |
| G2301 | Thbs1 | thrombospondin 1 | NM_011580 | 3E-20 | - |
| G4001 | Coro1C | coronin, actin binding protein | NM_014325 | 4E-07 | + |
| G4802 | Tubb2b | Tubulin beta | NM_023716 | 0.0001 | + |
| G6901 | Dclk1 | doublecortin-like kinase 1 | NM_019978 | 1E-06 | + |
| 3-Xenobiotic metabolism | |||||
| A0901 | Cyp2e1 | Cytochrome P450 2e1 | NM_021282 | 3E-44 | + |
| A2901 | Ephx1 | Epoxide Hydrolase 1 | NM_000120 | 4E-21 | + |
| A5701 | Gstp1 | glutathione S-transferase, pi 1 | NM_000852 | 1E-24 | + |
| A6601 | Gstt1 | glutathione S-transferase, theta 1 | NM_008185 | 1E-44 | - |
|
C0101 C1501 G1502 |
Txnrd1 | Thioredoxin reductase 1 | NM_001042523 | 1E-20 1E-14 3E-79 |
+ |
| C2901 | Cyp1b1 | Cytochrome P450 1b1 | NM_009994 | 0.0 | + |
| C3101 | Gstm5 | glutathione S-transferase, mu 5 | NM_010360 | 4E-51 | - |
| C3701 | Tnfa | tumor necrosis factor-alpha | NM_011659 | 0.00009 | + |
| G0101 | Psme4 | proteasome activator subunit 4 | NM_134013 | 4E-87 | + |
| G0701 | Mt2a | metallothionein 2A | NM_005953 | 7E-20 | - |
| G1201 | Ggt1 | gamma-glutamyltransferase 1 | NM_013430 | 3E-46 | + |
| G1501 | Txnrd2 | Thioredoxin reductase 2 | NM_006440 | 9E-19 | + |
| G2001 | Pdia4 | Disulfite Isomerase 4 | NM_004911 | 5E-46 | + |
| G2202 | Ahcy | S-adenosylhomocystein hydrolase | NM_016661 | 1E-10 | - |
| G2302 | Cyp2f2 | Cytochrome P450 2f2 | NM_007817 | 9E-25 | -- |
| G3302 | Cytb | Mesocricetus auratus cytochrome b | YP_003208313 | 9E-86 | ++ |
| 4-Apoptosis | |||||
| A4301 | Pik3r1 | phosphatidylinositol 3-kinase | NM_001024955 | 1E-80 | + |
| C1701 | Tp53 | tumor supressor p53 | U07182 | 6E-83 | - |
|
C7103 G3701 |
Bcl10 | B-cell CLL/lymphoma 10 | NM_003921 | 4E-37 5E-82 |
+ |
| G1503 | Nfkb1 | NF-κB | NM_008689 | 2E-102 | + |
| G2201 | Casp8 | Caspase 8 | NM_009812 | 6E-29 | - |
| G2305 | Eef1d | Eukaryotic translation elongation factor 1 delta | NM_029663 | 3E-17 | + |
| G3001 | Topors | topoisomerase I p53-binding | NM_134097 | 4E-10 | - |
|
G4101 G4102 |
Sh3kbp1 | SH3-domain kinase binding protein 1 | NM_031892 | 1E-09 0.0008 |
+ |
| G5301 | Cmyc | myelocytomatosis oncogene | AJ582076 | 2E-27 | - |
| 5-Lipidogenesis | |||||
| A0501 | Pla2g2d | phospholipase A2, group IID | NM_011109 | 0.00003 | + |
| A3003 | Dhcr7 | dehydrocholesterol reductase | NM_007856 | 3E-36 | + |
| C0701 | Scl27a1 | solute carrier family 27A1 | NM_198580 | 0.0 | + |
| C0901 | Acaa1 | Acetyl-CoA acyltransferase 1 | NM_130864 | 1E-77 | + |
| C4602 | Lpl | lipoprotein lipase | NM_008509 | 1E-37 | - |
| G0201 | Star | steroidogenic acute regulatory protein | NM_011485 | 7E-08 | - |
| 6-Protein conformation or transport | |||||
| A3801 | Rpn1 | ribophorin I | NM_133933 | 3E-14 | - |
| A4701 | Ergic3 | ERGIC and golgi 3 | NM_198398 | 6E-21 | + |
| A5302 | Gxylt1 | glycosyltransferase 8 domain containing 3 | NM_173601 | 0.0002 | + |
| C0601 | Ppia | peptidylprolyl isomerase A | NM_021130 | 7E-71 | + |
| C2001 | Grpel1 | GrpE-like 1, mitochondrial | NM_024478 | 1E-13 | + |
| C2201 | Fxn | frataxin | NM_008044 | 4E-33 | + |
| C3602 | Slc26a9 | solute carrier family 26, member 9 | NM_177243 | 1E-9 | + |
| C7501 | Slc13a3 | solute carrier family 13 member 3 | NM_054055 | 3E-29 | - |
| G1801 | Slc15a1 | Solute carrier family 15 member 1 | NM_053079 | 0.0002 | - |
| G3702 | Rpn2 | ribophorin II | NM_019642 | 3E-26 | - |
| G6101 | Nrbp1 | nuclear receptor binding protein 1 | NM_013392 | 4E-12 | + |
| G6601 | Slc6a8 | solute carrier family 6 member 8 | NM_005629 | 0.0001 | - |
| 7-Cell cycle | |||||
| A0401 | Ingap | Mesocricetus auratus islet neogenesis associated protein | U41738 | 4E-79 | - |
| A3002 | Cdkn2b | cyclin-dependent kinase inhibitor 2B | NM_004936 | 1E-9 | - |
| A4801 | Ppp1cc | protein phosphatase 1, catalytic subunit, gamma | NM_002710 | 2E-78 | + |
| A5702 | Sec11a | SEC11 homolog A | NM_014300 | 1E-19 | + |
| C4002 | Mapk4 | mitogen-activated protein kinase 4 | NM_002747 | 9E-26 | + |
| C4701 | Ccndbp1 | cyclin D-type binding-protein 1 | NM_010761 | 1E-17 | + |
|
C6301 C6302 |
Zw10 | ZW10 homolog centromere/kinetochore protein | NM_004724 | 1E-04 1E-05 |
- |
| 8-Other functions | |||||
| A3903 | Rexo2 | REX2, RNA exonuclease 2 homolog | NM_015523 | 6E-21 | - |
| A5403 | Hk2 | hexokinase 2 | NM_000189 | 4E-28 | + |
| A5602 | Pcbp2 | poly(rC)-binding protein 2 | NM_031989 | 1E-34 | + |
| A7301 | Lyrm4 | LYR motif containing 4 | NM_201358 | 0.0004 | - |
| C1801 | Itpripl2 | inositol 1,4,5-triphosphate receptor Interacting protein-like 2 | NM_001033380 | 4E-11 | + |
| C2401 | Tsn | translin | NM_011650 | 4E-10 | + |
| C4501 | Trip4 | thyroid hormone receptor interactor 4 | NM_016213 | 0.0006 | - |
| C7102 | Tbrg3 | transforming growth factor beta regulated gene 3 | NR_027799 | 0.001 | + |
| G0101 | Psme4 | proteasome activator subunit 4 | NM_014614 | 4E-87 | + |
| G0501 | Gatad2a | GATA zinc finger domain containing 2A | NM_001113346 | 8E-15 | + |
| G0901 | Hoxa10 | Homeobox A10 (HOXA10) | NM_008263 | 1E-47 | + |
| G1101 | Iap | Syrian hamster intracisternal A particle | M10134 | 3E-73 | + |
| G2303 | Zc3h12c | zinc finger CCCH type containing 12C | NM_001162921 | 7E-9 | - |
| G6801 | Fst | follistatin | NM_006350 | 3E-42 | - |
| Ribosomal proteins | |||||
| A3703 | Rpl37a | ribosomal protein L37a | NM_000998 | 0.001 | + |
|
A3901 C3901 |
Mrpl45 | mitochondrial ribosomal protein L45 | NM_025927 | 2E-10 6E-59 |
- |
| A3902 | Rps21 | ribosomal protein S21 | NM_001024 | 7E-14 | + |
| A4001 | ./. | mitochondrial 12S ribosomal RNA | X84390 | 0.0 | + |
| G2002 | Rpl27a | 60S ribosomal protein L27a | NM_000990 | 5E-18 | + |
| G3504 | Rpl10 | ribosomal protein L10 | NR_026898 | 4E-132 | + |
|
G3601 G3602 |
Rpl28 | ribosomal protein L28 | NM_009081 | 3E-22 8E-180 |
+ |
| G5701 | Rpl22 | ribosomal protein L22 | NM_000983 | 0.00009 | + |
The DEHP effect is identified by (+) for 2.0-fold over-expression, (++) for 10-fold over-expression, (-) for 2.0-fold under-expression and (--) for 10.0-fold under-expression for at least one dose of DEHP. The 37 Differential Display fragments showing no match after comparison with the RefSeq database were not listed in the table. Id (Identification Number) represents the internal reference of the sequence used before the identification of Differential Display sequence.
Transcription and signal transduction is another biological process targeted by DEHP treatment. We found 22 up-regulated genes, among which 3 were up-regulated more than 10-fold (rab1b, a Ras oncogen family member, Homeobox 1 and Forkhead P1). Heat- shock response related genes (hsp90, hsp70 and hsph1) and the genes involved in promoter methylation (mettl5, mett5d1) were up-regulated. On the other hand, 7 genes were down-regulated (CCAAT/enhancer binding protein delta, methyl-CpG binding domain protein 1, Map kinase 3, Protease subunit 5, SWI/SNF related actin dependent regulator of chromatin, ATP dependent helicase 4 and Forkhead A3).
Xenobiotic metabolism genes such as cytochromes and glutathione S-transferases were also found to be differentially expressed, indicating a mobilization of cellular defence and detoxication systems. An up-regulation of cyp1b1 and cyp2e1 was registered, whereas cyp2f2 was found to be down-regulated. Concerning GST, the Pi family was over-expressed while the Theta and Mu families were down-regulated.
Differential Display results confirmed the down-regulation of c-myc and showed down-regulation of p53. A down-regulation of pro-apoptotic genes (casp8, topors...) and an over-expression of anti-apoptotic genes (bcl10, nfkb1, sh3kbp1...) were also observed.
Expression of genes involved in the regulation of the cytoskeleton by qPCR
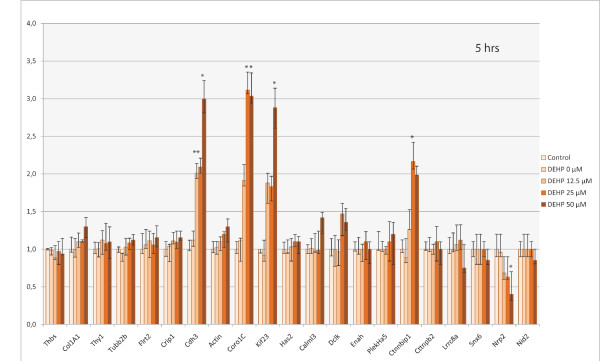
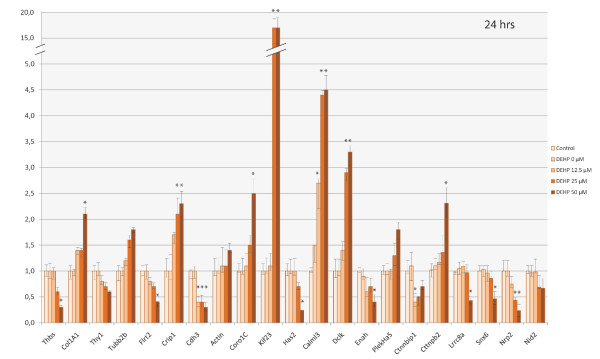
The mRNA level of the 21 genes involved in the regulation of the cytoskeleton that were identified as differentially expressed after 24 hrs was confirmed by qPCR. The expression of these genes was also studied after exposure to DEHP for 5 hrs. Out of the 21 genes, four (coro1C, kif23, cdh3 and cttnbip1) were significantly up-regulated by DEHP treatment after 5 hrs of exposure (Figure 2) and one (nrp2) was significantly down-regulated. A clear dose-response relationship was observed for these 5 genes. After 24 hrs, these changes were confirmed for 3 genes (up-regulation of coro1C and kif23, and down-regulation of nrp2; Figure 3). Nevertheless, the down- and up-regulation was more pronounced after 24 hrs than after 5 hrs of DEHP exposure, for nrp2 and kif23 respectively. For instance, in cells exposed to 50 μM of DEHP, Kif23 was up-regulated 17-fold at 24 hrs versus 3-fold at 5 hrs. After 24 hrs, 5 other genes were significantly up-regulated (col1A1, crip1, calml3, dclk, and cttnbp2) by a factor ranging from 2.0 to 4.5 with a dose-related effect (Figure 3). Eight other genes were significantly down-regulated (thbs1, flrt2, cdh3, has2, enah, ctnnbip1, lrrc8a and snx6), with an expression ratio between 0.2 and 0.5 (corresponding to 5 and 2 fold down-regulation respectively). All these genes were down-regulated in a dose-dependent manner, except for cdh3, enah, ctnnbip1, lrrc8a and snx6. A threshold was observed with the latter genes (12.5 μM for cdh3; 50 μM for enah, ctnnbip1, lrrc8a and snx6). Ctnnbip1 was significantly down-regulated only for the lowest dose of DEHP (12.5 μM).
Figure 2.
Representative qPCR results of differentially-expressed genes involved in cytoskeleton regulation (according to the GO process), identified by Differential Display after 5 hrs of SHE cell exposure to DEHP. These histograms show the ΔΔCt score normalized by gapdh mRNA level. Error bars represent the standard deviation of the ΔΔCt score. Only mRNA levels showing a two-fold increase or decrease at least, were considered indicative (*) of a change in gene expression.
Figure 3.
Representative qPCR results of differentially-expressed genes involved in cytoskeleton regulation (according to the GO process), identified by Differential Display after 24 hrs of SHE cell exposure to DEHP. These histograms show the ΔΔCt score normalized by gapdh mRNA level. Error bars represent the standard deviation of the ΔΔCt score. Only mRNA levels showing a two-fold increase or decrease at least, were considered indicative (*) of a change in gene expression.
Although they had been identified as differentially expressed in DD, five genes (thy1, tubb2b, β-actin, plekha5 and nid2) were not shown to be significantly over- or under-expressed by qPCR. Yet the expression profiles of these genes indicated a dose-related increase for tubb2b, β-actin and pleckha5 but below the qPCR 2.0-fold threshold. As for thy1 and nid2, the dose-related decrease was inferior to 0.5.
Expression of apoptosis-related genes, PPARs and CYP4 genes after DEHP treatment
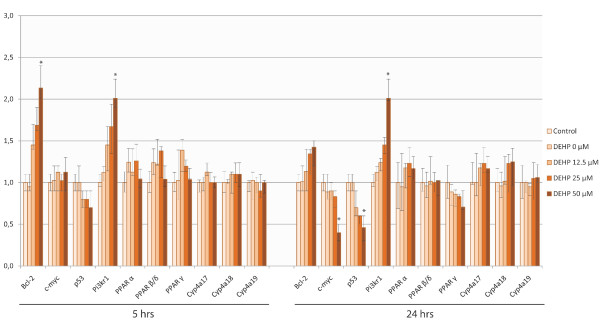
The expression level of bcl-2 and c-myc mRNA was used as controls of DEHP effects. An increased level of bcl-2 after 5 hrs of exposure and a decreased level of c-myc after 24 hrs (Figure 4) were observed according to qPCR, as expected.
Figure 4.
Expression level of bcl-2, c-myc, p53, pi3kr1, PPARs and CYP4 family gene mRNA after treatment of SHE cells with DEHP, using qPCR. These histograms show the ΔΔCt score normalized by the gapdh mRNA level. Error bars represent the standard deviation of the ΔΔCt score. Only mRNA levels showing a two-fold increase or decrease at least, were considered indicative (*) of a change in the gene expression. We observed a significant increase in the bcl-2 mRNA level after 5 hrs of exposure and a significant decrease in c-myc and p53 mRNA levels after 24 hrs of DEHP exposition. Pi3kr1 was found to be over-expressed for both lengths of exposure. None of PPARs or CYP4 genes was significantly over- or under-expressed after treatment.
p53 was down-regulated in a dose- and time-dependent manner; a significant decrease of the mRNA level was found after 24 hrs at 50 μM DEHP.
None of the PPAR genes was identified as being differentially expressed by DD after DEHP exposure. In order to check these results, we measured the mRNA level of PPARα, PPAR β/δ and PPAR γ, by qPCR using hamster specific primers. No change in the expression of these genes was observed by qPCR after 5 or 24 hrs of exposure with DEHP in our study conditions (Figure 4). The same verification was carried out for CYP4 genes. Neither Differential Display nor qPCR allowed us to identify significant expression changes compared to the control.
Discussion
The DDRT-PCR (Differential Display RT-PCR) technique was used in the present study to identify the differential mRNA expression patterns between control and DEHP-treated SHE cells. Indeed, this technique is still a method of choice for non-sequenced or partially-sequenced organisms and is able to identify nonabundant, rare or novel transcripts [36].
Using Differential Display, we found 122 genes whose expression was altered by DEHP treatment (79 up-regulated and 43 down-regulated). The concentrations studied (12.5, 25 and 50 μM) were in the range of concentrations that induced a morphological transformation of SHE cells, i.e. concentrations up to 77 μM for Mikalsen et al. [27] and in the range 25 μM-150 μM for Cruciani et al. [29].
We measured the mRNA level of genes involved in the regulation of the cytoskeleton using qPCR. This focus is justified by the fact that the modifications of cytoskeleton organization are early events in cell neoplastic process [37] and can be recorded in SHE cells after 7 days of exposure to carcinogenic agents in cell transformation assays. Morphological transformation affects a few percentage of the mixed population of SHE cells and all cell types [31]. From the present work, we can assume that the differentially-expressed genes measured in the first 24 hrs of exposure reflect the first targets of DEHP in the entire SHE cell population. The transcriptomic changes which were recorded correspond to the integrated mean of the cell responses significantly different in the exposed populations (p < 0.01), without consideration of cell specificity and sensitivity to DEHP. These significant expression changes in genes involved in cytoskeleton regulation, can be seen as early indicators of disturbances that will lead to cell transformation further in a few percentage of the most susceptible cells of the SHE population. The role of the cytoskeleton has been extensively studied in relation to invasion and metastasis, but little is known of its implication in the first stages of carcinogenesis. The identification of genomic changes associated with the triggering of cell transformation is useful from a mechanistic point of view and may be valuable in screening.
Effects on cytoskeleton-related genes
DEHP was shown to affect several functions related to the cytoskeleton. The genes involved in cytoskeleton regulation and identified by Differential Display are listed in table 2. To summarize, DEHP affects actin polymerisation and stabilization, as well as cell-to-cell and cell-to-matrix adhesion processes. The expression of genes involved in organelle transport, in cytoskeleton remodelling, or adhesion in response to external factors was also modified by DEHP. These results are in line with the recent findings of Posnack et al. [38] who identified disturbances in mechanical adhesion function and protein trafficking in rats cardiomyocytes exposed to DEHP.
Table 2.
List of genes involved in the regulation of the cytoskeleton and affected by DEHP
| 5 hrs | 24 hrs | |
|---|---|---|
| Up-regulated genes |
coro1C* (25 - 50 μM) kif23* (25 - 50 μM) cdh3* (12.5 - 25 - 50 μM) ctnnbip1* (25 μM) |
coro1C* (25 - 50 μM) kif23* (25 - 50 μM) col1a1* (50 μM) crip1* (25 - 50 μM) calml3* (12.5 - 25 - 50 μM) dclk1* (25 - 50 μM) cttnbp2* (50 μM) plekha5 tubb2b β-actin |
| Down-regulated genes | nrp2* (25 - 50 μM) |
nrp2* (25 - 50 μM) thbs* (50 μM) flrt2* (50 μM) cdh3* (12.5 - 25 - 50 μM) has2* (50 μM) enah* (50 μM) lrcc8a* (50 μM) snx6* (50 μM) ctnnbip1* (12.5 μM) nid2 thy1 |
This table summarizes the genes studied using qPCR. (*) indicates significant effects of DEHP (2.0-fold over- or under-expression) at concentration(s) specified in brackets. A trend for up- or down-regulation was found for the other genes (no concentration reported).
Actin polymerization and stabilization
To summarize the basic process, actin polymerization requires the Arp2/3 complex that needs to be stabilized by Enable Homolog (Enah) and is regulated by coronins. Enah is involved in the dynamic reorganization of the actin cytoskeleton, and stimulates nucleation and polymerization [39]. Coronins act on F-actin binding and bundling activities, but are able to inhibit the activity of Arp2/3 complex [40]. Actin polymers also require cortactin, which stabilizes nucleation sites for actin branching and elongation [41,42]. Crip1 facilitates actin filament bundling and stabilizes actin interaction with α-actinin too [43]. Linkage of actin polymers to adherens junctions, mainly composed of the transmembrane proteins cadherins, is insured through binding to α-catenin and β-catenin [44].
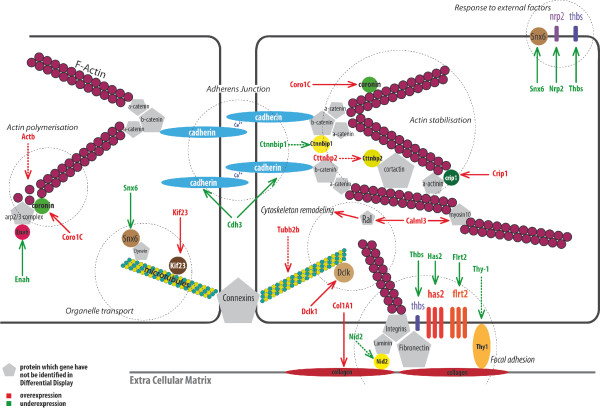
Based on the gene expression data generated, we have tried to synthesize the effects of DEHP on actin organisation and cell adhesion specifically (Figure 5; over-expression in red; under-expression in green). A 5- and 24-hrs exposure to DEHP over-expressed Coronin 1C (Coro1C), resulting in F-actin disassembly [45]. Disorganization was amplified by under-expression of Enah involved in actin nucleation and polymerization, and expression of Cttnbp2 that counteracts cortactin which is known to stabilize the actin network. On the other hand, the binding of actin filaments to cadherins through catenin links appears to be reinforced owing to under-expression of Ctnnbip1 (a β-catenin blocker) and over-expression of Crip1, which intensifies fixation to actinin. Globally, the effects of DEHP on actin cytoskeleton disturb actin polymerization while intensifying binding on actinin and catenins. Posnack et al. [38] explored DEHP effects on rats cardiomyocytes in a range of concentrations two and three orders of magnitude higher than here. They found an over-expression of actinin, α-catenin and N-cadherin in a concentration-dependent manner.
Figure 5.
Representative scheme of the genes affected by DEHP and involved in cytoskeleton regulation. Genes in red have been found to be over-expressed using Differential Display (DD) and genes in green under-expressed. Dotted lines represent genes identified by DD but whose expression was not significant in qPCR.
Cell-cell and cell-matrix adhesion
Cell-cell adhesion and cell-matrix adhesion were also affected by DEHP treatment. The decrease in the P-Cadherin (Cdh3) mRNA level after 24 hrs of exposure indicates that DEHP weakened cell-cell contact, after a transient increase at 5 hrs of exposure for all doses tested. Weakening of cell-matrix adhesion may result from a decrease in the Hyaluronan synthase 2 (has2) mRNA level [46] and in Thrombospondin (Thbs1), an adhesive protein that interacts with fibronectin, laminin, integrins and collagen [47]. Loss of cell adhesion may also be explained by over-expression of Coro1C because this gene negatively regulates cell-matrix adhesion through focal adhesion kinase (FAK)-mediated signalling [45]. Also, under-expression of Enah, which is known to be involved in the control of cellular adhesion by the recruitment of proteins containing SH3- domain [48], contributes to the loss of cell-cell adhesion.
In addition, DEHP may lessen extracellular matrix adhesion by reducing the expression level of a number of transmembrane proteins involved in cell-matrix connections: Fibronectin leucine rich 2 (Flrt2) and Leucine rich repeat 8A (Lrrc8a) [49], Nidogen 2, which connects laminin-1 to the matrix [50], and Thy-1, which mediates fibroblastic adhesion [51] and is Thbs1 expression-dependent [52]. On the other hand, DEHP effects reinforce the extra-cellular matrix through an over-expression of col1A1 increasing collagen. This effect may be seen as a compensatory reaction to the weakening of cell-to-matrix link proteins by DEHP. Sobarzo et al. [53] demonstrated an up-regulation of N-cadherin and α-catenin in rat testis after 2 and 7 days of DEHP treatment, suggesting also a deregulation of cell adhesion molecules in seminiferous tubules.
DEHP decreases the response to external factors, such as the Vascular Endothelial Growth Factor (VEGF) or the Epidermal Growth Factor (EGF) through under-expression of neuropilin 2 (nrp2) and sorting nexin 6 (snx6) respectively. Nrp2 is a membrane receptor capable of binding VEGF and semaphorins, therefore its under-expression may inhibit cell adhesion and migration via the loss of integrins [54]. Snx6 is able to interact with EGF receptor and Transforming Growth Factor (TGF)-β receptor [55]. Under-expression of snx6 and thbs1 may lead to decreased interaction with Latent TGF Binding Protein (LTBP) in the upstream of the TGF-β pathway contributing to the repression of the TGF-β signaling pathway [56,57]. Under-expression of TGF-β is known to decrease apoptosis in rodent hepatocytes treated with peroxisome proliferators (PPs) [24].
Organelle transport and cytoskeleton remodelling
DEHP also interferes with functions of microtubules (composed of α- and β-tubulin). Kif23, which encodes a kinesin protein, was highly over-expressed after 5 hrs and 24 hrs of DEHP exposure. Kif23 has been shown to transport membranous organelles and protein complexes from cell nucleus to cell periphery in a microtubule- and ATP-dependent manner. Doublecortin-like kinase (Dclk) is a microtubule-associated protein encoding a Ca2+/calmodulin-dependent kinase. Its activities on binding and microtubule polymerization facilitate cell motility by remodelling the microtubule cytoskeleton [58]. Over-expression of dclk at 24 hrs of DEHP treatment is in line with an increased trend in β-tubulin (tubb2b).
Calmoduline-like 3 (calml3) was over-expressed after 24 hrs of DEHP exposure. Calmodulin (CaM) is a calcium-binding protein that translates the Ca2+ signal into a wide variety of cellular processes, including the regulation of cytoskeleton remodelling acting with Caldesmon [59] or with Wnt pathway [60]. Calml3 is a CaM family member protein which increases cell motility by stabilizing and increasing myosin-10 for cell migration [61].
Other genes involved in signal transduction pathways and cytoskeleton regulation
We measured an over-expression level of phosphatidylinositol 3-kinase r1 (pi3kr1) using Differential Display and qPCR. Pi3k is a key signalling molecule in the PIP3 signalling transduction pathway and in actin reorganization and cell adhesion [62] and is able to regulate the synthesis of collagen I [63]. An activation of PI3K is also associated with a phosphorylation-dependent activation of Akt which contributes to tumorigenesis and metastasis [39]. The over-expression of pi3kr1 can be related to the under-expression of ctnnbip1 which interacts with β-catenin. In addition to the function of β-catenin in the actin cytoskeleton, its role in the regulation of Akt pathway activation [64] or in Wnt pathway regulation [65] is advanced. This protein forms part of a complex that captures growth and proliferation signals from the cell surface and is then activated to stimulate the expression of genes involved in cell proliferation. It would be worth studying β-catenin-dependent transcription in relation to carcinogenicity.
DEHP effects in the SHE model compared to rats and mice
While the expression of cyp1b1 and cyp2e1 was up-regulated and cyp2f2 under-expressed, no change in expression level of CYP4 genes was found using DD and qPCR after DEHP exposure in our experimental conditions. CYP4 genes are said to be involved in peroxisome proliferation [66]. Eveillard et al. [67] who studied the involvement of DEHP in lipidogenesis in rats, found a slight increase in the PPARα level after 21 days of oral exposure to DEHP (200 mg/kg.day). They registered a significant increase in CYP4 levels after 14 days [68] and after 21 days [67] of exposure. On the other hand, we found no increased mRNA level of CYP4 and PPAR genes in DEHP-treated SHE cells. This underlines that the genes expression changes noted in the present study are independent of PPARs induction. Eveillard et al. [67] found that induced expression of cyp2b10 by DEHP was also independent of PPARα induction but CAR dependent. No change in CAR expression was registered in SHE cells, which may explain why no change in cyp2b10 was noted. Our results are consistent with the study of Ren et al. [20] who identified DEHP regulated genes independent of PPARα and CAR in rats and mice.
In our study, lipogenesis and xenobiotic metabolism pathways were impacted by DEHP, but not in a major prior way. This may be explained by the lower sensitivity of the hamster model compared with rats and mice to peroxisome proliferators [69]. Indeed, the Syrian hamster model presents an intermediate response between rats or mice and humans who are known to be non-responsive to PP induction [24]. The hamster model, like humans, is less responsive to PP induction than rats and mice, which is an advantage for mechanistic studies of PP effects and for screening human chemical carcinogens.
On the other hand, three genes (Actin β, Lipoprotein lipase, and Acetyl-Coenzyme A acyltransferase 1a) and 5 gene isoforms (Glutathione S-transferase-pi, -mu, -theta, CCAAT/enhancer binding protein and Nuclear receptor subfamily 1) were commonly found in our study and those carried out by Eveillard et al. [67,68], suggesting a pattern of response specific to DEHP.
Takashima et al. [70] also found similar responses in DEHP-treated mice. Up-regulation of rab1b, a RAS oncogene family member involved in cellular signal transduction or survival, was found in the latter study and in the present one. β-Tubulin was clearly over-expressed in mice, a trend which was noted in our study. Some gene isoforms of cadherin, nidogen, cyp1 family genes or LIM domain were also impacted in the liver of mice exposed to DEHP [70].
DEHP effects on transcription factors
Other genes identified by Differential Display and involved in transcription and signal transduction pathways or apoptosis were also targeted by DEHP. A significant under-expression of p53 was found after 24 hrs of DEHP exposure using Differential Display and qPCR. This under-expression is in line with the anti- apoptotic effects of DEHP.
We confirmed the over-expression of bcl-2 after 5 hrs and the under-expression of c-myc after 24 hrs, events reported in a previous study on DEHP treated SHE cells in conditions similar to the present ones [26]. Map Kinases such as Mapk3, Mapk4 and Mapk15 were targeted by DEHP. Further investigations of Map Kinase pathways could be relevant due to their involvement in activities of transcription factors.
The G protein-coupled estrogen receptor (gper) was found to be over-expressed in Differential Display. Gper can be activated by estrogen-like compounds and its effect on cytoskeleton architecture has been reported [71]. Because of its implication in the regulation of MAPK [72] or TGF-β pathways [73], it would be worthwhile to investigate gper further.
Performances of DD
The confirmation of differentially-expressed genes by qPCR showed that the expression levels of more than 75% of genes identified by DD were confirmed by qPCR. A comparative table of the sensitivity of DD versus qPCR is given in additional file 1. qPCR is more likely to quantify subtle changes in the expression level of mRNAs at different concentrations while DD seems to be more sensitive but is less discriminating. To summarize, 35% of the genes identified as differentially expressed in DD gave the same response at the same DEHP concentrations with qPCR while 40% were detected by DD at a lower DEHP concentration than with qPCR.
Conclusion
Transcriptional responses of SHE cells to DEHP were studied in conditions inducing the cell neoplastic transformation, in order to identify gene expression changes in relation with effects of this non-genotoxic carcinogen. Functions impacted by DEHP were found to be PPAR-independent. Effects on cytoskeleton related genes indicated disturbances on actin polymerization and stabilization, cell-cell and cell-matrix adhesion and protein trafficking.
This is the first study that elucidates the genomic changes of DEHP on the organization of the cytoskeleton. Whether the expression changes of cytoskeleton-related genes identified here such as coro1C, nrp2, kif23, are specific to DEHP or to cell transforming agents more generally would require further studies. To answer, the gene sets identified as significantly over- or under-expressed in this study must be explored on other non-genotoxic carcinogens to identify biomarkers predictive of early events in the multistep carcinogenic process. Early disturbances in the expression of cytoskeleton-related genes should be considered good candidates.
Methods
Chemicals
DEHP (C.A.S. No. 117-81-7, purity 99%), purchased from Aldrich Chemicals (Gillingham, England) was dissolved in the DMSO solvent (C.A.S. No. 67.68.50). The latter was obtained from Sigma Aldrich (St Quentin Fallavier, France) and was used at a final concentration of 0.1%.
Nucleic acid stain Gelred purchased from Interchim (Montluçon, France) was used at a final concentration of 1:10000 (v/v).
All chemicals used for this study were electrophoresis grade or molecular biology grade. Their origin is specified in the following sections.
SHE cell culture and treatment
SHE cells were isolated from Syrian hamster embryos at day 13 of gestation using the procedure described by Pienta et al. [74] and in accordance with the modifications suggested by Elias et al. [75]. Differentiated tissues, such as eyes, heart and viscera were removed and the remaining tissues were dissociated by dispase (1.2 U/ml). Stock cells were preserved in liquid nitrogen.
After thawing, passage-2 primary cells were pre-cultured until they reached 80% confluency. The culture medium was Dulbecco's modified Eagle medium (DMEM) (Gibco, Invitrogen, Cergy Pontoise, France) supplemented with 10% foetal calf serum (FCS) (Hyclone, Brebières, France; lot N°#ASB28835), 1.5 g/L NaHCO3 at 37°C in a 10% CO2 humidified atmosphere and pH 7.0. No phenol red was added to the medium.
Cells used for the DEHP studies were sampled from a monolayer during the growing phase, 48 hrs after seeding. Cells were trypsinized and treated during replating with DEHP at concentrations of 0 μM (vehicle control), 12.5 μM, 25 μM and 50 μM in DMEM culture medium supplemented with 10% FCS. Cells were then incubated for 5 hrs and 24 hrs at 37°C in a 10% CO2 humidified atmosphere.
RNA isolation
Total RNA extractions were performed directly in the dish, using Nucleospin RNA II Extract Kit (Macherey Nagel, Hoerdt, France), according to the manufacturer's instructions. A DNAse I treatment was performed directly through the column used to collect RNAs and before the elution phase of DNA-free RNA.
RNA was quantified by spectrophotometry (Nanodrop, Labtech) measuring the A260/A280 ratio and its quality was ensured by electrophoresis using a 1% RNase-free agarose gel. Aliquots were stored at -80°C before use for Differential Display and Real-time PCR.
Anchored Reverse transcription (RT) and Differential Display
The Differential Display was performed as described by Liang et al. [35], with minor modifications concerning DD fragment revelation with GelRed.
For Differential Display, three separate RT reactions were performed with a different one-base anchored oligo-dT primer (H-dTA, H-dTC and H-dTG) to produce three different subsets of cDNA pools. The sequences of the anchored and the arbitrary primers are given as additional file 2. The RT reactions were carried out using 2 μL of each primer (50 μM) and 4 μg of total RNA. 8 μL of RevertAid M-MuLV Reverse Transcriptase 5x reaction buffer (Fermentas, Saint-Rémy-lès-Chevreuse, France), 1.5 μL of 10 mM dNTPs (Fermentas, Saint-Rémy-lès-Chevreuse, France) and up to 35 μL Nuclease-free water were added to each tube, mixed, then heated at 70°C for 3 min. Tubes were centrifuged and incubated on ice for 5 min, then 2 μL (40 U) of RNaseOUT Recombinant RNase Inhibitor (Invitrogen, Cergy Pontoise, France), 1 μL (200 U) of RevertAid M-MuLV RT (Fermentas, Saint-Rémy-lès-Chevreuse, France) and 2 μL of Nuclease-free water were added to each tube. Each tube was mixed by gentle pipetting then incubated in a thermocycler at 42°C for 1 h, followed by 95°C for 10 min. The tubes were then centrifuged and stored at -80°C until use.
Amplification was then performed using combinations of the three original anchored primers from the reverse transcription step and eighty arbitrary 13-mers (H-AP), giving a total of 240 amplification combinations. All reactions contained 2 μL of a 10x PCR buffer containing 25 mM of MgCl2, 1.6 μL of 1 mM dNTP mix, 1 U of Taq Polymerase (Euromedex, Souffelweyersheim, France) and the primer combination at a final concentration of 1 μM. Tubes were incubated for 5 min at 95°C. The next 40 cycles were 95°C for 30 s (denaturation), 40°C for 2 min (annealing), 72°C for 1 min (amplification). A final extension of 72°C for 10 min completed the cycle.
After thermocycling, PCR-amplified fragments were resolved in a 6% native polyacrylamide gel in 1 × TBE buffer (89 mM Tris base/89 mM boric acid/2 mM EDTA, pH 8.0), using 10 μL of PCR product mixed with 2 μl of loading buffer (0.05% xylene cyanol, 40% sucrose, 20 mM EDTA, pH 8.0). Gels were run at 100 V for 20 hrs, then the fragments were stained with GelRed 1X in water, for 30 min in the dark. Bands on the gel were revealed on a UV-transluminator. PCR products that showed differential expression between control and treated samples were identified with QuantityOne® 1-D analysis software (BioRad, Marne-la-Coquette, France).
Bands which were up- or down- regulated more than 2-fold, were selected and characterized in the next step of analysis. Differentially-expressed bands were excised, reamplified and their sizes were checked before cloning. To summarize, fragments of interest were recovered using a clean razor blade and extracted from the gel matrix by boiling in 200 μL of buffer (10 mM Tris/1 mM EDTA/1% SDS (w/v), pH 8.0) for 15 min. After overnight precipitation at -80°C, the eluted DNA was reamplified using the same primers and PCR conditions as the ones used in the DD-PCR step. Reamplified DNA was run in a 1.5% agarose gel containing 1X GelRed and recovered using NucleoSpin ® Extract II kit (Macherey Nagel, Hoerdt, France) before cloning.
Cloning was carried out using a TA Cloning Kit (pGEM-T, Promega, Charbonnières, France), according to the manufacturer's instructions. Plasmid DNA was extracted from the cultures using Nucleospin ® Plasmid QuickPure (Macherey Nagel, Hoerdt, France), according to the manufacturer's instructions and sequenced bidirectionally by the DNA sequencing service of MWG Operon (Ebersberg, Germany), using T7 and SP6 primers.
Identification of differentially-expressed genes
Sequences were compared with the National Centre of Biotechnology Information Gene Bank database (http://www.ncbi.nlm.nih.gov) using the tBLASTx algorithm and RefSeq mouse or Refseq human as a reference.
Confirmation of differentially-expressed sequences by Quantitative Real Time PCR (qPCR)
First-strand cDNA was synthesized from 2 μg of total RNA using VILO-Superscript™ III reverse transcriptase (Invitrogen, Cergy Pontoise, France) and random-hexamer primers. To summarize, 2 μg of total RNA was combined with 4 μL of 5X VILO™ reaction mix (containing RT buffer, MgCl2, dNTPs and random primers) and 2 μL of 10X enzyme mix (containing Superscript® III and RNase inhinitor). The final volume was adjusted to 20 μL and the reaction mix was incubated at 42°C for 60 min. Then, cDNAs were diluted 20-fold, according to the manufacturer's instructions, before qPCR amplifications. The oligonucleotides used as primers in the quantitative real time PCR assay are described in table 3. If possible, at least one primer in each pair spanned an exon-intron boundary. PCR was carried out using Fast SYBR®Green Master Mix (Applied Biosystem, Courtaboeuf, France). Amplifications were performed on a StepOnePlus Real-Time PCR system (Applied Biosystem, Courtaboeuf, France). Each qPCR reaction contained 10 μL of 2X Fast SYBR®Green Master Mix, 5 μL of primers, 2 μL of diluted cDNA and 3 μL of Nuclease-free water. Amplification parameters were set as follows: initial denaturation (95°C, 3 min), and then amplification (95°C, 3 s and 60°C 30s) for 40 cycles. Glyceraldehyde 3-phosphate dehydrogenase (gapdh) mRNA level was used as a housekeeping gene to normalize qPCR data. This gene was chosen because DEHP exposure did not affect its expression unlike β-actin which was also tested (data not shown). qPCR results were analyzed using the software provided with the thermocycler and DataAssist, using the ΔΔCt method [76]. Each validated primer pair used yielded a single peak of dissociation on the melting curve. The efficiency calculated by standard curve with five log-10 dilution points was between 0.95 and 1.05. A 2.0-fold threshold and a p-value of 0.05 were used to determine the significance of differential expression levels according to the standard parameters of DataAssist.
Table 3.
List of the primers used for real-time qPCR
| Genes | Accession N° | Primers sequence (5'-3') | |
|---|---|---|---|
| Housekeeping gene | |||
| gapdh | Glyceraldehyde 3-phosphate dehydrogenase | DQ403055 | F: CAATGACCCCTTCATTGACC R: GACAAGCTTCCCGTTCTCAG |
| Regulation of cytoskeleton | |||
| tubb2b | β-Tubulin | NM_023716 | F: GCAACATGAATGACCTGGTG R: ACCAGAGACCCAGCACAAAC |
| thy1 | Thy1 Antigen | NM_009382.3 | F: AAGGCCTCTGCCTGTAGTGA R: GAAGAGGCAGGTTGCAAGAC |
| actin | β-Actin | NM_007393 | F: CACCACCACAGCCGAGAG R: CCAGGGAGGAAGAGGATGC |
| col1a1 | Collagen α1 | NM_000088 | F: GGGTCATTTCCACATGCTTT R: TCCGGGTTTCAGAGTACCAC |
| thbs1 | Thrombospondin 1 | NM_011580 | F: CCAAAGCCTGCAAGAAAGAC R: CCTGCTTGTTGCAAACTTGA |
| plekha5 | Plekstrin homology A5 | NM_019012 | F: GTGCATCTGCCTGAAGACAA R: TGGGAACCTTTAACGACTGG |
| kif23 | Kinesin 23 | NM_024245.4 | F: CCTGAGCTTTCCTGACCAAG R: AGTTCCTTCTGGGTGGTGTG |
| has2 | Hyaluronan synthase 2 | NM_008216 | F: CGGAGGACGAGTCTATGAGC R: TTTTCCGGTGTTCCAAAAAG |
| flrt2 | Fibronectin leucine rich 2 | NM_201518 | F: ACCGCACTGTGGAAGATACC R: GCAAGACAACGAGCACAAAA |
| enah | Enabled homolog | NM_008680 | F: GCCTATGCTTCAGCACTTCC R: GGGCGATTGTCTTCTGACAT |
| dclk1 | Doublecortin like 1 | NM_019978 | F: AGCCTCCACCAGCTCAGTTA R: CCATACACATCGCTCCATTG |
| ctnnbip1 | Catenin β interacting protein 1 | NM_020248 | F: TTGGCTGCAGAAAGAAACCT R: CCAGCCAATCACAACCTTTT |
| crip1 | Cystein rich protein | NM_007763 | F: AGTCCAGAGCCTGCAACCTA R: GGAGTAGCAGGGATGATTGC |
| coro1c | Coronin | NM_014325 | F: GCAGAAGAGTGGTTCGAAGG R: TGATCAGGTCGCACTTCTTG |
| cdh3 | Cadherin 3 | NM_007665 | F: CACACGACCTCATGTTCACC R: CTGTACCTCATGGCCCACTT |
| calml3 | Calmodulin like 3 | NM_027416.3 | F: ATCGACAAGGATGGAAACG R: ATCTACCTCCTCGTCGCTCA |
| cttnbp2 | Cortactin binding protein 2 | NM_030249 | F: GACAAAGAAGGCTGGACTGC R: CTCACCCACGGAAATCCTTA |
| lrrc8a | Leucine rich repeat 8A | NM_177725 | F: AGAGCCCACTTACCCCAACT R: GTGTGCAGAAGCACGAGGTA |
| snx6 | Sorting nexin 6 | NM_021249 | F: CCAAGACCTGATTTTGATGCTTC R: CATGCATCGCAACTGTCTTC |
| nrp2 | Neuropilin 2 | NM_0010774 | F: ATAAGCACTGATGTCCCACTG R: GAGTTGCTCCAATCTCCTTCA |
| nid2 | Nidogen 2 | NM_008695 | F: CTCATTCAGTTGTGCCTGC R: ATAGCTGCCTCATGACATCG |
| Regulation of apoptosis | |||
| pi3kr1 | phosphatidylinositol 3-kinase (p85a) | NM_001077495 | F: CGAGCCCGACCGGAGGTGAA R: CGCACACTGCCGTCCGAGTT |
| bcl-2 | B-cell lymphoma 2 | AJ582074 | F: CGCAGAGATGTCCAGTCAGC R: CGAACTCAAAGAAGACCACAA |
| c-myc | cellular myelocytomatosis oncogene | AJ582076 | F: GACCCTGATTCGGACCTCTT R: CGACTCCACAGCCTTCTCTC |
| p53 | tumor supressor p53 | U07182 | F: ATGACGGAAGTTGTAAGAC R: TCGGATAAGATGCTGAGG |
| PPARs genes | |||
| ppar α | peroxisome proliferator activated receptor alpha | AY170844 | F: GTTTCTTTCGGCGAACTATT R: ACACGTGAGAATCTCTGCTT |
| ppar β/δ | peroxisome proliferator activated receptor beta/delta | AF486582 | F: TGCAAGATCCAGAAGAAGAA R: GTAGATGTGCTTGGAGAAGG |
| ppar γ | peroxisome proliferator activated receptor gamma | AB525757 | F: GGACCTCTCTATGATGGATG R: GGATGCAGGTTCTACTTTGA |
| CYP4 family genes | |||
| cyp4a17 | Cytochrome P450 4A17 | AJ555628 | F: ACCAGATGCCCTACACTACC R: GTGCGTAAATGGAGAGTACA |
| cyp4a18 | Cytochrome P450 4A18 | AJ555629 | F: ACCAGATGCCCTACACTACC R: GGCCATAAATGGAGATTGCA |
| cyp4a19 | Cytochrome P450 4A19 | AJ555630 | F: ACCAGATGCCCTACACTACC R: GTGCATAAATGGAGAGTGTG |
Authors' contributions
YL performed Differential Display, carried out qPCR analysis and was involved in the design of experiment. PP and FA were involved in the design of the experiment study and technical assistance. PV involved in the overall design and coordination of the study. All authors participated in the writing of the manuscript and approved it.
Supplementary Material
Comparative table of the sensitivity of DD versus qPCR. Comparisons between the ratio of bands intensity compared to the control on the gel using an image analysis software (QuantityOne® 1-D analysis software BioRad, Marne-la-Coquette, France) for Differential Display and the ΔΔCt score normalized by gapdh mRNA level after analysis with StepOne and DataAssist (Roche Applied Biosystem, Courtaboeuf, France) for qPCR.
Primers for Differential Display. List of the sequences of the anchored and the arbitrary primers used for the Differential Display experiments.
Contributor Information
Yann Landkocz, Email: yann.landkocz@univ-metz.fr.
Pascal Poupin, Email: poupin@univ-metz.fr.
Franck Atienzar, Email: franck.atienzar@ucb.com.
Paule Vasseur, Email: vasseur@univ-metz.fr.
Acknowledgements
This work was supported by the French Ligue Nationale contre le Cancer, the Region Lorraine, the Contrat Projet Etat-Region, the Ministry of Research and Technology, and the FEDER. The authors would like to acknowledge these sponsors for their support. Ms. C. Rast and M.A. Maire are gratefully acknowledged for assistance with the cell cultures. We warmly thank Tracy Carmona for her help in correcting the English of the paper.
References
- Heudorf U, Mersch-Sundermann V, Angerer J. Phthalates: Toxicology and exposure. Int J Hyg Environ Health. 2007;210:623–634. doi: 10.1016/j.ijheh.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Becker K, Göen T, Seiwert M, Conrad A, Pick-Fuss H, Müller J, Wittassek M, Schulz C, Kolossa-Gehring M. GerES IV: Phthalates metabolites and Bisphenol A in urine of German Children. Int J Hyg Environ Health. 2009;212(6):685–692. doi: 10.1016/j.ijheh.2009.08.002. [DOI] [PubMed] [Google Scholar]
- IARC. Nakajima T, Hopf NB, Schulte PA. Di(2-ethylhexyl) phthalate (DEHP) IARC Monographs. 2000;77 [Google Scholar]
- Sharpe RM. Hormones and testis development and the possible adverse effets on environmental chemicals. Toxicol Lett. 2001;120:221–232. doi: 10.1016/S0378-4274(01)00298-3. [DOI] [PubMed] [Google Scholar]
- Akingbemi BT, Ge R, Klinefelter GR, Zirkin BR, Hardy MP. Phthalate induced Leydig cell hyperplasia is associated with multiple endocrine disturbances. Proc Natl Acad Sci USA. 2004;101:775–780. doi: 10.1073/pnas.0305977101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latini G, Verroti A, De Felice C. Di-ethylhexyl phthalate and endocrine disruption: a review. Curr Drug Targ. 2004;4:37–40. doi: 10.2174/1568008043340017. [DOI] [PubMed] [Google Scholar]
- Pan G, Hanaoka T, Yoshimura M, Zhang S, Wang P, Tsukino H, Inoue K, Nakazawa H, Tsugane S, Takahashi K. Decreased serum free testosterone in workers exposed to high levels of di-n-butyl phthalate (DBP) and di-2-ethylhexyl phthalate (DEHP): a cross-sectional study in China. Environ Health Perspect. 2006;114:1643–1648. doi: 10.1289/ehp.9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker JD, Calafat AM, Hauser R. Di-(ethylhexyl)phthalate metabolites may alter thyroid hormone levels in men. Environ Health Perspect. 2007;115(7):1029–1034. doi: 10.1289/ehp.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick RL, Morrissey RE, Tomaszewski KE. Studies by the National Toxicology Program on di(2-ethylhexyl)phthalate. Toxicol Ind Health. 1987;3:99–118. doi: 10.1177/074823378700300208. [DOI] [PubMed] [Google Scholar]
- Lapinskas PJ, Brown S, Leesnitzer LM, Blanchard S, Swanson C, Cattley RC, Corton JC. Role of PPARα in mediating the effects of phthalates and metabolites in the liver. Toxicology. 2005;207:149–163. doi: 10.1016/j.tox.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Feige JN, Gelman L, Rossi D, Zoete V, Métivier R, Tudor C, Anghel SI, Grosdidier A, Lathion C, Engelborghs Y, Michielin O, Wahli W, Desvergne B. The endocrine disruptor monoethyl-hexyl-phthalate is a selective peroxisome proliferator-activated receptor γ modulator that promotes adipogenesis. J Biol Chem. 2007;282(26):19152–19166. doi: 10.1074/jbc.M702724200. [DOI] [PubMed] [Google Scholar]
- Erkekoğlu P, Rachidi W, De Rosa V, Giray B, Favier A, Hincal F. Protective effect of selenium supplementation on the genotoxicity of di(2-ethylhexyl)phthalate and mono(2-ethylhexyl)phthalate treatment in LNCaP cells. Free Radic Biol Med. 2010;49(4):559–66. doi: 10.1016/j.freeradbiomed.2010.04.038. [DOI] [PubMed] [Google Scholar]
- Erkekoğlu P, Rachidi W, Yüzügüllü OG, Giray B, Oztürk M, Favier A, Hincal F. Induction of ROS, p53, p21 in DEHP- and MEHP-exposed LNCaP cells-protection by selenium compounds. Food Chem Toxicol. 2011;49(7):1565–1571. doi: 10.1016/j.fct.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Erkekoğlu P, Rachidi W, Yüzügüllü OG, Giray B, Favier A, Oztürk M, Hincal F. Evaluation of cytotoxicity and oxidative DNA damaging effects of di(2-ethylhexyl)-phthalate (DEHP) and mono(2-ethylhexyl)-phthalate (MEHP) on MA-10 Leydig cells and protection by selenium. Toxicol Appl Pharmacol. 2010;248(1):52–62. doi: 10.1016/j.taap.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Doull J, Cattley R, Elcombe C, Lake B, Swenberg J, Wilkinson C, Williams G, van Gemert M. A cancer risk assessment of di(2-ethylhexyl)phthalate: application of the New U.S. EPA Risk Assessment Guidelines. Regul Toxicol Pharmacol. 1999;29:327–357. doi: 10.1006/rtph.1999.1296. [DOI] [PubMed] [Google Scholar]
- Voss C, Zerban H, Bannasch P, Berger MR. Lifelong exposure to di-(2-ethylhexyl)-phthalate induces tumors in liver ans testes of Sprague-Dawley rats. Toxicology. 2005;206(3):359–371. doi: 10.1016/j.tox.2004.07.016. [DOI] [PubMed] [Google Scholar]
- David RM, Moore MR, Finney DC, Guest D. Chronic toxicity of di(2-ethylhexyl)phthalate in rats. Toxicol Sci. 2000;55:433–443. doi: 10.1093/toxsci/55.2.433. [DOI] [PubMed] [Google Scholar]
- Melnick RL. Is peroxisome proliferation an obligatory precursor step in the carcinogenicity of di(2-ethylhexyl)phthalate (DEHP)? Environ Health Perspect. 2001;109:437–442. doi: 10.1289/ehp.01109437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Yamanoshita O, Asaeda N, Tagawa Y, Lee CH, Aoyama T, Ichihara G, Furuhashi K, Kamijima M, Gonzalez FJ, Nakajima T. Di(2-ethylhexyl)phthalate induces hepatic tumorigenesis through a peroxisome proliferator-activated receptor α-independent pathway. J Occup Health. 2007;49:172–182. doi: 10.1539/joh.49.172. [DOI] [PubMed] [Google Scholar]
- Ren H, Aleksunes LM, Wood C, Vallanat B, George MH, Klaassen CD, Corton JC. Characterization of peroxisome proliferator-activated receptor α-independent effect of PPARα activators in the rodent liver: di-(2-ethylhexyl) phthalate also activates the constitutive-activated receptor. Tox Sci. 2010;113(1):45–59. doi: 10.1093/toxsci/kfp251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RA. Evidence of cross talk between PPARα and p38 MAP Kinase. Tox Sci. 2002;68:270–274. doi: 10.1093/toxsci/68.2.270. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakajima T. PPARα- and DEHP-induced cancers. PPAR Res. 2008. pp. 1–12. [DOI] [PMC free article] [PubMed]
- Cruciani V, Rast C, Durant MJ, Alexandre S, Nguyen-Ba G, Vasseur P. Morphological transformation and inhibition of intercellular communication of Syrian hamster embryo cells by hepatic peroxisome proliferators. Mutat Res Fundam Mol Mech Mut. 1997;379(Suppl 1):S195. [Google Scholar]
- James NH, Roberts RA. Species differences in response to peroxisome proliferators correlate in vitro with induction of DNA synthesis rather than suppression of apoptosis. Carcinogenesis. 1996;17(8):1623–1632. doi: 10.1093/carcin/17.8.1623. [DOI] [PubMed] [Google Scholar]
- Roberts RA, James NH, Hasmall SC, Holden PR, Lambe K, Macdonald N, West D, Woodyatt NJ, Whitcome D. Apoptosis and proliferation in non-genotoxic carcinogenesis: species differences and role of PPARα. Toxicol Lett. 2000;112-113:49–57. doi: 10.1016/s0378-4274(99)00243-x. [DOI] [PubMed] [Google Scholar]
- Maire MA, Rast C, Vasseur P. Di-(ethylhexyl)phthalate (DEHP) increases Bcl-2/Bax ratio and modifies c-myc expression in Syrian hamster embryo (SHE) cells. Toxicol Lett. 2005;158(3):237–245. doi: 10.1016/j.toxlet.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Mikalsen SO, Holen I, Sanner T. Morphological transformation and catalase activity of Syrian hamster embryo cells treated with hepatic peroxisome proliferators, TPA and nickel sulphate. Cell Biol Toxicol. 1990;6(1):1–14. doi: 10.1007/BF00135022. [DOI] [PubMed] [Google Scholar]
- LeBoeuf RA, Kerckaert GA, Aardema MJ, Gibson DP, Brauninger R, Isfort RJ. The pH 6.7 Syrian hamster embryo cell transformation assay assessing the carcinogenic potential of chemicals. Mutat Res. 1996;356(1):85–127. doi: 10.1016/0027-5107(95)00199-9. [DOI] [PubMed] [Google Scholar]
- Cruciani V, Rast C, Alexandre S, Nguyen-Ba G, Vasseur P. Peroxisome proliferator-induced transformation of Syrian hamster embryo cells: influence of experimental procedures. Toxicol In Vitro. 1999;13:445–457. doi: 10.1016/S0887-2333(99)00016-8. [DOI] [PubMed] [Google Scholar]
- Alexandre S, Rast C, Maire MA, Orfila L, Vasseur P. ZnCl2 induces Syrian hamster embryo (SHE) cell transformation. Toxicol Lett. 2003;143:77–87. doi: 10.1016/s0378-4274(02)00488-5. [DOI] [PubMed] [Google Scholar]
- Isfort RJ, Kerckaert G, Anderson NL, LeBoeuf RA. Two-dimensional gel electrophoresis analysis of Syrian hamster embryo cells: morphological transformation is not cell type specific. Electrophoresis. 1992;13(11):855–861. doi: 10.1002/elps.11501301187. [DOI] [PubMed] [Google Scholar]
- Berwald Y, Sachs L. In vitro cell transformation with chemicals carcinogens. Nature. 1963;200:1182–1184. doi: 10.1038/2001182a0. [DOI] [PubMed] [Google Scholar]
- Combes R, Balls M, Curren R, Fischbach M, Fusenig N, Kirkland D, Lasne C, Landolph J, LeBoeuf R, Marquardt H, McCormick J, Müller L, Rivedal E, Sabbioni E, Tanaka N, Vasseur P, Yamasaki H. In: The report and recommendations of ECVAM Workshop XX. Balls M, editor. Vol. 27. 1999. Cell transformation assays as predictors of human carcinogenicity; pp. 745–767. ATLA. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liang P, Meade JD, Pardee AB. A protocol for differential display of mRNA expression using either fluorescent or radioactive labelling. Nat Prot. 2007;2(3):457–470. doi: 10.1038/nprot.2007.46. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Recent advances in differential display. Curr Opin Immunol. 1995;7:274–280. doi: 10.1016/0952-7915(95)80015-8. [DOI] [PubMed] [Google Scholar]
- Pollack R, Osborn M, Weber K. Patterns of organization of actin and myosin in normal and transformed cultured cells. Proc Nat Acad Sci USA. 1975;72(3):994–998. doi: 10.1073/pnas.72.3.994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posnack NG, Lee NH, Brown R, Sarvazyan N. Gene expression profiling of DEHP-treated cardiomyocytes reveals potential causes of phthalate arrhythmogenicity. Toxicology. 2011;279:54–64. doi: 10.1016/j.tox.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts A, Van Troys M, Ampe C. The actin cytoskeleton in normal and pathological cell motility. Int J Biochem Cell Biol. 2004;26:1890–1909. doi: 10.1016/j.biocel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Rosentreter A, Hofmann A, Xavier CP, Stumpf M, Noegel AA, Clemen CS. Coronin 3 involvement in F-actin-dependent processes at the cell cortex. Exp Cell Res. 2007;313:878–895. doi: 10.1016/j.yexcr.2006.12.015. [DOI] [PubMed] [Google Scholar]
- Weed SA, Parsons JT. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 2001;20:6418–6434. doi: 10.1038/sj.onc.1204783. [DOI] [PubMed] [Google Scholar]
- Daly RJ. Cortactin signalling and dynamic actin networks. Biochem J. 2004;382:13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TC, Singleton C, Fraley TS, Greenwood JA. Cystein-rich protein 1 (CRP1) regulates actin filament bundling. BMC Cell Biol. 2005;6:45. doi: 10.1186/1471-2121-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4(2):118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- Samarin SN, Koch S, Ivanov AI, Parkos CA, Nusrat A. Coronin 1C negatively regulates cell-matrix adhesion and motility of intestinal epithelial cells. Biochem Biophys Res Commun. 2010;391(1):394–400. doi: 10.1016/j.bbrc.2009.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Knudson W, Knudson CB, Ishiguro N. Antisense inhibition of hyaluronan synthase-2 in human osteosarcoma cells inhibits hyaluronan retention and tumorigenicity. Exp Cell Res. 2005;307:194–203. doi: 10.1016/j.yexcr.2005.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, Guo NH, Rodrigues RG, Kaiser J, Roberts DD. Pro-adhesive and chemotactic activities of thrombospondin-1 for breast carcinoma cells are mediated by α3β1 integrin and regulated insulin-like growth factor-1 and CD98. J Biol Chem. 1999;274(16):11408–11416. doi: 10.1074/jbc.274.16.11408. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski AV, Gertler FB, Loureiro JJ. Function and regulation of Ena/VASP proteins. Trends Cell Biol. 2003;13:386–392. doi: 10.1016/S0962-8924(03)00130-2. [DOI] [PubMed] [Google Scholar]
- Lacy SE, Bönnemann CG, Buzney EA, Kunkel LM. Identification of FLRT1, FLRT2 and FLRT3: A novel family of transmembrane leucine-rich repeat proteins. Genomics. 1999;62:417–426. doi: 10.1006/geno.1999.6033. [DOI] [PubMed] [Google Scholar]
- Kohfeldt E, Sasaki T, Göhring W, Timpl R. Nidogen-2: A new basement membrane protein with diverse binding properties. J Mol Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- Barker TH, Grenett HE, MacEwen MW, Tilden SG, Fuller GM, Settleman J, Woods A, Murphy-Ullrich J, Hagood JS. Thy-1 regulates fibroblast focal adhesion, cytoskeletal organization and migration through modulation of p190 ThoGAP and Rho GTPase activity. Exp Cell Res. 2004;295:488–496. doi: 10.1016/j.yexcr.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Rege TA, Hagood JS. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB J. 2006;20:1045–1054. doi: 10.1096/fj.05-5460rev. [DOI] [PubMed] [Google Scholar]
- Sobarzo CM, Lustig L, Ponzio R, Denduchis B. Effect of di-(2-ethylhexyl)phthalate on N-cadherin and catenin protein expression in rat testis. Reprod Tox. 2006;22:77–86. doi: 10.1016/j.reprotox.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Bielenberg DR, Pettaway CA, Takashima S, Klagsbrun M. Neuropilins in neoplasms: expression, regulation, and function. Exp Cell Res. 2006;312:584–593. doi: 10.1016/j.yexcr.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- Öklu R, Hesketh R. The latent transforming growth factor β binding protein (LTBP) family. Biochem J. 2000;352:601–610. doi: 10.1042/0264-6021:3520601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZQ, Cao WH, Xie JJ, Lin J, Shen ZY, Zhang QY, Shen JH, Xu LY, Li EM. Expression and prognostic significance of THBS1, Cyr61 and CTGF in esophageal squamous cell carcinoma. BMC Cancer. 2009;9:291–299. doi: 10.1186/1471-2407-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PT, Gleeson JG, Corbo JC, Flanagan L, Walsh CA. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20(24):9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai CM, Gu Z. Caldesmon phosphorylation in actin cytoskeletal remodeling. Eur J Cell Biol. 2006;85:305–309. doi: 10.1016/j.ejcb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Wang Q, Symes AJ, Kane CA, Freeman A, Nariculam J, Munson P, Thrasivoulou C, Masters JR, Ahmed A. A novel role for Wnt/Ca2+ signaling in actin cytoskeleton remodeling and cell motility in prostate cancer. PloS One. 2010;5(5):1–11. doi: 10.1371/journal.pone.0010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RD, Strehler EE. Calmodulin-like protein enhances myosin-10 translation. Biochem Biophys Res Commun. 2008;369:654–659. doi: 10.1016/j.bbrc.2008.02.056. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- Reif S, Lang A, Lindquist JN, Yata Y, Gäbele E, Scanga A, Brenner DA, Rippe RA. The role of focal adhesion kinase-phosphatidylinositol 3-kinase-Akt signaling in hepatic stallate cell proliferation and type I collagen expression. J Biol Chem. 2003;278(10):8083–8090. doi: 10.1074/jbc.M212927200. [DOI] [PubMed] [Google Scholar]
- Espada J, Galaz S, Sanz-Rodríguez F, Blázquez-Castro A, Stockert JC, Bagazgoitia L, Jaén P, González S, Cano A, Juarranz A. Oncongenic H-Ras and PI3K signaling can inhibit E-Cadherin-dependent apoptosis and promote cell survival after photodynamic therapy in mouse keratinocytes. J Cell Physiol. 2009;219:84–93. doi: 10.1002/jcp.21652. [DOI] [PubMed] [Google Scholar]
- Wang X, Goode EL, Fredericksen ZS, Vierkant RA, Pankratz VS, Liu-Mares W, Rider DN, Vachon CM, Cerhan JR, Olson JE, Couch FJ. Association of genetic variation in genes implicated in the β-Catenin destruction complex with risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(8):2101–2108. doi: 10.1158/1055-9965.EPI-08-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DR, Elcombe CR. Induction of Acyl-Coa oxidase and Cytochrome-P450iva1 Rna in rat primary hepatocyte culture by peroxisome proliferators. Biochem J. 1991;280:249–253. doi: 10.1042/bj2800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveillard A, Mselli-Lakhal L, Mogha A, Lasserre F, Polizzi A, Pascussi JM, Guillou H, Martin PGP, Pineau T. Di-(2-ethylhexyl)-phthalate (DEHP) activates the constitutive androstane receptor (CAR): a novel signalling pathway sensitive to phthalates. Biochem Pharmacol. 2009;77:1735–1746. doi: 10.1016/j.bcp.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Eveillard A, Lasserre F, de Tayrac M, Polizzi A, Claus S, Canlet C, Mselli-Lakhal L, Gotardi G, Paris A, Guillou H, Martin PGP, Pineau T. Identification of potential mechanisms of toxicity after di-(2-ethylhexyl)-phthalate (DEHP) adult exposure in the liver using a system biology approach. Tox App Pharmacol. 2009;236:282–292. doi: 10.1016/j.taap.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Lake BG, Rumsby PC, Price RJ, Cunninghame ME. Species differences in hepatic peroxisome proliferation, cell replication and transforming growth factor-beta 1 gene expression in the rat, Syrian hamster and guinea pig. Mutat Res Fundam Mol Mech Mut. 2000;448:213–225. doi: 10.1016/S0027-5107(99)00238-9. [DOI] [PubMed] [Google Scholar]
- Takashima K, Ito Y, Gonzalez FJ, Nakajima T. Different mechanisms of DEHP-induced hepatocellular adenoma tumorigenesis in Wild-type and Pparα-null mice. J Occup Health. 2008;50:169–180. doi: 10.1539/joh.L7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JA, Graeber T, Frackelton AR Jr, Kim M, Schwarzbauer JE, Filardo EJ. Coordinate regulation of estrogen-mediated fibronectin matrix assembly and epidermal growth factor receptor transactivation by G protein-coupled receptor, GPR30. Endocrinology. 2009;23:1052–1064. doi: 10.1210/me.2008-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz ER, Barton M. Signaling, physiological functions and clinical relevance of the G protein-coupled estrogen receptor GPER. Prostaglandins Other Lipid Mediat. 2009;89(3-4):89–97. doi: 10.1016/j.prostaglandins.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleuser B, Malek D, Gust R, Pertz HH, Potteck H. 17-β-estradiol inhibits transforming growth factor-β signaling and function in breast cancer cells via activation of extracellular signal-regulated kinase throught the G protein-coupled receptor 30. Mol Pharmacol. 2008;74:1533–1543. doi: 10.1124/mol.108.046854. [DOI] [PubMed] [Google Scholar]
- Pienta RJ, Poiley JA, Lebherz WB III. Morphological transformation of early passage golden Syrian hamster embryo cells derived from cryopreserved primary cultures as a reliable in vitro bioassay for identifying diverse carcinogens. Int J Cancer. 1977;19:642–655. doi: 10.1002/ijc.2910190508. [DOI] [PubMed] [Google Scholar]
- Elias Z, Poirot O, Daniere MC, Terzetti F, Marande AM, Dzwigaj S, Pezerat H, Fenoglio I, Fubini B. Cytotoxic and transforming effects of silica particles with different surface properties in Syrian hamster embryo (SHE) cells. Toxicol In Vitro. 1989;14:409–422. doi: 10.1016/s0887-2333(00)00039-4. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparative table of the sensitivity of DD versus qPCR. Comparisons between the ratio of bands intensity compared to the control on the gel using an image analysis software (QuantityOne® 1-D analysis software BioRad, Marne-la-Coquette, France) for Differential Display and the ΔΔCt score normalized by gapdh mRNA level after analysis with StepOne and DataAssist (Roche Applied Biosystem, Courtaboeuf, France) for qPCR.
Primers for Differential Display. List of the sequences of the anchored and the arbitrary primers used for the Differential Display experiments.