Summary
Background
The centromere (CEN) DNA-kinetochore complex is the specialized chromatin structure that mediates chromosome attachment to the spindle and is required for high-fidelity chromosome segregation. Although kinetochore function is conserved from budding yeast to humans, it was thought that transcription had no role in centromere function in budding yeast, in contrast to other eukaryotes including fission yeast.
Results
We report here that transcription at the centromere facilitates centromere activity in the budding yeast Saccharomyces cerevisiae. We identified transcripts at CEN DNA and found that Cbf1, which is a transcription factor that binds to CEN DNA, is required for transcription at CEN DNA. Chromosome instability of cbf1Δ cells is suppressed by transcription driven from an artificial promoter. Furthermore, we have identified Ste12, which is a transcription factor, and Dig1, a Ste12 inhibitor, as a novel CEN-associated protein complex by an in vitro kinetochore assembly system. Dig1 represses Ste12-dependent transcription at the centromere.
Conclusions
Our studies reveal that transcription at the centromere plays an important role in centromere function in budding yeast.
Introduction
In eukaryotic cells, all duplicated chromosomes are segregated equally to daughter cells during mitotic cell division. To make the event a success, a highly conserved architecture consisting of centromere (CEN) DNA, kinetochores, microtubules, and microtubule-organizing centers (MTOCs), must be properly assembled during each cell cycle [1].
The centromere of budding yeast Saccharomyces cerevisiae (S. cerevisiae), which is known as a point centromere [2], has been well studied, and S. cerevisiae is a good model organism in which to investigate how kinetochore proteins assemble onto CEN DNA [3, 4]. Each CEN DNA of S. cerevisiae is an approximately 125-bp region that has three conserved centromere DNA elements CDEI, CDEII, and CDEIII [5]. CDEI includes the CACRTG region, where a homodimer of Cbf1 (helix-loop-helix family) associates [5, 6]. CDEII is a 78- to 86-bp sequence that is about 90% AT rich [5]. This element is thought to be important for histone and/or Ndc10 interaction [7–9]. The 25-bp CDEIII contains a conserved CCG motif that is crucial for forming the CBF3 complex (comprising Ndc10, Cep3, Ctf13, and Skp1) [10]. More than 70 kinetochore proteins have been identified in S. cerevisiae [3, 4]. The kinetochore comprises three layers that connect between CEN DNA and microtubules in a hierarchical manner: a DNA-binding layer, a linker layer, and a microtubule (MT)-binding layer. The CEN localization of most kinetochore proteins depends on the CBF3 complex in S. cerevisiae. Recent evidence suggests that kinetochores are disassembled during CEN DNA replication in early S phase but are reassembled in late S phase [11].
In diverse eukaryotic species such as the fission yeast Schizosaccharomyces pombe (S. pombe), plants, and metazoans, regional CEN DNAs are much longer and more complex than point CEN DNA in budding yeast [2]. S. pombe CEN regions are large, repetitive structures that range from 35 kb to 110 kb in length [12]. The CEN regions in humans and Drosophila melanogaster are composed of repetitive satellite elements, which are from 200 kb to 4 Mb long [13]. RNA interference (RNAi), a gene-silencing pathway triggered by double-stranded RNA, corresponds to the formation and the maintenance of centromeric heterochromatin in regional CEN DNA. These species use the ribonuclease III (RNase III) endonuclease Dicer and the effector protein Argonaute to generate small interfering RNAs (siRNAs) [14]. In contrast, the budding yeast S. cerevisiae, which has point CEN DNA, does not contain RNAi genes such as Dicer and presumably does not make use of the RNAi pathway [15].
Here, we have shown that transcription at the centromere makes a direct contribution to centromere function in budding yeast. The transcription factor Cbf1 contributes to CEN transcripts whereas Dig1 inhibits Ste12-dependent CEN transcripts. Although strong transcription by an artificial promoter deactivates centromere activity [16, 17], we found that a certain level of transcriptional activity is required for centromere function.
Results
Identification of CEN3-associated proteins
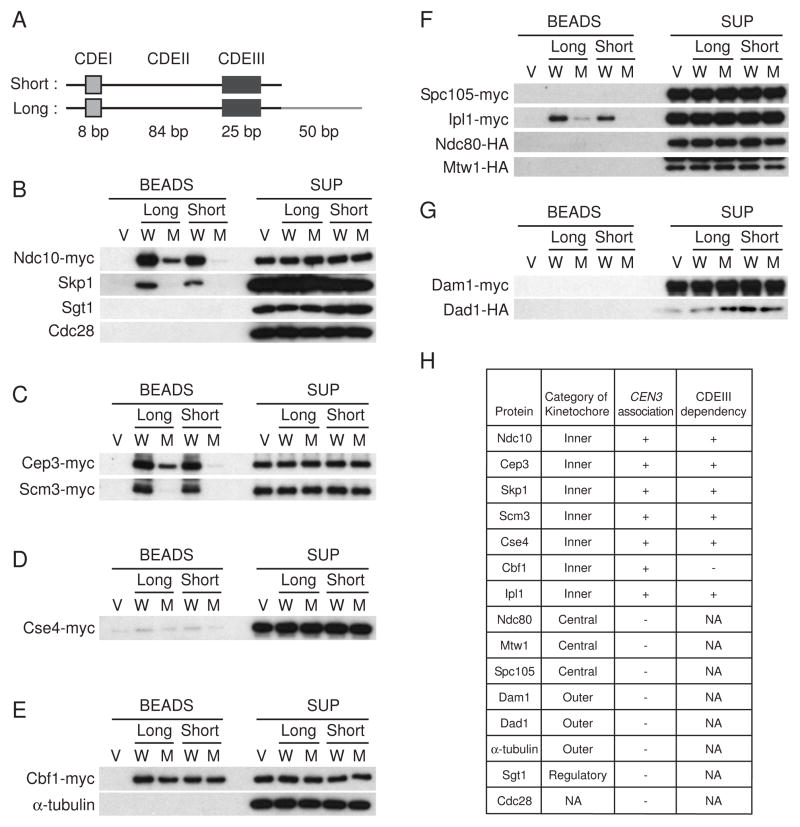
To identify novel proteins that associate with CEN DNA, an in vitro assembly system was used (Figure S1). Espelin et al. [18] found that Ndc10 binding with DNA extends beyond CDEIII. Thus, we prepared two CEN3 constructs (Figure 1A). The short version of CEN3 DNA, which is 134-bp long, consists of only CDEI, CDEII, and CDEIII. The long version, which is 184-bp long, contains the additional Ndc10 binding site. A mutant CEN3 containing a point mutation in the CCG motif in CDEIII, which is deficient in centromere activity [10, 19], was used as a negative control. To increase the binding capacity, we used plasmids containing 8 tandem copies of CEN3 (Figure S1A) [20]. To investigate whether CEN DNA and protein complexes are assembled properly, we first tested whether CBF3 components (Ndc10, Skp1, and Cep3) would bind to CEN DNA (Figures 1B and 1C). Indeed, Ndc10, Skp1, and Cep3 all bound to both the long and the short versions of wild-type CEN DNA. In contrast, these proteins did not bind to vector only or to the CEN3 mutant beads, except that Ndc10 and Cep3 were loaded at a low level onto the long version of the mutant CEN3 due to the extended CBF3. Sgt1 and Cdc28, which are not components of the kinetochore [21], were used as negative controls, and no signals were detected (Figure 1B). These results indicated that the core-kinetochore complex was assembled specifically in this in vitro system. Cse4 (the yeast homologue of human centromere-specific histone H3 variant CENP-A) and Scm3, which is required for Cse4 incorporation into the centromeric chromatin [22], bound to both the long and the short versions of CEN3 DNA but not to the CEN3 mutant DNA (Figures 1C and 1D; Figure S2). These results therefore suggest that localization of Cse4 and Scm3 with CEN DNA depends on the CBF3 complex, which is consistent with the previous finding that Cse4-CEN3 association was decreased in ndc10-1 mutants [23]. The association of Cbf1 with CEN3 DNA was not altered by the mutation of the CCG motif in CDEIII (Figure 1E), which is also consistent with the previous finding that Cbf1 binds to CDEI. We did not detect any CEN association with Spc105, Ndc80, or Mtw1 (the central kinetochore proteins); Dam1 or Dad1 (the outer kinetochore proteins); or α-tubulin (Figures 1E–1G). We found that Ipl1 (the yeast Aurora kinase) did associate with wild-type CEN3 DNA but not with the mutant CEN3 DNA (Figure 1F), implying that Ipl1 functions closely to the core-kinetochore CEN DNA complex.
Figure 1. Initial kinetochore assembly in vitro.
(A) The 117-bp CEN3 DNA consists of three conserved DNA elements, designated CDEI, CDEII, and CDEIII. CDEI consists of a conserved 8-bp sequence incorporating a 6-bp palindrome, which is important for Cbf1 association. Approximately 93% of the CDEII sequence is made up of A:T base pairs. CDEIII has a size of 25 bp and includes the completely conserved sequence CCG. As a negative control in the in vitro kinetochore assembly system, the CCG sequence of CDEIII was replaced to CCC. The short version of CEN3 DNA has a length of 134 bp, containing only CDEI, CDEII, and CDEIII. The long version of CEN3 DNA has a length of 184 bp, containing the same sequences plus an Ndc10-binding site (see Espelin et al., 1997 [18]). (B–G) Protein extracts from cyclic cells were incubated with magnetic beads coupled to plasmid DNA. Vector (V) or plasmid containing 8 tandem copies of wild-type (W) or mutant (M) CEN3 was used. After 4 min at room temperature with highly concentrated protein extract, the beads were collected and washed (Figure S1B). Proteins bound to the beads were analyzed by 4–15% SDS-PAGE and immunoblotting. The protein extracts derived from the tagged strains were as follows: (B) Ndc10-myc Ndc80-HA Mtw1-HA (Y2046); (C) Cep3-myc Scm3-myc Mif2-myc (Y2049); (D) Cse4-myc (Y2044); (E) Cbf1-myc Isw1-HA (Y2083); (F) Spc105-myc Ipl1-myc Ndc80-HA Mtw1-HA (Y2047); and (G) Dam1-myc Stu2-myc Dad1-HA (Y2048). (H) Summary of kinetochore assembly in 4 min. NA: Not applicable.
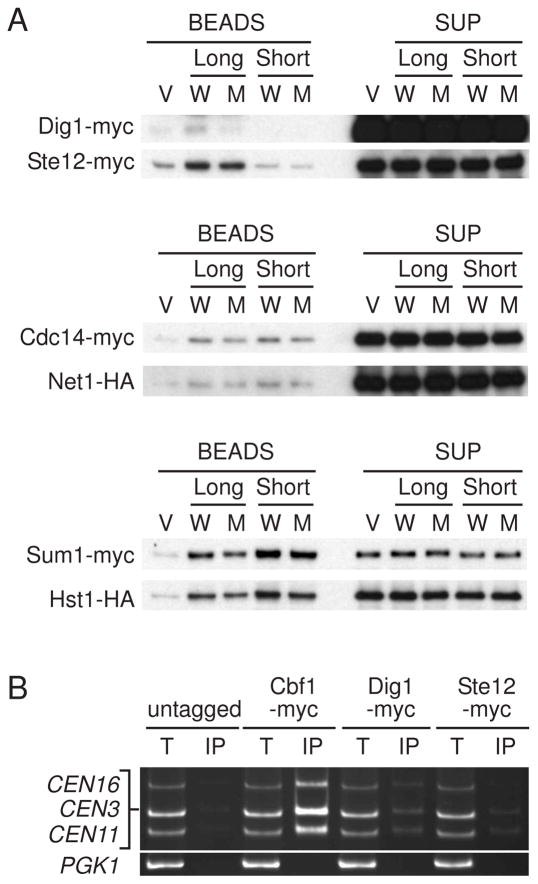
Next we performed mass spectrometry to identify CEN-associating proteins. Dig1-Ste12, Sum1-Hst1, and Cdc14-Net1 complexes were identified in the CEN DNA magnetic beads by two independent mass spectrometry analyses as well as Cbf1 (Figure S3; data not shown). The specificity of these results was confirmed by performing immunoblotting of the elution from the CEN DNA magnetic beads. All of these proteins were associated with both wild-type and mutant CEN3 DNA, except that Dig1 was loaded at a low level onto mutant CEN3 DNA. (Figure 2A; Figure S4). Sum1-Hst1 is a part of the Sum1/Rfm1/Hst1 complex that represses meiotic genes during vegetative growth via histone deacetylation by Hst1 (Homologue of Sir Two) [24, 25]. Cdc14-Net1 is the core subunit of the RENT complex (Net1, Sir2, and Cdc14) involved in nucleolar silencing and telophase exit [26]. Ste12 is a transcription factor that is controlled by a mitogen-activated protein (MAP) kinase cascade in mating pheromone and invasive growth pathways [27]. Dig1 is an inhibitor of Ste12 [28, 29]. Ste12 and Dig1 were associated with the long version of CEN3 but not the short version in the in vitro assembly system (Figure 2A; Figure S4), indicating that the binding region of these proteins exists outside of CDEIII. In fact, we found a Ste12-binding site, which is called a pheromone response element [PREs; TGAAAC(A/G)] [30], in the pericentromeric region of CEN3 (Figure 3A). The chromatin immunoprecipitation (ChIP) assay showed that Cbf1, Ste12, and Dig1 coimmunoprecipitated specifically with CEN DNA (Figure 2B), indicating that these proteins associate with CEN DNA in vivo.
Figure 2. Dig1 and Ste12 are bound to centromeric chromatin.
(A) Dig1-Ste12, Cdc14-Net1 and Sum1-Hst1 are bound to centromeric chromatin in vitro. In vitro kinetochore assembly system was executed as described in Figure 1. The protein extracts derived from the tagged strains were as follows: Dig1-myc Ste12-myc (Y2052); Cdc14-myc Net1-HA (Y2050); and Sum1-myc Hst1-myc (Y2051). (B) Anti-myc chromatin immunoprecipitation (ChIP) assays were performed from log-phase cells. Total lysate (T) and coimmunoprecipitated DNA (IP) were analyzed by PCR with primers specific to centromeric regions of chromosomes III, XI, and XVI and to a noncentromeric region (PGK1). The yeast strains used were as follows: untagged (YPH499), Cbf1-myc (Y2053), Dig1-myc (Y2054), and Ste12-myc (Y2055).
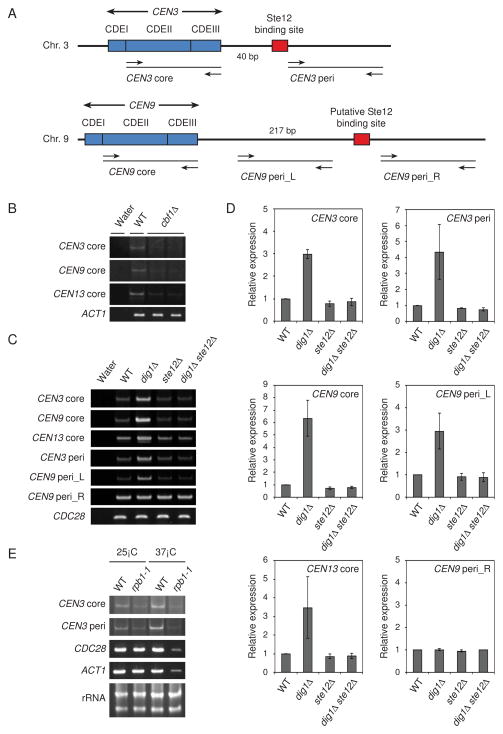
Figure 3. Centromeric transcripts in Saccharomyces cerevisiae.
(A) Schematic representations of the CEN3 and CEN9 regions. Arrows mark the location of primers. The Ste12 binding site of CEN3 (TGAAACG) is located 41–47 bp from CDEIII. The putative Ste12 binding site of CEN9 (TGTAACA) is located 218–224 bp from CDEIII. (B) Transcription derived from CEN3, CEN9, and CEN13 was detected in the wild-type but not in the cbf1Δ mutants. RT-PCR analyses were performed from log phase total RNA in wild-type (YPH499) and cbf1Δ mutants (Y1987 and Y1988). ACT1 was used as a loading control. (C) Accumulation of transcripts derived from the core or pericentromeric regions in the dig1Δ mutant was determined with RT-PCR. CDC28 was used as a loading control. The yeast strains used were as follows: wild-type (YPH499), dig1Δ (Y1979), ste12Δ (Y1985), and dig1Δ ste12Δ (Y2056). (D) Quantification of transcripts in (C). Relative expression levels were calculated by dividing CDC28 expression level. (E) RNA polymerase II is required for the CEN3 transcripts. RT-PCR analyses were performed from log phase total RNA in wild-type (YF7) and the largest subunit mutant of RNA polymerase II (rpb1-1) (YF38). CDC28 and ACT1 were used as positive controls. Ribosomal RNA was used as a loading control.
Identification of transcription at CEN DNA
These observations made us consider the possibility that transcription occurs from or through the centromere. Therefore, we tested whether transcripts occurred at the centromere in wild-type cells. Core centromeric transcripts of CEN3, CEN9, and CEN13 were detected by reverse transcriptase PCR (RT-PCR) in the wild-type strain but not in cbf1Δ mutants (Figure 3B), indicating that Cbf1 is required for centromeric transcription. We also examined whether the core or pericentromeric transcripts of CEN3 or CEN9 were decreased in ste12Δ mutants or/and increased in dig1Δ mutants (Figures 3C and 3D). To detect the transcription, we designed primers upstream and downstream of the Ste12 binding site (Figure 3A). Interestingly, deletion of DIG1 increased the core centromeric expression of CEN3, CEN9, and CEN13 (Figures 3C and 3D). Similarly, one of the pericentromeric transcripts of CEN9 (CEN9 peri_L), which is derived from the left side of the putative Ste12 binding site, was high in dig1Δ mutants (Figures 3C and 3D). The core centromeric expression of CEN3, CEN9, and CEN13 was slightly decreased in the ste12Δ mutants and the increased expression in the dig1Δ mutants was abolished by deletion of STE12 (Figures 3C and 3D). This suggests that Ste12 has a positive role in basal CEN transcription but that full Ste12 transcription induction is repressed by Dig1. We also tested the transcription derived from the right side of the Ste12 binding site. The pericentromeric transcripts of CEN3 (CEN3 peri) were elevated in dig1Δ mutants and the elevated expression was abolished by deletion of STE12, as the case of the core centromeric transcripts (Figures 3C and 3D). In contrast, there were no significant differences in expression between wild-type and dig1Δ strain in the pericentromeric transcripts of CEN9 (CEN9 peri_R). Therefore, these results suggest that Dig1 inhibits core and pericentromeric transcription regulated by Ste12.
In S. pombe, RNA polymerase II is required for pericentromeric transcription mediated by the siRNA interference [31]. We thus examined whether the largest subunit (Rpb1) of RNA polymerase II is responsible for core and pericentromeric transcription using the temperature-sensitive mutant rpb1-1 (Figure 3E). In wild-type cells, both core and pericentromeric CEN3 transcripts were clearly detectable, as were CDC28 and ACT1 transcripts. As expected, CDC28 and ACT1 transcripts were greatly reduced in rpb1-1 mutant cells (Figure 3E). In addition, CEN3 core and pericentromeric transcripts were greatly reduced when the cells were shifted to the non-permissive temperature (37°C), which indicates that Rpb1 is required for both core and pericentromeric transcription of CEN3.
The role of centromeric transcription
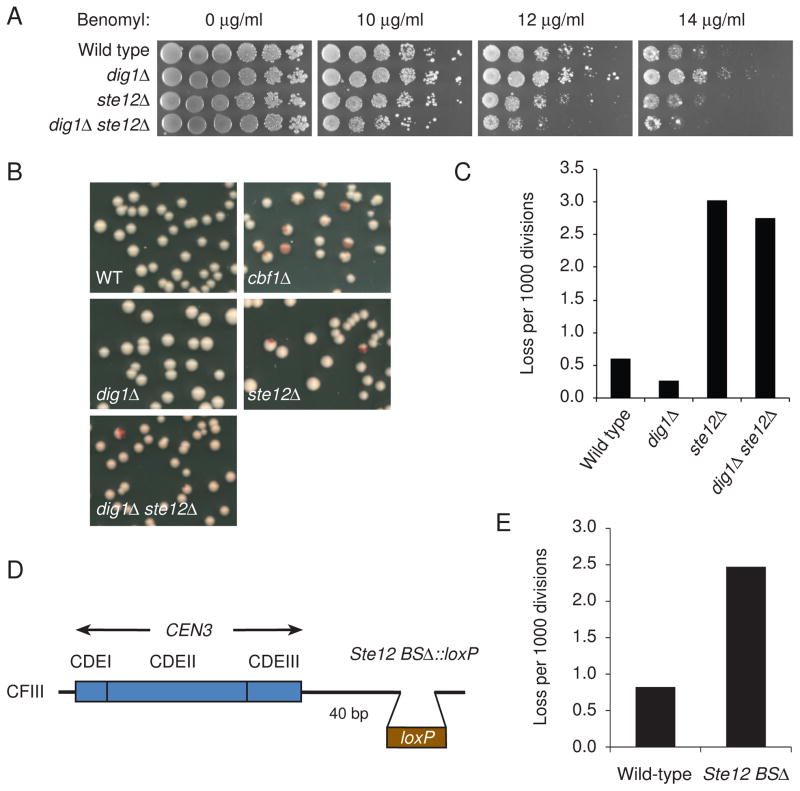
Loss of Cbf1 causes chromosome loss and sensitivity to benomyl (a microtubule-depolymerizing drug) [32] (Figure 4B). To test whether deletion of STE12 or DIG1 influences centromere function, we investigated traits common to kinetochore mutants, such as benomyl sensitivity and infidelity of chromosome segregation (Figures 4A–4C). ste12Δ cells were mildly sensitive to benomyl and exhibited a substantial chromosome missegregation phenotype. Interestingly, dig1Δ cells were resistant to benomyl and had an increased fidelity of chromosome segregation than wild-type cells. Furthermore, dig1Δ ste12Δ cells were highly sensitive to benomyl and had a dramatic increase in chromosome missegregation, similar to the phenotype of ste12Δ cells. These observations indicate that both Ste12 and Dig1 affect the fidelity of chromosome segregation and revealed that the increased transcription at CEN DNA, at least more than wild-type level, is required for high fidelity of chromosome segregation. Next, we deleted the Ste12 binding site near CEN3 on the chromosome fragment (Figure 4D), and found that the resulting Ste12 binding site deletion mutant exhibited a chromosome missegregation phenotype similar to those in ste12Δ or dig1Δ ste12Δ mutant cells (Figure 4E). Together, these results suggest that a certain level of transcriptional activity at CEN DNA is required for centromere function, whereas it was previously shown that a high level transcription through CEN3 impairs centromere function [17].
Figure 4. Centromeric transcripts contribute to centromere function.
(A) Benomyl resistance of dig1Δ cells and benomyl sensitivity of ste12Δ and dig1Δ ste12Δ cells. Yeast cells were spotted in 5-fold dilutions from 5 × 104 cells per spot on YPD plates containing benomyl. The plates were incubated at 23°C for 5 days and photographed. The yeast strains used were as follows: wild-type (YPH499), dig1Δ (Y1978), ste12Δ (Y1985), and dig1Δ ste12Δ (Y2056). (B) cbf1Δ, ste12Δ, and cbf1Δ ste12Δ mutants sectoring phenotypes. Each strain includes a single SUP11-marked chromosome III fragment containing CEN3. (C) Chromosome loss rate in null mutants was determined by half-sector analysis. Wild type: 18 half-sectored colonies/30,214 total colonies; dig1Δ: 6/22,972; ste12Δ: 44/14,567; dig1Δ ste12Δ: 40/14,575. The yeast strains used in B and C were as follows: wild-type (Y14), cbf1Δ (Y2011), dig1Δ (Y2016), ste12Δ (Y2060 and Y2061), and dig1Δ ste12Δ (Y2062 and Y2063). (D) Schematic diagram showing deletion of the Ste12 binding site (Ste12 BSΔ) on the chromosome fragment. (E) Increased chromosome missegregation in Ste12 BSΔ. Chromosome loss rate was determined by half-sector analysis. Wild-type (Y14): 7 half-sectored colonies/8,550 total colonies; Ste12 BSΔ (Y2082): 20/8,085.
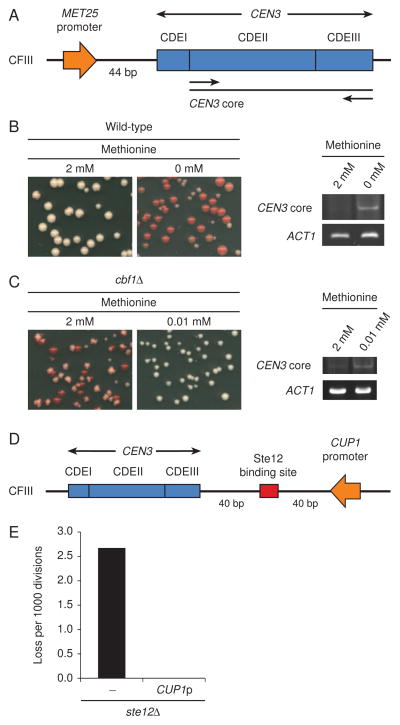
To confirm this result, we constructed a strain in which the core CEN3 transcripts on the chromosome fragment are controlled by the methionine-repressible MET25 promoter (Figure 5A). When the cells are grown in medium containing methionine (2 mM), transcription from the MET25 promoter is repressed. In contrast, when cells are grown in medium without or containing limited methionine (0.01 mM), strong transcription is induced from the MET25 promoter, resulting in about 20-fold induction [33]. Wild-type cells, in the absence of methionine or in the presence of limited methionine, showed significant chromosome missegregation (Figure 5B; data not shown), indicating that a high level of transcription through CEN3 impairs centromere function, as was previously shown [17].
Figure 5. Centromeric transcripts induced by an artificial promoter suppress the chromosome missegregation phenotype of cbf1Δ and ste12Δ.
(A) A schematic diagram showing the integration of the MET25 promoter in front of CDEI on the chromosome fragment. (B) Overexpressed transcripts impair centromere function. PMET25-CEN3 (Y392) cells were precultured in minimal (SD) medium plus 2 mM methionine (to repress the MET25 promoter) and lacking uracil (to keep the chromosome fragments). Colony color assay was performed after plating the cells onto SD plates, limiting the amount of adenine and adding the indicated concentration of methionine. RT-PCR analysis was performed from the total RNA taken after 3 h of induction. (C) Centromeric transcripts from a MET25 promoter suppress the sectoring phenotype of cbf1Δ mutants. cbf1Δ PMET25-CEN3 (Y1990) cells were examined as described in (B). The medium containing 0.01 mM methionine was used as a derepressing condition. (D) Schematic diagram showing the integration of the CUP1 promoter in front of the Ste12 binding site on the chromosome fragment. (E) Yeast cells were precultured in SD medium lacking tryptophan (to maintain the chromosome fragments). Colony color assay was performed after plating the cells onto SD plates, limiting the amount of adenine. ste12Δ (Y2060): 15 half-sectored colonies/5,615 total colonies; ste12Δ PCUP1-CEN3 (Y2080); 0/12,622.
If the hypothesis that cbf1Δ cells have reduced fidelity of chromosome segregation due to loss of core centromeric transcription is true, then a high level of transcription from the MET25 promoter should be able to suppress the chromosome missegregation phenotype of cbf1Δ cells. Because cbf1Δ cells failed to grow in the methionine-free medium [32], the cells were grown in the medium containing 0.01 mM methionine to induce transcription from the MET25 promoter. Indeed, the transcripts derived from the MET25 promoter suppressed the chromosome missegregation phenotype of cbf1Δ cells (Figure 5C). We also constructed a cbf1Δ strain in which transcription through CEN3 is induced by a CUP1 promoter. Similarly, transcripts derived from the CUP1 promoter partially suppressed the chromosome missegregation phenotype of cbf1Δ cells (Figure S5).
Next, we examined whether the same was true for ste12Δ. We constructed a ste12Δ strain in which the peri-CEN3 transcripts are controlled by a CUP1 promoter (Figure 5D). Transcription from the CUP1 promoter suppressed chromosome instability of the ste12Δ mutants (Figure 5E). Together, these results strongly suggest that centromeric transcription contributes to centromere function.
Discussion
In this study, we have identified transcripts at CEN DNA regulated by transcriptional factors both Cbf1 and Ste12 in an RNA polymerase II-dependent manner (Figure 6). Moreover, the silencing factors such as Hst1, Cdc14, and Sir1 are also part of the macromolecular complex of the kinetochore. These findings are reminiscent of transcriptional regulation at the regional centromere in higher eukaryotes.
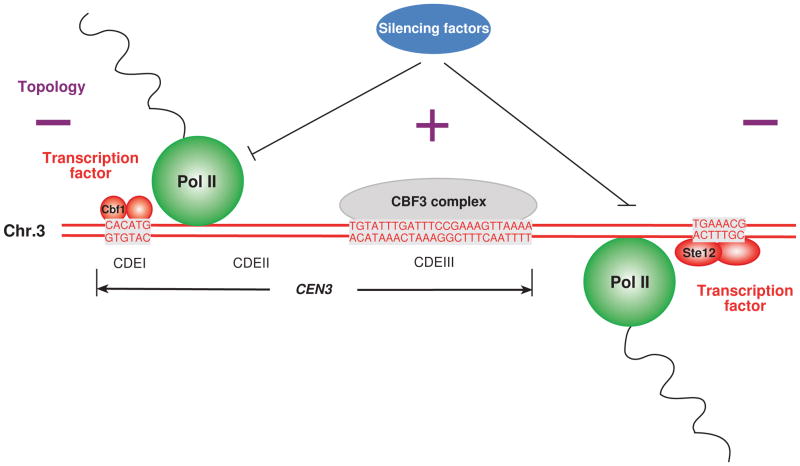
Figure 6. Transcription at the centromere plays an important role in centromere function.
CEN3, which is 117 bp, is composed of three regions, CDEI, CDEII, and CDEIII. The sequence of CDEIII, which binds to CBF3 (the core kinetochore complex), and Cbf1 and Ste12 (transcription factors) binding sites are shown in grey boxes. Cbf1 and Ste12 contribute to transcription at the centromere. On the other hand, silencing factors, such as Sir1, Hst1, or Cdc14, may inhibit the transcription. RNA polymerase II is required for core- and peri-centromeric transcription. Predicted DNA topology is shown as plus (positive DNA supercoils) or minus (negative DNA supercoils). Intriguingly, topological analysis of yeast minichromosomes revealed that functional centromeres induce positive DNA supercoils [40]. Transcription at the centromere may generate proper topology of CEN DNA.
Transcription at the centromere is important for centromere function
We identified novel CEN-associated proteins such as Dig1 and Ste12 by our in vitro kinetochore assembly system. The CEN DNA-protein complex contains only the inner kinetochore proteins (Figure 1H). Both Cbf1 and Ste12, which are known transcription factors [30, 32–34], associate with centromeric chromatin and help maintain mitotic chromosome stability. Our results strongly suggest that both Cbf1 and Ste12 regulate centromeric transcripts in an RNA polymerase II-dependent manner. Moreover, Dig1 inhibits centromeric transcripts. We have discovered a putative Ste12 binding site (TGAAACG) at peri-CEN3 and another (TGTAACA) at peri-CEN9 (Figure 3A). Using ChIP assays, we have shown that Ste12 associates with other CEN DNA (Figure 2B) and found putative Ste12 binding sites at pericentromeric regions in all 16 chromosomes (Table S1), suggesting that core or pericentromeric transcription by Ste12 is conserved in all chromosomes. Interestingly, there are multiple putative Ste12 binding site around some CENs (Table S1). Ste12 inducible genes usually have more than one PRE in their promoter regions [35]. Pericentromeric regions regulated by Ste12 appear to be similar to the typical promoter region of Ste12 inducible genes. Moreover, genome-wide analyses by Harbison et al. [36] and Tachibana et al. [37] revealed that other transcriptional regulatory codes, which are specific DNA sequences that induce or repress gene expression, exist in some pericentromeric regions (Table S2). Thus, additional transcription factors might contribute to centromeric transcription.
The possible role of transcription at CEN DNA
CEN DNA might have evolutionarily originated from a promoter region. Hemmerich et al. [38] illustrated the analogy between the yeast centromere and the MET promoter. We also found a similarity between the yeast pericentromere and the promoter regulated by Ste12. It is tempting to speculate that the kinetochore assembly system on CEN DNA resembles the access of transcription or silencing factors on the promoter DNA.
What is the role of transcription at the centromere? Dynamic topological changes of DNA are known to occur during the transcription process [39]. A recent finding suggests that positive supercoiling is a general feature of centromeric nucleosomes in eukaryotic cells, although H3 nucleosomes induce negative supercoils [40]. Therefore, CEN transcription might be responsible for the proper topology of CEN DNA (Figure 6).
Comparison with RNAi machinery
In regional CEN DNA of higher eukaryotes, RNAi machinery is required for gene silencing in the assembly of pericentromeric heterochromatin. Budding yeast S. cerevisiae lacks the RNAi machinery, although other budding yeast species, such as Saccharomyces castellii and Candida albicans, has the machinery [15]. The level of transcription derived from the pericentromere is much higher than that from the core region of CEN DNA in S. pombe [41, 42]. Transfer RNA (tRNA) genes are thought to be the insulators that mark the distinct chromatin domains within the centromere. It has recently been reported that transcripts from the central core domain of fission yeast centromere are degraded by the exosome [42]. Increased level of transcription including CEN3 region was also detected in exosome mutants in S. cerevisiae [43]. Considering these previous results, our findings raise the possibility that a certain level of transcription at CEN DNA is important for centromere function, which is evolutionally conserved, and thus, RNAi in higher eukaryotes might be needed only to control levels of transcripts at the centromere.
The RNA-silencing pathway in S. pombe contributes to heterochromatin formation and maintenance at centromeres [44]. In this process, chromodomain protein Swi6 recruits Clr3 histone deacetylase as a silencing factor and Epe1 transcriptional activator as an anti-silencing factor. The balance between the opposing activities of these proteins is essential for the determination of the transcriptional status at centromeres [44]. Sir1, which is the budding yeast silencing protein, is a functional component of centromeric chromatin [45]. We have found several silencing factors, such as Hst1-Sum1 and Cdc14-Net1, as CEN-associated proteins (Figure 2A). Therefore, transcriptional regulation, which is the balance of expression levels, is important to positively or negatively maintain the centromeric nucleosome (Figure 6). It will be interesting to decipher how centromeric transcription is regulated by these factors and how kinetochore assembly is regulated by centromeric transcription.
Experimental Procedures
Yeast strains
Table S3 presents the genotypes of yeast strains used for this study. Details of strain construction are provided in Supplemental Experimental Procedures.
Plasmids
Table S4 lists the plasmids used in the kinetochore assembly system in vitro. The construction of tandem CEN3 plasmids is depicted in Figure S1. To construct the wild-type and mutant single copies of CEN3 plasmids, 134-bp (short) or 184-bp (long) CEN3 fragments were connected between the XhoI and SalI sites of pBluescriptII SK(+).
Preparation of CEN3 beads
Plasmid DNA was covalently modified with biotin with photoprobe long arm biotin reagents (SP-1020, Vector Laboratories, Burlingame, CA) by thermal coupling (95°C, 30 min). Next, 60 μg of biotinylated plasmid DNA was incubated with 3 mg of streptavidin-coated paramagnetic beads (Dynabeads M-280 Streptavidin, Invitrogen Dynal AS, Oslo, Norway) in a buffer containing 1 M NaCl, 10 mM HEPES-KOH (pH 7.6) and 1 mM EDTA using gentle rotation at room temperature for 8 h. The beads were washed and equilibrated in a buffer containing 300 μl of 10 mM HEPES-KOH (pH 7.6) and 1 mM EDTA.
Preparation of highly concentrated protein extracts
Highly concentrated protein extracts were prepared as previously described [20] with some minor modifications. In brief, yeast cells grown in YPD at 25°C were harvested and washed twice with cell wash buffer (20 mM HEPES-KOH (pH 7.8), 1 M Sorbitol) at 4°C and once with 10 volumes of lysis buffer (100 mM HEPES-KOH (pH 7.8), 0.8 M Sorbitol, 50 mM potassium glutamate, 10 mM MgOAc, and 2 mM EDTA). The cells were resuspended in 0.25 volume (per weight) of lysis buffer containing 4 mM DTT and 4× protease inhibitor cocktail tablet (Roche, Indianapolis, IN). Yeast popcorn was prepared by dropping the cell suspension into liquid nitrogen. The frozen cells were disrupted in the electronic mortar grinder (RM100, Retsch, Germany) with liquid nitrogen for 1 h. After thawing the broken yeast powder, potassium glutamate was added to the homogenate to give a final concentration of 300 mM. The homogenate was incubated at 4°C for 30 min with gentle agitation and ultracentrifuged in a Beckman SW55 rotor at 33,000 rpm for 30 min at 4°C. The supernatant was also ultracentrifuged in a Beckman SW55 rotor at 55,000 rpm for 1 h at 4°C. The recovered supernatant was aliquotted, frozen in liquid nitrogen, and stored at −80°C. Typically, 10–15 g of the cell pellet was processed, and the protein concentration was from 50 to 100 mg/ml.
In vitro kinetochore assembly system
The in vitro kinetochore assembly system is based on the loading assay described elsewhere [20, 46]. Briefly, 35 μl of reaction buffer (57 mM HEPES-KOH [pH 7.6], 714 mM Sorbitol, 23 mM MgOAc, 5.7 mM EGTA, 2.3 mM DTT, 6.9 mM ATP, 46 mM creatine phosphate, 1.6 units of creatine phosphokinase, 1.1× protease inhibitor cocktail tablet, 5.1 μg/μl poly(dI-dC)•poly(dI-dC)) (Sigma-Aldrich, St. Louis, MO) was mixed with 40 μl of highly concentrated protein extracts and preincubated on ice for 10 min before addition of the Dynabeads. Next, 20 μl of 10 μg/μl bead suspension was placed on a magnetic separator, and 15 μl of supernatant was removed, leaving the beads in the tube. The preincubated mixture was added to the beads and mixed well by pipetting. The total volume of 80 μl of mixture was incubated using gentle rotation for 4 min at room temperature. The reaction was stopped by the addition of 9 vol (720 μl) of ice-cold wash buffer (50 mM HEPES-KOH (pH 7.6), 75 mM potassium glutamate, 1 mM EGTA, 5 mM MgOAc, 10% glycerol, 0.2% Triton X-100, 1 mM DTT, and 1× protease inhibitor cocktail tablet). After magnetic separation, the beads were washed 5 times with 800 μl of ice-cold wash buffer. The first wash also contained 0.1 μg/μl poly(dI-dC)•poly(dI-dC). Thereafter, the beads were washed once in 25 μl of 1% SDS, and the eluate was isolated by a magnetic separator. Finally, a 5-μl portion of Dynabeads (BEADS) or the supernatant (SUP) was loaded onto an SDS-polyacrylamide gel.
Supplementary Material
Acknowledgments
We thank V. Measday for her helpful comments; Y. Kawasaki for his technical advice; members of the Katsumi and Risa Kitagawa laboratories for stimulating conversation and advice; and P. Hieter, S. Buratowski, S.G. Oliver, and R. Deshaies for their generous gifts of reagents. This work was supported by the Cancer Center Support Grant CA21765 from the National Cancer Institute, by NIH grant GM68418, and by the American Lebanese Syrian Association Charities (ALSAC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kitagawa K, Hieter P. Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- 2.Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–718. doi: 10.1016/s0959-437x(02)00351-9. [DOI] [PubMed] [Google Scholar]
- 3.Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- 4.McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- 5.Hegemann JH, Fleig UN. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- 6.Mellor J, Jiang W, Funk M, Rathjen J, Barnes CA, Hinz T, Hegemann JH, Philippsen P. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 1990;9:4017–4026. doi: 10.1002/j.1460-2075.1990.tb07623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 8.Keith KC, Fitzgerald-Hayes M. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere dna around a cse4p variant nucleosome. Genetics. 2000;156:973–981. doi: 10.1093/genetics/156.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 11.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge JF. Centromeric chromatin in fission yeast. Front Biosci. 2008;13:3896–3905. doi: 10.2741/2977. [DOI] [PubMed] [Google Scholar]
- 13.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 15.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins KA, Castillo AR, Tatsutani SY, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jehn B, Niedenthal R, Hegemann JH. In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol Cell Biol. 1991;11:5212–5221. doi: 10.1128/mcb.11.10.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasaki Y, Kim HD, Kojima A, Seki T, Sugino A. Reconstitution of Saccharomyces cerevisiae prereplicative complex assembly in vitro. Genes Cells. 2006;11:745–756. doi: 10.1111/j.1365-2443.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- 21.Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol Cell. 1999;4:21–33. doi: 10.1016/s1097-2765(00)80184-7. [DOI] [PubMed] [Google Scholar]
- 22.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Hajra S, Ghosh SK, Jayaram M. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation. J Cell Biol. 2006;174:779–790. doi: 10.1083/jcb.200603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon AK. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Seraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 28.Cook JG, Bardwell L, Kron SJ, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 29.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 30.Chou S, Lane S, Liu H. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 32.Cai M, Davis RW. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- 33.Kent NA, Tsang JS, Crowther DJ, Mellor J. Chromatin structure modulation in Saccharomyces cerevisiae by centromere and promoter factor 1. Mol Cell Biol. 1994;14:5229–5241. doi: 10.1128/mcb.14.8.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellor J, Rathjen J, Jiang W, Barnes CA, Dowell SJ. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 1991;19:2961–2969. doi: 10.1093/nar/19.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su TC, Tamarkina E, Sadowski I. Organizational constraints on Ste12 cis-elements for a pheromone response in Saccharomyces cerevisiae. FEBS J. 2010;277:3235–3248. doi: 10.1111/j.1742-4658.2010.07728.x. [DOI] [PubMed] [Google Scholar]
- 36.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tachibana C, Yoo JY, Tagne JB, Kacherovsky N, Lee TI, Young ET. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25:2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemmerich P, Stoyan T, Wieland G, Koch M, Lechner J, Diekmann S. Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc Natl Acad Sci U S A. 2000;97:12583–12588. doi: 10.1073/pnas.97.23.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 40.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 42.Choi ES, Stralfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. Identification of Noncoding Transcripts from within CENP-A Chromatin at Fission Yeast Centromeres. The Journal of biological chemistry. 2011;286:23600–23607. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. The EMBO journal. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 45.Sharp JA, Krawitz DC, Gardner KA, Fox CA, Kaufman PD. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 2003;17:2356–2361. doi: 10.1101/gad.1131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seki T, Diffley JF. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci U S A. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.