Abstract
The rapid pace of discoveries in tumor biology, imaging technology, and human genetics hold promise for an era of personalized oncology care. The successful development of a handful of new targeted agents has generated much hope and hype about the delivery of safer and more effective new treatments for cancer. The design and conduct of clinical trials has not yet adjusted to a new era of personalized oncology and so we are more in transition to that era than in it. With the development of treatments for breast cancer as a model, we review the approaches to clinical trials and development of novel therapeutics in the prior era of population oncology, the current transitional era, and the future era of personalized oncology.
Keywords: Clinical Trials as Topic [E05.318.760.535], Individualized Medicine [E02.574], Antineoplastic Agents [D27.505.954.248], Breast Neoplasms [C04.588.180]
Introduction
If it were not for the great variability among individuals medicine might as well be a science and not an art. William Osler, 1892
Medicine has always been personalized. Cancer specialists have long appreciated that each patient presents with her/his own unique clinical history, prognosis, treatment tolerance, and supportive care needs. Physicians have always focused on what is best for each individual patient1. Advances in both biology and information technology have brought ‘personalization’ forward as a new buzzword in healthcare. For the last two decades, laboratory scientists, clinicians, and epidemiologists have applied technological advances so that we now recognize that just as patients differ in how they are affected by their diseases, cancers have unique natural histories with distinctive biology. Tumors once described solely by their organs of origin now comprise subsets with different biological drivers and clinical outcomes. Artful humanistic skills remain important to personalizing cancer care, but science-based tools increasingly guide the design of a patient’s individualized treatment plan.
Advances in cancer care with potential for widespread impact are made through clinical trials. These studies provide structure to the selection of patients, control the delivery of standard and test interventions, and provide reliable estimates of the therapeutic benefit of the interventions tested when they are delivered to patients similar to those enrolled and treated on the study protocol. Acknowledging the limitations to the generalizability of clinical trial results, they remain the primary means by which advances in science and drug development can be translated into advances in patient care throughout health systems.
Technological innovations and scientific advances in understanding cancer at the molecular level have accelerated discovery and development of diagnostics and therapeutics. Today an oncologist can perform some impressive feats that were just exciting new ideas in research 10 years ago. At the start of the day’s clinic, the clinician could cure a woman with HER2 gene-amplified breast cancer by adding trastuzumab to standard adjuvant chemotherapy. Just after lunchtime, the same oncologist can review a report on results of polymerase chain reaction tests on DNA in the peripheral blood of an asymptomatic patient with chronic myelogenous leukemia and if necessary, change the patient’s prescription from imatinib to dasatinib or nilotinib to maintain the patient’s clinical remission. At the end of the day, a patient with K-ras mutated metastatic colorectal cancer can be saved unnecessary toxicity and cost by discussing not to proceed with cetuximab or panitumumab treatments.
Scientists and clinicians celebrate these advances. They get excited by the potential for new technologies and insights to lead to more life-changing breakthroughs for patients. But observers outside of cancer research argue that these successes are too small and too few2, 3. Despite new technologies for drug discovery, novel cancer therapeutics typically fail to complete clinical development4, 5. Although we savor the promise of a new era of personalized oncology, we are more transitioning into that era than truly there.
Progress to personalized oncology requires advances in the design and conduct of clinical trials. The ultimate goal for personalized oncology is to maximize the therapeutic index for treating or curing cancer in each patient. In medical oncology specifically, this means selecting the right drug and administering it at the dose that produces the maximum efficacy and with the least toxicity every time, for every patient6. During this period of transition from population-based to personalized oncology, cancer investigators are facing two concurrent and sometimes competing challenges: 1) conducting clinical trials of new, potentially more effective therapeutic interventions, more quickly than ever before and 2) transforming the clinical trials infrastructure and designs from those suited for the era of population oncology to those necessary in the era of personalized oncology.
To illustrate how advances in laboratory and clinical science have propelled us into the current transitional period and how clinical trials must evolve to lead us into the era of personalized oncology this article will focus on systemic therapy options for treatment of invasive breast cancer in the adjuvant and advanced disease settings (Table 1). Among solid tumors, breast cancer treatment arguably has made some of the greatest advances during the previous three decades. During that period, breast cancer was approached as a homogeneous disease, except for the recognition of hormone responsive or unresponsive tumors identified by expression of the estrogen receptor. The goal was to reduce suffering and death from this disease. The strategy entailed public health approaches such as raising awareness, developing screening and early detection methods to reduce the risk for death from breast cancer, and studying surgical and adjuvant therapy methods to improve cure rates while reducing morbidity. Treatments were developed first by testing for safety and signs of efficacy in cohorts of patients with advanced disease with intent to then test promising drugs in earlier stages of disease to increase cure rates. The focus of this strategy was to improve outcomes for the entire population of women at risk for or who developed breast cancer and so we refer to this period as the era of population oncology.
TABLE 1.
Oncology Care and Clinical Trials in the Eras of Population Oncology, Transition, and Personalized Oncology
| Population Oncology | Transitional Era | Personalized Oncology | |
|---|---|---|---|
| Screening | population-wide risk reduction | population-wide approaches modified for at-risk sub-populations | individualized risk estimation & programs adapted to individual risk |
| Diagnosis | organ-of-origin/histology-based | organ-of-origin, histology, & some molecular markers | primarily molecular marker-based |
| Staging | anatomic extent of disease | anatomic extent with some molecular risk profiling | primarily molecular-risk based |
| Treatment Determination | typically organ-of-origin & stage-based | organ-of-origin & stage-based with some implementation of molecular markers | primarily molecular marker-based |
| Assessment Intervals | based on clinical evaluation/exam findings | based on routine interval imaging | early, frequent serial assessments by imaging, circulating tumor cells and other marker assessments |
| Early Phase Clinical Trials | oriented to maximum tolerated dose | oriented to “optimum biologic dose” | determine range of tolerable & active doses |
| Mid-Phase Clinical Trials | histology & prior treatment based eligibility; typically single arm non-comparator trials | histology & prior treatment based eligibility; some marker-based screening; some randomized, controlled trials | some trials histology & prior treatment based eligibility with rapid, serial assessments; many with eligibility restricted to tumor marker subsets |
The landmark studies of trastuzumab, for patients with a specific sub-type of breast cancer, signaled the beginning of the current transitional period in cancer clinical trials. We provide a framework for understanding how clinical trial designs and operations need to adapt in order for us to deliver more scientifically based personalized cancer care and describe some promising innovations that will likely usher us into the era of personalized oncology.
Clinical Trials in the Era of Population Oncology
Perspective on clinical trials and progress in breast cancer care
The experiences of our grandmothers with breast cancer illustrate the critical importance of clinical trials to progress in cancer care. The senior author has already described the disappointment and agony his grandmother experienced after being diagnosed with breast cancer in 19651. She underwent radical mastectomy, chest wall radiation associated with severe skin toxicity, developed chronic lymphedema and developed a second primary tumor in the contralateral breast. This resulted in another radical mastectomy, more radiation and within a few more years, progressive metastases, debility, ineffective systemic hormonal therapy, and death preceded by much suffering.
Twenty-five years later, the junior author’s grandmother (E.L.) had Stage II breast cancer identified on screening mammography. She underwent lumpectomy with clean margins, and resected axillary lymph nodes revealed no evidence of metastases. She received adjuvant radiation therapy based on the results of the National Surgical Adjuvant Breast and Bowel Project (NSABP) trial B-067, 8. Near the completion of the planned 25 fractions of radiation she developed uncomfortable skin toxicity. Her radiation oncologist encouraged her to complete the 25 fraction course and additional fractions focused on the tumor site as a “boost”, an emerging standard of care based on yet-to-be published clinical trials. Without evidence of a survival benefit for the boost approach and weary from the treatment, she elected to complete 25 fractions and not to proceed with further radiotherapy. Twenty years later, she remains free from tumor recurrence and shortly after being interviewed for this article, she attended a jazz concert.
E.L. was cured of her disease with less morbidity than the previous standard-of-care, modified radical mastectomy. Her suitability for this treatment approach (her age, stage, and absence of co-morbidities), guidance on the execution of the surgery (achieving an adequate margin for breast conserving surgery), the dose and schedule of her radiation treatment (50 Gy in 25 fractions) and prognostic expectations for her freedom from local recurrence (90% over 8 years) were based on multiple clinical trials culminating in NSABP B-06 and subsequent analyses of data collected in that trial. Her individualized treatment plan included a decision not to proceed with the tumor site “boost” or to receive adjuvant hormonal therapy. These decisions were not scientifically-based, but rather were based on E.L.’s unwillingness to experience short-term toxicities of additional therapy and intuition that her disease was likely cured.
Paclitaxel for breast cancer, archetype drug of the population oncology era
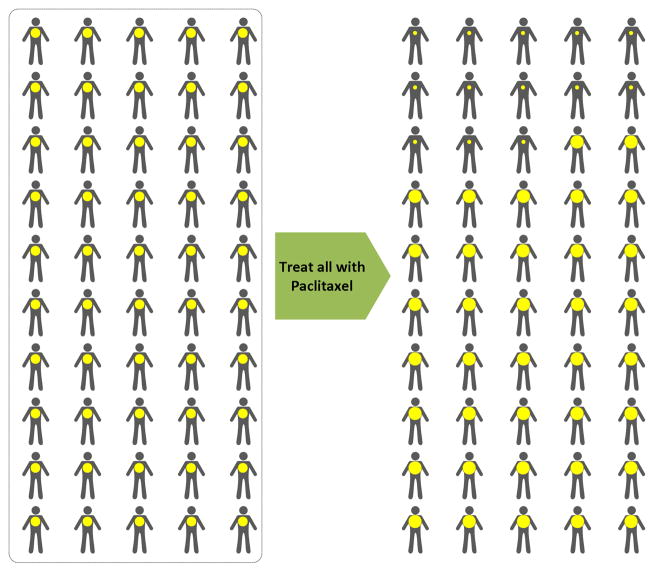
At the time E.L. confronted her breast cancer, paclitaxel was under active development as a novel agent for systemic treatment of advanced breast cancer. The first major publication, characteristic of clinical trials of that period, reported a single arm study of 25 patients with metastatic breast cancer that progressed despite previous systemic therapy9. No agents were known to be effective on disease at this advanced state; therefore a comparator arm was not necessary. Having even a small fraction of patients with objective evidence of tumor shrinkage (the response rate), would be sufficient evidence of drug effect to warrant further study. Standardized methods of assessing and reporting toxicities and treatment responses were used. In this study, 3 patients developed complete responses and another 11 showed objective evidence of tumor shrinkage. Two other similarly designed clinical trials were conducted at other institutions with minor differences in the range of doses tested, the supportive care delivered, and the number of prior therapies allowed for eligible patients10, 11. Both studies used the same 24 hour drug infusion and response rates of 30–57% were observed. A subsequent phase 3 trial, of 3-hour infusions of paclitaxel, randomized advanced breast cancer patients to receive 135 or 175mg/m2 12. Again, with no known standard of care for such patients, there was no standard comparator or placebo control arm. The overall objective response rate of 26% in the multi-center setting with evidence of dose-response effect was sufficient to justify approval of paclitaxel by the United States Food and Drug Administration for treatment of metastatic disease after previous systemic therapy. As authors of that study concluded, “The full impact of this novel agent in the treatment of breast cancer is being evaluated in large trials that use combination chemotherapy and involve earlier stages of disease.”
In the era of population oncology, this approach to the development of a novel anticancer agent was reasonable and efficient. Testing for safe dosing and evidence of anticancer activity was conducted in cohorts of willing patients for whom there was no established standard of care. The patient population was defined by the organ of origin, the extent of disease, and the previous treatment history (Fig. 1). The findings from studies in this population were extrapolated to similar populations of patients in community oncology practice. Concurrently, pharmacokinetics and pharmacodynamics data from these trials informed the further development of the drug in populations of patients with other diseases and in combination regimens administered to breast cancer patients with curable, less extensive disease. With additional clinical trials, paclitaxel became a standard agent in the treatment of metastatic breast cancer. Paclitaxel also became a component of effective adjuvant therapy regimens designed to cure early stage breast cancer13, 14. Representative of clinical trials in the Era of Population Oncology, these studies enrolled almost 6,200 patients to demonstrate a small but statistically significant reduction in risk for disease recurrence as no methods were available to identify patients likely to benefit. A more recent paper from the current Transitional Era suggests that the benefit of adding paclitaxel as a component of adjuvant chemotherapy is enjoyed only by a subset of patients that can be characterized by the specific tumor marker Her-2 described in the next section15.
FIG 1.
Clinical Trial In Era of Population Oncology
Clinical Trials and Diagnostics Development in the Transitional Era
Trastuzumab, archetype drug of the transitional era
During the 1990’s, as paclitaxel was developed for other indications and disease settings, the testing of a novel breast cancer treatment, trastuzumab, heralded the beginning of the current transitional era. This monoclonal antibody binds to Her-2, a surface receptor aberrantly expressed in 25–30% of breast cancer patients. Her-2 expression was associated with a poor prognosis, and laboratory studies demonstrated that Her-2 expression played a direct role in aggressive tumor growth. A monoclonal antibody that binds this protein stopped the growth of tumor cells expressing Her-2 in vitro and demonstrated evidence of synergistic effects when combined with chemotherapy. In the initial phase I16 and phase II monotherapy trials17, the drug could be safely administered to achieve serum concentrations in patients that inhibited growth of Her-2 over-expressing tumors in animal models. Few patients experienced acute toxicities other than infusion reactions.
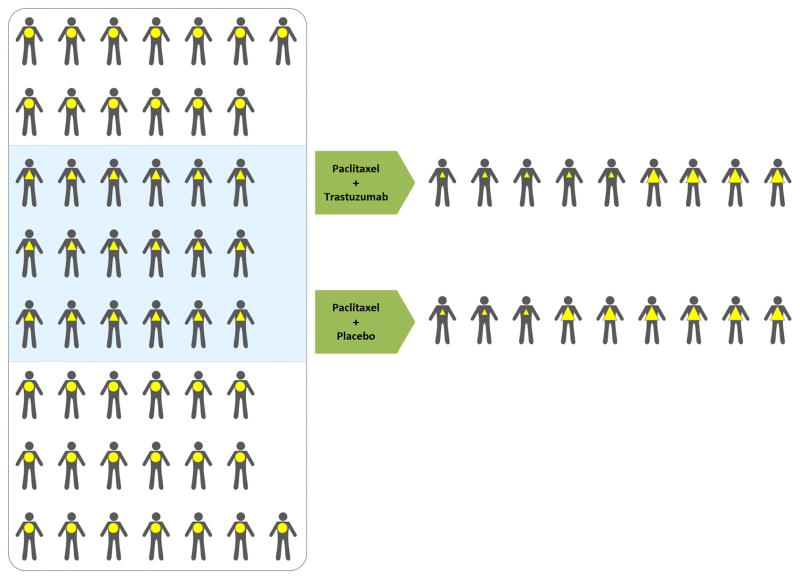
These findings spurred enthusiasm for the 3-point concept of “targeted therapy”: 1) develop a drug to interfere with the function of a molecule readily identified in cancer cells but not healthy tissues, 2) know that this target plays a critical role in the abnormal growth and/or invasiveness of the tumor, and 3) expect that the relative specificity of the drug for the target molecule and the target molecule for tumor cells rather than normal cells would achieve dramatic improvements in the therapeutic index over standard cancer chemotherapy. Establishing this new paradigm for cancer drug development, patients were selected for the phase II trials based on criteria to enhance the likelihood of successful clinical development. First, patients typically had recent failure of previous systemic therapy (patients with evidence of active disease progression rather than indolent disease). Second, patients with metastases that could be convincingly assessed by serial CT imaging were preferentially enrolled (although bone metastases are common in breast cancer, their response to therapy is difficult to measure reproducibly). Third, to be enrolled patients had to have evidence of tumor Her-2 overexpression detected with an immunohistochemical reagent that bound the same epitope of Her-2 as trastuzumab itself. With this selective approach, single arm phase II trials demonstrated objective response rates higher than expected for a typical advanced breast cancer patient population, even considering the more aggressive natural history of Her-2 expressing breast cancer.
The successful completion of randomized phase III trials of trastuzumab validated the targeted anti-cancer therapy paradigm. In combination with chemotherapy in the treatment of metastatic disease18 and subsequently in the adjuvant therapy setting19, 20, the addition of trastuzumab to standard treatment improved median survival in patients who had Her-2 overexpressing breast cancer. Recall that, by the original immunohistochemical (IHC) assay criteria, 25–30% of all breast cancer patients have Her-2 overexpressing tumors (Fig. 2). If these trials had been performed in all advanced breast cancer patients, with and without Her-2 overexpression, they would likely have had negative results. For example, in the study of patients with metastatic disease and 2+ or 3+ staining of the tumor for Her-2 expression, the response rate was 50% and one-year mortality was 22% for the trastuzumab treated arm and 32% and 33% respectively for the control arm (Fig. 2). By extrapolation21, without selection based on Her-2 expression, the response rate would have been 37% and one-year mortality rate 30%. The difference between the treatment and control arms with this number of patients would not have been statistically significant In this case; the pre-selection of only patients with Her-2 over-expressing tumors enabled detecting the therapeutic benefit. Without focusing the clinical trial on the correct subset of the breast cancer patient population an effective therapy might well have been discarded.
Fig 2.
First Clinical Trial In The Transitional Era
Developing therapeutics for biomarker-defined sub-populations consistently
One of the most striking elements of the successful development of trastuzumab was that it converted a biomarker for negative prognosis (Her-2) into a biomarker predictive for benefit from the drug. A fundamental challenge for the transitional era has been the concurrent development of anti-cancer therapeutics and biomarkers. The question remains for all agents developed with the “targeted therapy” concept - can the sub-population most likely to benefit from therapy be readily and reliably identified?
For trastuzumab the target disrupted by the drug truly drives progression of disease and/or resistance to other therapies. For many other promising agents in development in the early part of the transitional era, this has not been the case. Many cancer therapeutics programs were based on specific targets that appeared to be important in cultured tumor cells, animal models, and/or human disease. In some cases this evidence may not have been as strong as for Her-2. An exemplary case is the development of marimastat, an inhibitor of several matrix metalloproteinases, enzymes thought to be important in tumor invasion and metastasis. However, confirmation of the importance of the target during the early clinical development was lacking. Despite the limited evidence, marimastat and other metalloproteinase inhibitors proceeded to phase III trials only then to demonstrate convincingly that they did not have therapeutic benefit22. Such failures have been more common than successes such as trastuzumab5, not due to failure of the drug as much as to failure to verify the importance of the drug target in cancer biology.
Even with successes such as trastuzumab, when an assay determines the sub-population of patients who are prescribed a life-saving drug, the reliability of the assay becomes crucial23. Ideally the test should identify all the patients in whom the drug has therapeutic benefits and exclude those in whom it does not. Although the IHC assay for detecting overexpression of Her-2 defined a population of patients for whom trastuzumab was clearly helpful, this test is imperfect. There are some patients who would benefit from trastuzumab but have a negative test and there are others who test positive but do not benefit from trastuzumab. Indeed, one retrospective study demonstrated that detection of Her-2 overexpression by either IHC or FISH was not necessary for patients to benefit from the addition of trastuzumab to adjuvant chemotherapy24. As a single retrospective study this has not changed clinical practice recommendations but has led to further research to determine how best to use trastuzumab and to discover new tumor biomarkers that identify patients likely to benefit from the drug. Consequently, an important aspect of developing therapeutics based on biomarker testing is to recognize that the assay method that proves a drug can be effective may not be a “gold standard” for identifying all the patients who will and won’t benefit from the therapy.
During the transitional era new therapeutics such as imatinib (and more recently nilotinib and dasatinib) for CML and trastuzumab for Her-2 expressing breast cancer have brought significant benefits to sub-populations of cancer patients. To replicate these successes in other diseases, efforts are increasingly focused on development of drugs with companion diagnostics that identify patients likely to benefit from treatment and study designs to confirm efficacy of a treatment in a subset of patients 25–30. Excitement for these approaches has been building with the recently regulatory approvals of vemurafenib for advanced melanoma that harbors a BRAF V600E mutation and crizotinib in non small cell lung cancer with an EML4-ALK translication. Yet, we are not routinely delivering on the promise of targeted therapy as for some agents that have been recently approved (for example erlotinib in the treatment of pancreas cancer), the benefits are very small, and the subsets of the population that derive the greatest benefit have not been identified.
When we can routinely identify sub-populations that will benefit from a treatment our current concepts of cancer therapy will change. Throughout the population-based era to the present, we have approached treatment of tumors based on the organ of origin. Again, the development of trastuzumab demonstrates our transition to the era of personalized oncology. Although Her-2 was originally considered a target for breast cancer therapy, adding trastuzumab to the standard treatment for advanced gastroesophageal adenocarcinomas prolonged survival in those patients whose tumors overexpressed Her-231. This proves the concept that in the future, tumors will likely be treated based on their specific molecular alterations rather than organ of origin. Clinical trials in the era of personalized oncology will have to select patients in a manner to determine efficiently the sub-population most likely to benefit, regardless of the site of disease origin.
Advances in understanding tumor biology
The optimism for a future of personalized therapy derives from the great strides scientists have made in understanding tumor biology, especially the molecular factors driving different cancers. These discoveries have provided valuable insights for how to approach cancer therapeutics development and individualized therapy. An example is the study of gene expression in breast tumors.
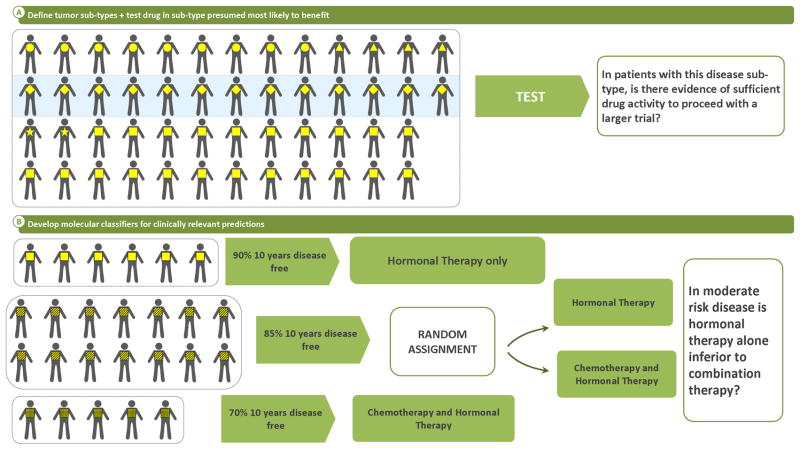
DNA microarrays, “gene chips”, have enabled the simultaneous study of the expression of thousands of genes in a tumor at the time of biopsy or resection. In the past decade, two fundamental applications of this technology have advanced our understanding of breast cancer therapeutics. The first generated a common gene-expression-profile-based molecular sub-classification system32. Through study of genes relevant to tumor growth, Sorlie, Perou, et al. generated a novel, reproducible categorization system for breast cancer that has begun replacing the widely used categories defined by IHC staining (eg. “hormone receptor positive/Her-2 negative”). The new classification demonstrates different natural histories for basal-like, ERBB2+ (Her-2 expressing), normal-breast-like, and luminal (estrogen-receptor-expressing) sub-types of breast cancer. The basal-like category comprises tumors described as “triple negative” (not expressing estrogen or progesterone receptors or Her-2) and those occurring in women who carry BRCA1 mutations. The gene expression pattern in these basal-like tumors is so distinct that it is now recognized as a separate disease entity and is being treated very differently from the rest of breast cancer33–36. Similarly the other categories have differing prognoses and are associated with sensitivity and resistance to different therapies for breast cancer. Tumor gene-expression profiling has therefore identified sub-populations of breast cancer not previously recognized to be related. Since these tumors share collections of “driving forces” among several aberrantly functioning gene pathways, and therefore are likely to respond to similar therapeutic interventions, ongoing studies can now collect these sub-populations into a single clinical trial. One example is a current study of the polyADP-ribose polymerase inhibitor, veliparib (NCT 00892736), in which patients with BRCA 1 or 2 mutations or the triple-negative phenotype are all eligible based on the expectation that basal-like tumors are likely to benefit from this treatment (Fig 3A). Without requiring patients to have tumor gene-expression profiling, these methods have refined the design of clinical trials to test therapeutic interventions with likelihood for greater impact on specific subsets of breast cancer patients more so than was possible during the development of paclitaxel or trastuzumab.
Fig 3.
Current Clinical Trial Strategies
The second application of expression array analysis of breast cancer has been the prognostic evaluation of luminal subtypes of breast cancer. For many patients with this disease who do not have regional lymph node metastases, hormonal therapies may be sufficient curative treatment. Although commonly added, systemic chemotherapy may provide no additional benefit. Two predictor algorithms for refining prognosis and identifying the subset of patients who do not benefit from adding chemotherapy were developed through iterative testing. One method (Oncotype Dx™, Genomic Health, Redwood City, CA) uses formalin-fixed paraffin-embedded tissue37, while the other (Mammaprint™, Agendia B.V., Amsterdam, The Netherlands) requires tumor tissue freshly preserved after resection for RNA extraction38. One recently completed trial, TAILORx (Trial Assigning Individualized Options for Treatment) for Oncotype Dx™39 (Fig. 3B) and one ongoing study, MINDACT (Microarray in Node-negative Disease may Avoid ChemoTherapy trial) for Mammaprint™40, have been designed to confirm prospectively the safety of withholding chemotherapy from patients predicted by these genome profiles to be at low-risk of recurrence. These studies have also been designed to refine the genomic tests to expand their ability to predict other clinical outcomes.
The impact of human genome sequencing on advances in oncology.
Many novel technologies were developed during the course of the Human Genome Project. Many interrelated methods can be applied to the personalization of cancer care. In oncology, the major areas of active investigation are focused on: the patient’s (germline) genome, the tumor genome, and tumor gene-expression.
Germline genome
The intrinsic variation in our germline DNA comprises millions of single nucleotide polymorphisms and other variations in genome sequence that make each of us unique individuals. Some of these polymorphisms can contribute to interindividual differences in risk for developing a particular cancer, differences in our metabolism of envrionmental carcinogens or anticancer drugs, differences in our bodies’ responses to the drugs, and our bodies’ tolerance to treatment and ability to mount an immune response to cancer. So analysis of the germline genome has much to offer in understanding how best to individualize prevention, screening, and treatment strategies for an individual patient.
Tumor genome
Tumors arise from alterations to the germline DNA in individual cells that lead to the changes in cell function that are manifest as the hallmarks of cancer41. Identifying these somatic changes to the germline sequence in the tumor can facilitate development of targeted therapies and selection of patients most likely to benefit from a particular treatment. Prior to development of technologies for rapid DNA sequencing, study of changes in DNA structure through microscopic examination of tumor chromosomes (cytogenetics) was helpful to sub-categorizing hematologic malignancies, particularly leukemias. These cytogenetic markers are now routinely used to establish the diagnosis, determine the prognosis and select treatment for patients with myeloid leukemias. Current molecular genetic technologies such as Fluorescence In Situ Hybridization (FISH) enable more sensitive and specific evaluation of leukemia and solid tumor cells. FISH is used to detect amplification of the gene encoding Her-2, ERBB2 in breast cancer and in lung cancer detects the translocation that fuses ALK to EML4 and predicts responsiveness to the kinase inhibitor crizotinib.
Sequencing specific regions of the tumor genome can detect alterations not detectable by cytogenetic analysis or FISH. For example identifying particular mutations that confer resistance, such as KRAS mutations and cetuximab, or sensitivity, such as BRAF V600E mutations and vemurafenib, to specific therapeutics can guide selection of treatment. The next generation of sequencing technology should enable more comprehensive characterization of the numerous additional tumor genome alterations. Some of these are the “driver” mutations that promote growth and metastasis. These are considered the primary targets for therapeutic intervention. Other mutations arise through the rapid growth and genetic instability of tumor cells. Although these “passenger” mutations might not be necessary to initiate or sustain tumor growth, they can confer resistance to specific therapies and affect the growth/metastasis profile of the tumor. Alternatively, passenger mutations can be simply genetic alterations that are detectable but have no bearing on the natural history of the cancer.
Tumor genome expression profiling
Microarrays of sequence fragments from genes can be used to monitor relative differences in mRNA expression (increases or decreases relative to a reference- normal tissue, other tumors, etc.) of thousands of genes simultaneously within tumor cells. Aside from informing tumor biology in general, the most clinically relevant use to date has been in gene expression profiling to subcategorize breast cancers and refine prognosis in breast cancers that by all currently available means would be considered intermediate risk. Other applications, such as examining profiles or changes in profiles to predict sensitivity/resistance to specific cancer therapies have been proposed but have not been validated in the clinical setting42–45.
Advances in understanding human biology and genetic variation
Some of the initial predictions of how the completion of human genome sequencing would affect medical practice in 2011 have come true46, but those promising to affect medical decision-making directly and routinely remain unfulfilled. Most of the published genotype/phenotype association studies in the oncology literature have reported on inadequately sized samples or incompletely justified hypotheses47–49. More recently, genome-wide association studies50–55 have identified genetic risk factors for developing breast cancer. These common genetic polymorphisms only subtly affect individual patients’ risks56, 57, but other genetic variants can play more significant roles in the treatment outcomes for breast cancer. For example, women homozygous for alleles that lead to defective CYP2D6 protein production may receive no benefit from tamoxifen therapy58. These patients are unable to convert tamoxifen to its most potent estrogen receptor-modulating metabolite59. This discovery was made after all clinical trials that proved the value of tamoxifen in breast cancer care were completed. Retrospective studies demonstrated that including CYP2D6-deficient patients in studies of tamoxifen efficacy likely diminished the estimated efficacy of tamoxifen. If women who were CYP2D6-deficient were excluded from tamoxifen trials, the drug would likely have shown even greater efficacy than was demonstrated by the original data.
The cost of genome sequencing is declining exponentially. Within the next five years, sequencing a patient’s entire genome is projected to cost approximately $1,00060. Although having genome sequence information will become increasingly useful over the course of an individual’s lifespan, in the immediate future, the volumes of genome data will overwhelm our capacity to interpret the data or use them in validated, reliable ways except in few cases (Fig. 4).
Fig 4.
Viewing Patient and Disease Individuality In the Era of Personalized Oncology
The ready availability of genome sequences will challenge the delivery of personalized healthcare beyond oncology. Except for identical twins, each individual differs from every other by millions of variations in the genome. Identifying which of these genetic variants can and will affect clinical outcomes can be proved in two ways: 1) studying enough individuals with the same genetic variants and similar clinical scenarios to determine the relationship empirically, or 2) conducting serial mechanistic studies to demonstrate the effects of a genetic variant on gene or protein function and then to predict the clinical significance of that altered function. But once a gene variant-clinical outcome relationship is proved, current principles of evidence-based medicine require a randomized prospective confirmation of the hypothesis for treatment decision-making as a standard to justify widespread implementation. For example, answering the question of whether CYP2D6 genotype information is useful in making a clinical decision to use tamoxifen in post-menopausal women with breast cancer is a crucial proof-of-concept for pharmacogenomics and oncology. In this case, the gene variants are common and expected to have major effects on clinical outcomes. But a prospective test of whether treating a woman with normal CYP2D6 function with tamoxifen and a woman with diminished CYP2D6 activity with an aromatase inhibitor yields the same benefit in recurrence free survival will be expensive and not in the commercial interest of any drug-maker. It is a challenge to prove the value of a set of common genetic variants in this one clinical setting. Setting these expectations of proof and applying current clinical trial methods for every use of germline genomic data in patient care is unrealistic. These standards of proof threaten to stifle our ability to deliver personalized cancer care, especially in cases of rare genetic variants found in only small fractions of the population or in less critical clinical circumstances. To deliver on the promise of human genome sequencing for personalizing therapy will require more innovative data collection and analysis methods than the randomized prospective clinical trial. One novel approach has been serial empiric testing described in detail in the next section61, 62.
Advances in the collection and analysis of clinical data
The advances in tumor biology, human biology and genetic variation described above would not have been possible without concomitant advances in information technology. The storage and interpretation of the volumes of data generated in microarray and genome sequencing studies were based on new methods of analysis. These were made possible with software that exploited declining costs in hardware for data storage and high-speed central processors. These technologies likewise, can be applied to and advance the conduct and analysis of cancer clinical trial data.
Clinical trials in other fields of medicine have overcome fundamental challenges through novel methods of trial design and data analysis63. One major current challenge to the conduct of oncology clinical trials is the reliance on the standardized Response Evaluation Criteria in Solid Tumors (RECIST)64. This categorical system of imaging-determined tumor responses to therapy is used for calculating response rate and progression-free survival in clinical trials. Newer, functional imaging techniques have potential to advance the development of new drugs and to identify earlier in the course of an individual patient’s treatment whether or not her/his disease is responding65–67. However, these new techniques are expensive and how readily they can be standardized across institutions remains to be determined. With CT imaging as the current community standard for clinical trial imaging68, advances in the reliability and reproducibility of CT digital imaging data, using more quantitative information from each patient could accelerate the pace at which new drugs are developed69.
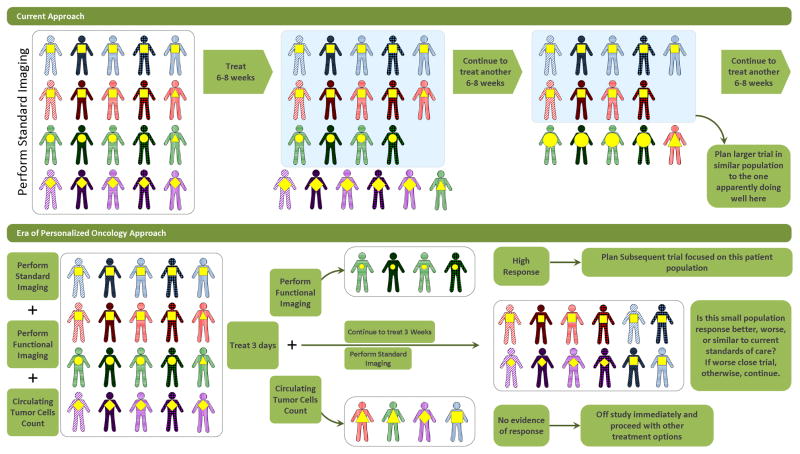
Computationally intensive techniques of studying drugs, a growing field referred to as pharmacometrics70–72, is an innovative approach to advancing oncology clinical trials. In 2009, two seminal papers were published describing the successful development and potential uses of statistical models of tumor growth in humans73, 74. Using tumor size measurements and treatment histories from patients enrolled in a phase II trial of capecitabine and a phase III trial of 5-fluorouracil, Claret, et al., developed a mathematical model of tumor growth and patient survival in advanced colorectal cancer (CRC)73. Similarly, Wang, et al., collected longitudinal tumor growth and patient survival data from 3,398 non-small cell lung cancer (NSCLC) patients enrolled in 4 clinical trials submitted to the FDA74. In both cases, the major factors predicting survival were the baseline sum of the longest dimensions of the measured tumors and the fractional change in tumor size at the end of two cycles of therapy (7 weeks in CRC and 8 weeks in NSCLC). These statistical models that describe the typical history of disease progression in a population have important downstream applications for clinical trials in the era of personalized oncology. First, these models suggest that using quantitative changes in tumor size rather than categorical endpoints may shorten the time to completion of an informative clinical trial75, 76. Second, these models have a dynamic structure that can incorporate information from newer imaging technologies such as the digital measurement of tumor volume69, 77 to further speed the assessment of novel therapeutics. Third, individuals who significantly deviate from the typical projected pattern of disease progression can be identified early in a clinical trial as “outliers” and studied intensively to understand better the clinical and tumor molecular features that might explain their observed response to treatment (Fig. 5). At the present time these models are most useful as informative tools in the early clinical trials of develop novel agents and combinations. Although there is cause for optimism in the efficiencies that could be achieved with such models, they have not yet been sufficiently well studied to substitute for the typical clinical benefit endpoints such as overall survival used in confirmatory phase III trials.
Fig 5.
Potential Clinical Trial Strategies in the Era of Personalized Oncology/Quicker Assessment of Disease response
Delivering effective personalized therapy without requiring clinical trials is another potential advance enabled by information technology and pharmacometrics. The concepts and tools required to use individual patients’ therapy experiences to inform treatment decisions in subsequent patients have already been implemented in pediatric oncology. At Children’s Hospital of Philadelphia, researchers have developed basic information technology tools and integrated pharmacometrics methods to give physicians real-time information on how an individual patient’s metabolism of a drug (methotrexate) compares with the rest of the population of previously treated patients. A mathematical model that describes the serial concentrations of serum methotrexate measured in patients previously treated with the drug serves as the “back-end” of the system. At the “front-end” is a visual display, what the researchers call “the drug-specific dashboard.” The dashboard displays collected data such as serum methotrexate concentrations from the individual patient, a population average based on the mathematical model, and the projected course of methotrexate elimination for the individual patient, based on the mathematical model. The back-end of the system also collects each new individual patient’s data. A research team periodically reviews the collected data and modifies the model to improve its performance for future patients. This interactive decision support system can guide physicians on a safe course of further management during each patient’s cycle of therapy61. For example based on the displayed data, a physician might adjust the dose or schedule of the folinic acid therapy to be administered. Currently these decisions are made empirically, based on established nomograms. With the new information technology, the nomogram can be revised over time to improve its performance as more patients’ data are entered into the system.
These same investigators and others in their field78 have undertaken new efforts to link diverse data sets across the entire translational research enterprise. The basic concept has two elements: 1) to connect information relevant to human patient cancer therapy from other fields of medicine and experience treating other diseases (horizontal integration) and 2) to connect information from in vitro assays, animal models, individual patients, and populations (vertical integration) to develop more mechanistic understanding of determinants of treatment success and failure. This systems pharmacology approach offers the potential to make treatment decisions for patients based on the most relevant mechanistic and empiric data without relying on randomized phase III clinical trials. This methodology does periodically need to be subjected to testing, but some of these questions are best answered with a prospective clinical trial while others might best be answered by continuous, modest improvement in information technology engineering. For example, whether guiding leucovorin rescue with a dashboard/decision-support tool really yields better outcomes of safety and efficacy is a question that can be tested in a clinical trial comparing patients treated with the dashboard technique versus a cohort treated with current standards of care. But to determine whether including a specific set of genetic polymorphisms in the mathematical model improves the performance of the dashboard/decision-support tool is better addressed through repeated in silico testing on existing data sets.
Obviously, this approach is currently limited and inappropriate for almost all cases of treatment decision-making today, but in the era of personalized oncology such information, not involving clinical trials, will be an important element of developing and delivering personalized cancer therapy. Clinical trials will then play a key role in comparing entire processes of personalized care across cohorts of patients rather than assessing each detail within each element of a process.
Clinical Trials in the Era of Personalized Oncology
The organization and completion of clinical trials of targeted therapies and molecular diagnostic tests are important rate-limiting steps to advancing personalized cancer care. The United States Food and Drug Administration has already described the strain which this “Critical Path” of drug development has been under for all fields of medicine6, 79. Strikingly, compared to the rest of medicine, oncology has been one of the worst under-performers in bringing new agents through the Critical Path 4, 5. In the introduction we explained that the transition into the era of personalized oncology raises two concurrent and competing challenges: 1) to conduct clinical trials of new, potentially more effective therapeutic interventions, more quickly than ever before and 2) to transform clinical trial infrastructure and designs from those suited for the era of population oncology to those necessary for the era of personalized oncology. A primary concern for this era is to allocate effectively the resources for conducting this research, and delivering to cancer patients the benefits of these trials more efficiently. We will truly enter the era of personalized oncology when clinical trials are routinely characterized by improvements in the 4 I’s: Involvement, Informativeness, Innovation, and Interconnectedness.
Involvement
Currently only 3–5% of cancer patients participate in clinical trials80. Many factors contribute to this low participation rate, but the most prominent are the financial disincentives to patients and community oncologists, and the redundant and inconsistent administrative burdens on clinical investigators for opening and conducting clinical trials81. To remove barriers to accrual and increase patient and clinician participation will require a coordinated efforts from the American Society of Clinical Oncology (ASCO), the National Cancer Institute, the U.S. Food & Drug Administration (FDA), and the Centers for Medicare & Medicaid Services (CMS).1
A basic need is for increased federal funding of cancer clinical trials and adequate reimbursement for all routine clinical care costs sustained by patients enrolled on trials. Managing these patients is time consuming and, complex. Oncologists should be reimbursed at a higher level for the greater time and effort devoted to providing care for patients in the context of clinical trials1. Although CMS and many private insurers cover routine care costs of clinical trials, patients can be left with prohibitive co-payments or in Employer Retirement Income Security Act (ERISA) plans, lack of coverage altogether. In states where care delivered in phase II clinical trials is covered, participation by patients in trials is significantly higher82. Elimination of co-payments for patients who enroll in clinical trials and amendment of ERISA and federal benefits plans to provide coverage for clinical trial participation would quickly increase the number of patients willing to participate in clinical trials.
Protection of human subjects who volunteer is essential to clinical research. The Institute of Medicine (IOM) and ASCO recently studied the array of federal laws such as the Health Insurance Portability and Accountability Act (HIPAA), the regulatory policies administered by CMS officials, and clinical trials oversight by the U.S. Food and Drug Administration. Concurrently the National Cancer Institute completed an operations analysis of typical clinical trial development. Opening a phase III trial in an NCI cooperative group now requires an average of 769 steps, 36 approvals, and 2.5 years from formal concept review to study opening83 -longer than it took to author, conduct, and publish the first CALGB trial.1 That first trial was opened in 1956 and compared continuous versus intermittent dosing of methotrexate and 6-mercaptopurine in the treatment of acute leukemia. The protocol was only 8 pages in length but included clear eligibility and exclusion criteria, required pre-treatment testing, plans for randomization and treatment, a description of anticipated toxicities and recommended supportive care measures, and provisions for central histomorphology review and response evaluation. The study enrolled 168 patients and was published in December 195884. With this striking accumulation of operational inefficiencies since that time, the IOM/ASCO report identified consistently ambiguous rules inconsistently interpreted as a major impediment to research. The NCI analysis determined an excess of regulatory requirements and “non-valued added” steps that contributed little to patient safety. These delays were imposed by no doubt well intentioned reviewers who were not accountable for timelines for efficient opening and completion of the clinical trial protocols. The NCI Operational Efficiency Working Group has established and is testing new policies to reduce the time to open and complete new clinical trials. Across national borders the International Conference on Harmonization has helped to make interpretation of regulations for U.S., European, and Asian agencies responsible for approving new medicines more consistent.
Informativeness
Clinical trials can be more informative through improvements in study design and in the collection and analysis of data generated by the trials. In the development of novel cancer therapeutics, there is a bias toward reporting positive results and interpreting small, non-comparative clinical trials as supporting further development and testing. This was evident in a recent analysis of combination chemotherapy trials published in 2001–200285. More than 360 phase II trials that combined more than 1 anticancer agent were published. More than 16,000 patients enrolled in these studies. Of these trials 72% concluded that the tested combination warranted further study. In the subsequent 5 year period only 10 tested combinations resulted in a phase III trial that established a new or improved standard-of-care treatment option. These facts beg two questions, 1) could more have been learned in the original trials before declaring them to be “positive”? and 2) would the collective clinical trials system have made more significant advances during this period if better designed clinical trials had enrolled more patients more rapidly? That is, could clinical trials have generated more useful information, more quickly?
The patients who volunteer to participate in early clinical trials of novel anticancer therapeutics are typically seeking therapeutic benefits, even if the likelihood of benefit is low86, 87. Thus, phase II oncology trials have typically tested the hypothesis that a new agent is worth further study, frequently leading to phase III trials, without disproving the null hypothesis (i.e., that the drug doesn’t work very well) 88. Conducting so many of these studies has come at the cost of not having optimized doses, schedules, and patient selection tools to improve the performance of our therapeutics. In the era of personalized oncology, administration of a new drug to each patient will be an opportunity to learn more about the drug and its therapeutic index. Therefore, regardless of the primary study endpoint, every trial should generate information to better guide development of therapeutics and their more effective use.
An information-generating approach to clinical studies led to an increase in cure rates of pediatric acute lymphoblastic leukemia from 21% in 1966 to 86% in 1997 without the introduction of a single new drug89. In the earlier part of the transitional era, the limitation on informativeness of individual studies not only was the study design90, but also the challenges to storing, sharing, and analyzing the data collected. The National Cancer Institute launched the Cancer Biomedical Informatics Grid (caBIG) project in 2004 as a pilot effort to set and promote uniform methods of data collection and storage across the cancer research enterprise. The goals of this project are: “to connect scientists and practitioners through a shareable and interoperable infrastructure, to develop standard rules and a common language to more easily share information, and to build or adapt tools for collecting, analyzing, integrating, and disseminating information associated with cancer research and care.”91 This sharing of information (while maintaining individual patients’ confidentiality) to support innovative research should usher in the era of personalized oncology.
Innovation and Interconnectedness
Innovation refers to the testing and implementation of novel approaches (clinical trial designs and operations, funding mechanisms, resource utilization, data collection, data analysis, etc.) to developing more effective therapeutics more efficiently than existing methods. Interconnectedness refers to more cooperative efforts across institutions, industry, and organizations to conduct clinical trials that will have the greatest impact on cancer care more efficiently. The breadth and technical complexity of new technologies that could advance personalized oncology care demands a more interconnected approach to the development of diagnostics and therapeutics for two reasons. First, as new promising technology emerges it can be expensive and difficult to replicate and standardize techniques much beyond the laboratory where the technology was initially developed. In this era of international overnight shipping, it now makes more sense to validate a novel technology and estimate its potential impact on care before committing additional resources to mass producing the necessary equipment and reagents to make that technology more widely available. Consortia of institutions that can standardize acquisition, processing and shipping of patient specimens may interconnect with each having a lab that specializes in different methods of specimen analysis. Such interconnected facilities may expedite development of personalized cancer therapeutics more efficiently than single centers. Second, as therapeutics are developed to treat small subsets of individual disease populations, the operations to perform trials in isolation with old methods become inefficient, almost untenable. An example is the development of the ALK (anaplastic lymphoma kinase) inhibitor crizotinib. The development of this agent has focused on the approximately 5% of all non-small cell lung cancer patients who have tumors harboring a chromosomal translocation that abnormally activates the ALK gene. Patients have tumor tissue screened for the ALK translocation at a central laboratory and only those subjects who have the translocation confirmed are eligible to participate in the study. Enrollment in this study has appealed to patients because of the encouraging frequency and magnitude of tumor shrinkage for those carrying the translocation. In fact, drug/tumor marker pairs with such strong relationships have been suggested to warrant an alternative, accelerated path to FDA approval92.
Despite the promise for crizotinib, most patients who provide tumor to screen for therapy do not have the translocation. These patients have depleted their stored tissue, have typically devoted a week or more withholding initial treatment for metastatic disease waiting for the result and may miss the opportunity to receive alternative investigational treatments as a result. In a more interconnected environment, patients without one marker might then be directed to other trials that might be more appropriate. Two examples of innovative, more interconnected trials are the MINDACT (Microarray In Node negative Disease may Avoid ChemoTherapy)93 and I-SPY 2 (Investigation of Serial studies to Predict Your therapeutic response with imaging and molecular analysis 2)94 trials currently enrolling breast cancer patients.
The MINDACT trial culminates years of efforts to interconnect expert centers in Europe to advance the use of new gene expression predictors to select patients for adjuvant chemotherapy in node-negative breast cancer. The original validation study of the Mammaprint™ gene predictor was conducted retrospectively on tissues collected from patients treated at the Netherlands Cancer Institute95. Shortly thereafter, a European consortium performed a second, larger validation study with samples from patients treated at 5 independent centers in France, Sweden, and the United Kingdom. The Mammaprint™ assay was performed on these samples in blinded fashion by researchers at Agendia, the company developing and marketing Mammaprint™, and the results were analyzed by another group in Belgium38. This multi-center effort quickly confirmed the claims of the original publications and established a network for conducting first a prospective pilot study for technical processing, shipment, and analysis of tumor tissue in the central laboratory40, and now the confirmatory, prospective trial. This study will not only determine the clinical utility of Mammaprint™, but also collect data necessary to extend our understanding of the interaction between tumor gene expression, disease prognosis, and outcomes from adjuvant chemotherapy93.
MINDACT is a clinical trial of a diagnostic/prognostic tool. Due to unmet medical needs for many cancer patients, most trials are directed to the most pressing need for innovation, i.e., the development of therapeutics96. The I-SPY 2 trial has emulated the interconnectedness of MINDACT with an innovative approach to new drug development and concurrent biomarker development94. The interconnectedness of the trial entails multiple centers with experience working together to conduct standardized tumor biopsy/sample collection, centers using the same platforms for sample tracking and integration with data from serial imaging during the course of neoadjuvant therapy, a template for treatment administration that adapts to addition and testing of new agents, and a central administrative infrastructure to ensure the appropriate selection of agents for testing, a structured method for determining when a new agent should cease further study, and when an agent has demonstrated sufficient likelihood of potential benefit to proceed to the next phase of testing.
This infrastructure was built in I-SPY 1, a previous clinical study, and has now been leveraged to compare the efficacy of novel drugs in combination with standard chemotherapy compared to standard therapy alone. This series of phase 2 trials uses adaptive design methods that will “graduate” regimens with a higher likelihood for being more effective than standard therapy to subsequent testing. Aside from this novel structure, the study innovates in 2 additional ways: 1) by changing the traditional approach to developing adjuvant therapy and 2) by integrating biomarker development into the initial testing of novel agents. The traditional approach to developing adjuvant therapy is demonstrated by paclitaxel and trastuzumab as described above. These agents were first tested in patients who had a good performance status, but otherwise had the most advanced metastatic disease. The treatments were then tested in progressively earlier phases of advanced disease before being determined safe and active enough to warrant testing in the curative intent, adjuvant setting. As an alternative approach, I-SPY 2 tests novel agents in the neoadjuvant treatment setting and focuses on the pathologic complete response rate as the primary endpoint of interest. Serial imaging and determination of molecular changes in tumors during the interval from diagnostic biopsy to curative resection provide a screening, validation and qualification process so that when a new agent is graduated to further testing the sponsor of the drug will have estimates of the specificity and sensitivity of molecular and imaging markers in predicting benefit from the novel agent. Further innovations include the availability of this infrastructure to various sponsors based on a new drug meeting objective, pre-specified criteria and the manageable patient accrual rate of the trial centers. Even the funding structure, through the Foundation for the NIH Biomarkers Consortium is unusual as an academia/government/industry partnership focused on accelerating development of personalized cancer therapy.
Conclusion
Nearly 110 years since Osler first articulated the idea that understanding the variability among individuals could make medical care more science than art we are reaching the era of personalized oncology. It has been advances in laboratory science, new molecular and information technologies, and some early case examples in chronic myeloid leukemia and Her-2 expressing breast cancer that make this seem possible. It will be equally innovative advances in the design and conduct of clinical trials that will deliver us into the era of personalized oncology.
Footnotes
DISCLOSURES: Dr. Maitland was supported by K23CA124802. The authors have no conflicts of interest relevant to this article.
Contributor Information
Michael L. Maitland, Email: mmaitlan@medicine.bsd.uchicago.edu, Section of Hematology/Oncology, Associate Director, Committee on Clinical Pharmacology and Pharmacogenomics, University of Chicago.
Richard L. Schilsky, Email: rschilsk@medicine.bsd.uchicago.edu, Section of Hematology/Oncology, Deputy Director, Comprehensive Cancer Center, University of Chicago.
References
- 1.Schilsky RL. Personalizing cancer care: American Society of Clinical Oncology presidential address 2009. J Clin Oncol. 2009;27:3725–3730. doi: 10.1200/JCO.2009.24.6827. [DOI] [PubMed] [Google Scholar]
- 2.Leaf C. Why we’re losing the war on cancer (and how to win it) Fortune. 2004;149:76–82. 84–76, 88. passim. [PubMed] [Google Scholar]
- 3.Porter R. Offering Resistance. New York Times Book Review; 1997. [Google Scholar]
- 4.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 5.DiMasi JA, Grabowski HG. Economics of new oncology drug development. J Clin Oncol. 2007;25:209–216. doi: 10.1200/JCO.2006.09.0803. [DOI] [PubMed] [Google Scholar]
- 6.Woodcock J. The prospects for “personalized medicine” in drug development and drug therapy. Clin Pharmacol Ther. 2007;81:164–169. doi: 10.1038/sj.clpt.6100063. [DOI] [PubMed] [Google Scholar]
- 7.Fisher B, Bauer M, Margolese R, et al. Five-year results of a randomized clinical trial comparing total mastectomy and segmental mastectomy with or without radiation in the treatment of breast cancer. N Engl J Med. 1985;312:665–673. doi: 10.1056/NEJM198503143121101. [DOI] [PubMed] [Google Scholar]
- 8.Fisher B, Redmond C, Poisson R, et al. Eight-year results of a randomized clinical trial comparing total mastectomy and lumpectomy with or without irradiation in the treatment of breast cancer. N Engl J Med. 1989;320:822–828. doi: 10.1056/NEJM198903303201302. [DOI] [PubMed] [Google Scholar]
- 9.Holmes FA, Walters RS, Theriault RL, et al. Phase II trial of taxol, an active drug in the treatment of metastatic breast cancer. J Natl Cancer Inst. 1991;83:1797–1805. doi: 10.1093/jnci/83.24.1797-a. [DOI] [PubMed] [Google Scholar]
- 10.Reichman BS, Seidman AD, Crown JP, et al. Paclitaxel and recombinant human granulocyte colony-stimulating factor as initial chemotherapy for metastatic breast cancer. J Clin Oncol. 1993;11:1943–1951. doi: 10.1200/JCO.1993.11.10.1943. [DOI] [PubMed] [Google Scholar]
- 11.Seidman AD, Tiersten A, Hudis C, et al. Phase II trial of paclitaxel by 3-hour infusion as initial and salvage chemotherapy for metastatic breast cancer. J Clin Oncol. 1995;13:2575–2581. doi: 10.1200/JCO.1995.13.10.2575. [DOI] [PubMed] [Google Scholar]
- 12.Nabholtz JM, Gelmon K, Bontenbal M, et al. Multicenter, randomized comparative study of two doses of paclitaxel in patients with metastatic breast cancer. J Clin Oncol. 1996;14:1858–1867. doi: 10.1200/JCO.1996.14.6.1858. [DOI] [PubMed] [Google Scholar]
- 13.Henderson IC, Berry DA, Demetri GD, et al. Improved outcomes from adding sequential Paclitaxel but not from escalating Doxorubicin dose in an adjuvant chemotherapy regimen for patients with node-positive primary breast cancer. J Clin Oncol. 2003;21:976–983. doi: 10.1200/JCO.2003.02.063. [DOI] [PubMed] [Google Scholar]
- 14.Mamounas EP, Bryant J, Lembersky B, et al. Paclitaxel after doxorubicin plus cyclophosphamide as adjuvant chemotherapy for node-positive breast cancer: results from NSABP B-28. J Clin Oncol. 2005;23:3686–3696. doi: 10.1200/JCO.2005.10.517. [DOI] [PubMed] [Google Scholar]
- 15.Hayes DF, Thor AD, Dressler LG, et al. HER2 and response to paclitaxel in node-positive breast cancer. N Engl J Med. 2007;357:1496–1506. doi: 10.1056/NEJMoa071167. [DOI] [PubMed] [Google Scholar]
- 16.Baselga J, Tripathy D, Mendelsohn J, et al. Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- 17.Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 18.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 19.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 20.Joensuu H, Kellokumpu-Lehtinen PL, Bono P, et al. Adjuvant docetaxel or vinorelbine with or without trastuzumab for breast cancer. N Engl J Med. 2006;354:809–820. doi: 10.1056/NEJMoa053028. [DOI] [PubMed] [Google Scholar]
- 21.Buyse M. Breaking from Tradition in the Design of Phase II/III Clinical Trials. American Society of Clinical Oncology Annual Meeting; Orlando. 2005. [Google Scholar]
- 22.Sparano JA, Bernardo P, Stephenson P, et al. Randomized phase III trial of marimastat versus placebo in patients with metastatic breast cancer who have responding or stable disease after first-line chemotherapy: Eastern Cooperative Oncology Group trial E2196. J Clin Oncol. 2004;22:4683–4690. doi: 10.1200/JCO.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 23.Schilsky RL. Target practice: oncology drug development in the era of genomic medicine. Clin Trials. 2007;4:163–166. doi: 10.1177/1740774507076807. discussion 173–167. [DOI] [PubMed] [Google Scholar]
- 24.Paik S, Kim C, Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358:1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- 25.Freidlin B, McShane LM, Korn EL. Randomized clinical trials with biomarkers: design issues. J Natl Cancer Inst. 2010;102:152–160. doi: 10.1093/jnci/djp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janes H, Pepe MS, Bossuyt PM, Barlow WE. Measuring the performance of markers for guiding treatment decisions. Ann Intern Med. 2011;154:253–259. doi: 10.1059/0003-4819-154-4-201102150-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang W, Freidlin B, Simon R. Biomarker-adaptive threshold design: a procedure for evaluating treatment with possible biomarker-defined subset effect. J Natl Cancer Inst. 2007;99:1036–1043. doi: 10.1093/jnci/djm022. [DOI] [PubMed] [Google Scholar]
- 28.Lazar AA, Cole BF, Bonetti M, Gelber RD. Evaluation of treatment-effect heterogeneity using biomarkers measured on a continuous scale: subpopulation treatment effect pattern plot. J Clin Oncol. 2010;28:4539–4544. doi: 10.1200/JCO.2009.27.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sargent DJ, Conley BA, Allegra C, Collette L. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23:2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 30.Song X, Pepe MS. Evaluating markers for selecting a patient’s treatment. Biometrics. 2004;60:874–883. doi: 10.1111/j.0006-341X.2004.00242.x. [DOI] [PubMed] [Google Scholar]
- 31.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 32.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 34.Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26:2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- 35.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Toft DJ, Cryns VL. Minireview: Basal-like breast cancer: from molecular profiles to targeted therapies. Mol Endocrinol. 2011;25:199–211. doi: 10.1210/me.2010-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 38.Buyse M, Loi S, van’t Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 39.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 40.Mook S, Bonnefoi H, Pruneri G, et al. Daily clinical practice of fresh tumour tissue freezing and gene expression profiling; logistics pilot study preceding the MINDACT trial. Eur J Cancer. 2009;45:1201–1208. doi: 10.1016/j.ejca.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Baggerly K. Disclose all data in publications. Nature. 2010;467:401. doi: 10.1038/467401b. [DOI] [PubMed] [Google Scholar]
- 43.Baggerly KA, Coombes KR. What Information Should Be Required to Support Clinical “Omics” Publications? Clin Chem. 2011 doi: 10.1373/clinchem.2010.158618. [DOI] [PubMed] [Google Scholar]
- 44.Baggerly KA, Coombes KR, Neeley ES. Run batch effects potentially compromise the usefulness of genomic signatures for ovarian cancer. J Clin Oncol. 2008;26:1186–1187. doi: 10.1200/JCO.2007.15.1951. author reply 1187–1188. [DOI] [PubMed] [Google Scholar]
- 45.Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collins FS. Shattuck lecture--medical and societal consequences of the Human Genome Project. N Engl J Med. 1999;341:28–37. doi: 10.1056/NEJM199907013410106. [DOI] [PubMed] [Google Scholar]
- 47.Gambaro G, Anglani F, D’Angelo A. Association studies of genetic polymorphisms and complex disease. Lancet. 2000;355:308–311. doi: 10.1016/s0140-6736(99)07202-5. [DOI] [PubMed] [Google Scholar]
- 48.Hirschhorn JN, Altshuler D. Once and again-issues surrounding replication in genetic association studies. J Clin Endocrinol Metab. 2002;87:4438–4441. doi: 10.1210/jc.2002-021329. [DOI] [PubMed] [Google Scholar]
- 49.Maitland ML, Ratain MJ, Cox NJ. Interpreting P values in pharmacogenetic studies: a call for process and perspective. J Clin Oncol. 2007;25:4513–4515. doi: 10.1200/JCO.2007.12.7803. [DOI] [PubMed] [Google Scholar]
- 50.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 54.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 55.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wacholder S, Hartge P, Prentice R, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng W, Wen W, Gao YT, et al. Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women. J Natl Cancer Inst. 2010;102:972–981. doi: 10.1093/jnci/djq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. Jama. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 60.Gravitz L. Technology Review: 10 Emerging Technologies 2009: $100 Genome. MIT Technology Review; 2009. [Google Scholar]
- 61.Barrett JS, Mondick JT, Narayan M, Vijayakumar K, Vijayakumar S. Integration of modeling and simulation into hospital-based decision support systems guiding pediatric pharmacotherapy. BMC Med Inform Decis Mak. 2008;8:6. doi: 10.1186/1472-6947-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dombrowsky E, Jayaraman B, Narayan M, Barrett JS. Evaluating performance of a decision support system to improve methotrexate pharmacotherapy in children and young adults with cancer. Ther Drug Monit. 2011;33:99–107. doi: 10.1097/FTD.0b013e318203b41e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma MR, Maitland ML, Ratain MJ. Other paradigms: better treatments are identified by better trials: the value of randomized phase II studies. Cancer J. 2009;15:426–430. doi: 10.1097/PPO.0b013e3181b9c5d5. [DOI] [PubMed] [Google Scholar]
- 64.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 65.Hillner BE, Siegel BA, Liu D, et al. Impact of positron emission tomography/computed tomography and positron emission tomography (PET) alone on expected management of patients with cancer: initial results from the National Oncologic PET Registry. J Clin Oncol. 2008;26:2155–2161. doi: 10.1200/JCO.2007.14.5631. [DOI] [PubMed] [Google Scholar]
- 66.Day SE, Kettunen MI, Gallagher FA, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 67.Shields AF, Grierson JR, Dohmen BM, et al. Imaging proliferation in vivo with [F-18]FLT and positron emission tomography. Nat Med. 1998;4:1334–1336. doi: 10.1038/3337. [DOI] [PubMed] [Google Scholar]
- 68.Dinan MA, Curtis LH, Hammill BG, et al. Changes in the use and costs of diagnostic imaging among Medicare beneficiaries with cancer, 1999–2006. JAMA. 2010;303:1625–1631. doi: 10.1001/jama.2010.460. [DOI] [PubMed] [Google Scholar]
- 69.Maitland ML. Volumes to learn: advancing therapeutics with innovative computed tomography image data analysis. Clin Cancer Res. 2010;16:4493–4495. doi: 10.1158/1078-0432.CCR-10-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattaram VA, Bonapace C, Chilukuri DM, et al. Impact of pharmacometric reviews on new drug approval and labeling decisions--a survey of 31 new drug applications submitted between 2005 and 2006. Clin Pharmacol Ther. 2007;81:213–221. doi: 10.1038/sj.clpt.6100051. [DOI] [PubMed] [Google Scholar]
- 71.Gobburu JV, Lesko LJ. Quantitative disease, drug, and trial models. Annu Rev Pharmacol Toxicol. 2009;49:291–301. doi: 10.1146/annurev.pharmtox.011008.145613. [DOI] [PubMed] [Google Scholar]
- 72.Gobburu JV, Marroum PJ. Utilisation of pharmacokinetic-pharmacodynamic modelling and simulation in regulatory decision-making. Clin Pharmacokinet. 2001;40:883–892. doi: 10.2165/00003088-200140120-00001. [DOI] [PubMed] [Google Scholar]
- 73.Claret L, Girard P, Hoff PM, et al. Model-based prediction of phase III overall survival in colorectal cancer on the basis of phase II tumor dynamics. J Clin Oncol. 2009;27:4103–4108. doi: 10.1200/JCO.2008.21.0807. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y, Sung C, Dartois C, et al. Elucidation of relationship between tumor size and survival in non-small-cell lung cancer patients can aid early decision making in clinical drug development. Clin Pharmacol Ther. 2009;86:167–174. doi: 10.1038/clpt.2009.64. [DOI] [PubMed] [Google Scholar]
- 75.Bruno R, Claret L. On the use of change in tumor size to predict survival in clinical oncology studies: toward a new paradigm to design and evaluate phase II studies. Clin Pharmacol Ther. 2009;86:136–138. doi: 10.1038/clpt.2009.97. [DOI] [PubMed] [Google Scholar]
- 76.Karrison TG, Maitland ML, Stadler WM, Ratain MJ. Design of phase II cancer trials using a continuous endpoint of change in tumor size: application to a study of sorafenib and erlotinib in non small-cell lung cancer. J Natl Cancer Inst. 2007;99:1455–1461. doi: 10.1093/jnci/djm158. [DOI] [PubMed] [Google Scholar]
- 77.Mozley PD, Schwartz LH, Bendtsen C, Zhao B, Petrick N, Buckler AJ. Change in lung tumor volume as a biomarker of treatment response: a critical review of the evidence. Ann Oncol. 2010 doi: 10.1093/annonc/mdq051. [DOI] [PubMed] [Google Scholar]
- 78.Berg JM, Rogers ME, Lyster PM. Systems biology and pharmacology. Clin Pharmacol Ther. 2010;88:17–19. doi: 10.1038/clpt.2010.69. [DOI] [PubMed] [Google Scholar]
- 79.United States Department of Health and Human Services FaDA. Innovation or Stagnation: Challenge and Opportunity on the Critical Path to New Medical Products. 2004. [Google Scholar]
- 80.Murthy VH, Krumholz HM, Gross CP. Participation in cancer clinical trials: race, sex-, and age-based disparities. Jama. 2004;291:2720–2726. doi: 10.1001/jama.291.22.2720. [DOI] [PubMed] [Google Scholar]
- 81.Schilsky RL. Accrual to cancer clinical trials in the era of molecular medicine. Sci Transl Med. 2011;3:75cm79. doi: 10.1126/scitranslmed.3001712. [DOI] [PubMed] [Google Scholar]
- 82.Gross CP, Murthy V, Li Y, Kaluzny AD, Krumholz HM. Cancer trial enrollment after state-mandated reimbursement. J Natl Cancer Inst. 2004;96:1063–1069. doi: 10.1093/jnci/djh193. [DOI] [PubMed] [Google Scholar]
- 83.Dilts DM, Cheng SK, Crites JS, Sandler AB, Doroshow JH. Phase III clinical trial development: a process of chutes and ladders. Clin Cancer Res. 2010;16:5381–5389. doi: 10.1158/1078-0432.CCR-10-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frei E, Holland J, Schneiderman M, et al. A Comparative Study of Two Regimens of Combination Chemotherapy in Acute Leukemia. Blood. 1958;13:1126–1148. [PubMed] [Google Scholar]
- 85.Maitland ML, Hudoba C, Snider KL, Ratain MJ. Analysis of the yield of phase II combination therapy trials in medical oncology. Clin Cancer Res. 2010;16:5296–5302. doi: 10.1158/1078-0432.CCR-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agrawal M, Grady C, Fairclough DL, Meropol NJ, Maynard K, Emanuel EJ. Patients’ decision-making process regarding participation in phase I oncology research. J Clin Oncol. 2006;24:4479–4484. doi: 10.1200/JCO.2006.06.0269. [DOI] [PubMed] [Google Scholar]
- 87.Catania C, De Pas T, Goldhirsch A, et al. Participation in clinical trials as viewed by the patient: understanding cultural and emotional aspects which influence choice. Oncology. 2008;74:177–187. doi: 10.1159/000151365. [DOI] [PubMed] [Google Scholar]
- 88.Ratain MJ. Phase II oncology trials: let’s be positive. Clin Cancer Res. 2005;11:5661–5662. doi: 10.1158/1078-0432.CCR-05-1046. [DOI] [PubMed] [Google Scholar]
- 89.Pui CH, Evans WE. Acute lymphoblastic leukemia. N Engl J Med. 1998;339:605–615. doi: 10.1056/NEJM199808273390907. [DOI] [PubMed] [Google Scholar]
- 90.Ratain MJ, Karrison TG. Testing the wrong hypothesis in phase II oncology trials: there is a better alternative. Clin Cancer Res. 2007;13:781–782. doi: 10.1158/1078-0432.CCR-06-2533. [DOI] [PubMed] [Google Scholar]
- 91.Institute NC. [Accessed December 7, 2010];caBIGR Mission and Goals. Available at: http://cabig.cancer.gov/caBIGstory/mission/
- 92.McClellan M, Benner J, Schilsky R, et al. An accelerated pathway for targeted cancer therapies. Nat Rev Drug Discov. 2011;10:79–80. doi: 10.1038/nrd3360. [DOI] [PubMed] [Google Scholar]
- 93.Bogaerts J, Cardoso F, Buyse M, et al. Gene signature evaluation as a prognostic tool: challenges in the design of the MINDACT trial. Nat Clin Pract Oncol. 2006;3:540–551. doi: 10.1038/ncponc0591. [DOI] [PubMed] [Google Scholar]
- 94.Barker AD, Sigman CC, Kelloff GJ, Hylton NM, Berry DA, Esserman LJ. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86:97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 95.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 96.Davis JC, Furstenthal L, Desai AA, et al. The microeconomics of personalized medicine: today’s challenge and tomorrow’s promise. Nat Rev Drug Discov. 2009;8:279–286. doi: 10.1038/nrd2825. [DOI] [PubMed] [Google Scholar]