Abstract
We report here that FTO (fat mass and obesity-associated protein) exhibits efficient oxidative demethylation activity of abundant N6-methyladenosine (m6A) residues in RNA in vitro. FTO knockdown with siRNA led to an increased level of m6A in mRNA, whereas overexpression of FTO resulted in a decreased level of m6A in human cells. We further show that FTO partially colocalizes with nuclear speckles, supporting m6A in nuclear RNA as a physiological substrate of FTO.
Obesity has become a serious world-wide public health concern. Fat mass and obesity-associated (FTO) protein has been shown to affect human obesity and energy homeostasis1–6. Since the first connection with obesity was made through genome-wide association studies, FTO has become a prominent target of research in the field. However, the physiological substrate and function of FTO remains unknown, which prevents molecular level understanding of the mechanism and pathway of the FTO-mediated regulation.
FTO belongs to the non-heme FeII/α-KG-dependent dioxygenase AlkB family proteins that also includes ABH1 to ABH87,8. Among them, ABH1, ABH2, and ABH3 have been shown to oxidatively demethylate N-methylated DNA/RNA bases9,10, and ABH8 catalyzes the hydroxylation of hypermodified tRNA wobble uridine11,12. FTO has previously been shown to oxidatively demethylate m3T and m3U in single-stranded DNA (ssDNA) and single-stranded RNA (ssRNA) in vitro7,13, although the observed activity is exceedingly low compared to those of the other AlkB family proteins14. The recent crystal structure of FTO confirms the substrate preference of FTO for single-stranded nucleic acids15. Therefore, modified RNAs are plausible substrate candidates for FTO.
Since the physiological substrates of the AlkB family proteins might not be limited to N1-or N3-modified purines or pyrimidines, as in the case of ABH811,12, we turned our attention to N6-methyladenosine (m6A) (Fig. 1a), which is the dominating methylated base in mRNA with a frequency of 3–5 m6A per mRNA molecule in mammalian cells16–19. Although the exact role of m6A in mRNA is largely unknown, it has been proposed to affect mRNA processing and export from nucleus to cytoplasm19.
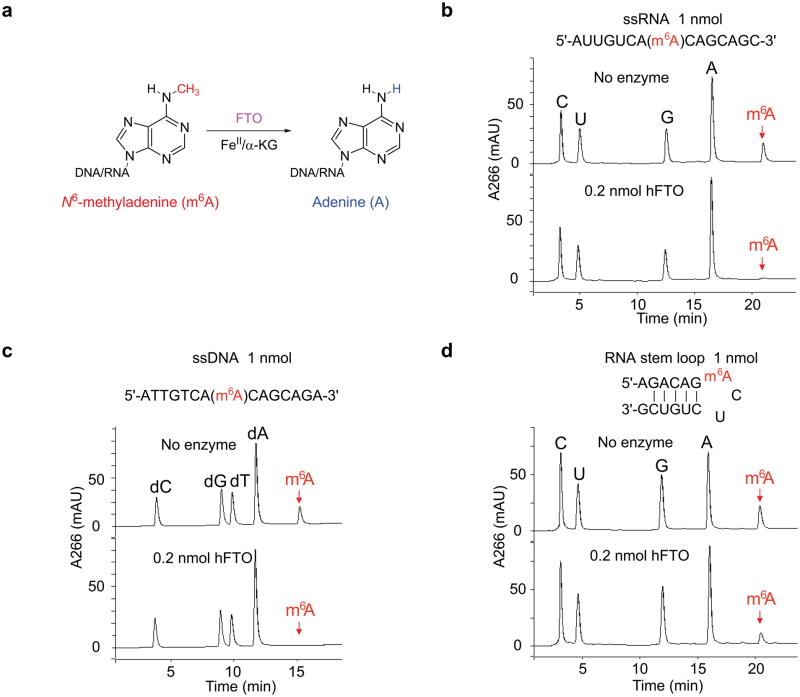
Figure 1. Oxidative demethylation of m6A in nucleic acids by FTO.
(a) Proposed oxidative demethylation of m6A to A in DNA and RNA by FTO in the presence of iron(II) and α-KG. (b, c) As shown by HPLC profiles of digested substrates, 20 mol% of FTO can completely demethylate m6A in ssRNA and ssDNA in 3 h at room temperature at neutral pH. (d) About 40% demethylation of m6A in a RNA stem loop could be observed under the same reaction conditions.
We synthesized ssRNA and ssDNA with site-specifically incorporated m6A (Supplementary Scheme 1 and Supplementary Fig. 1). After treating an 8-mer ssDNA containing m6A (1 nmol) with an equal amount of recombinant human FTO (Supplementary Fig. 2) at pH 7.0 at room temperature overnight, we observed a loss of 14 Dalton in mass indicating that a demethylation process had occurred (Supplementary Fig. 3). To further investigate the demethylation activity of FTO towards m6A in DNA and RNA, we synthesized a series of 15-mer ssDNA and ssRNA containing a central m6A base. These single-stranded oligonucleotides were treated with 20 mol% of FTO at pH 7.0 at room temperature for 3 h. We then digested the oligonucleotides into nucleosides using nuclease P1 and alkaline phosphatase, and separated the products on a C18 column by HPLC. The HPLC trace profile indicated that m6A in ssDNA and ssRNA was completely converted to adenosine upon FTO treatment (Fig. 1b,c). We further tested a short m6A-containing RNA using a known sequence of cellular bovine prolactin (bPRL) mRNA, in which the position of m6A had been mapped18,19. While this short RNA sequence can form a stem loop secondary structure, we still observed 40% of demethylation of m6A under the same conditions (Fig. 1d). We tested m6A-containing dsDNA and dsRNA with negligible activity observed under the same reaction conditions (0.2 equivalent of FTO for 3 h). After treatment with one molar equivalent of FTO at 16 °C overnight, we observed a demethylation yield of 40% for dsDNA and 24% for dsRNA, respectively (Supplementary Fig. 4).
To confirm that the observed demethylation requires the oxidative function of FTO, we tested m6A demethylation using several FTO mutants with active site mutations. Arg316 is a key residue for stabilizing the binding of α-KG. We expressed and purified the FTO R316Q mutant protein (Supplementary Fig. 2), and found that this R316Q mutation abolished 80% of the wild-type activity towards m6A demethylation in vitro (Supplementary Fig. 5). We subsequently constructed and tested two double mutant FTO proteins, H231A/D233A, in which two iron(II) ligands were mutated, and R316Q/R322Q, in which two α-KG ligands were mutated (Supplementary Fig. 2). Both of these double mutants completely lost m6A demethylation activity (Supplementary Fig. 5). These results confirmed that FTO catalyzes oxidative demethylation of m6A in an iron(II)- and α-KG-dependent manner.
We determined detailed reaction kinetics of FTO on a 15-mer m6A-containing ssRNA at pH 7.0 at 20 °C (Supplementary Fig. 6). The results are summarized in Supplementary Table 1 in comparison to our previously published kinetic data on m3U in ssRNA at pH 6.0. FTO exhibits a pH-dependent oxidative demethylation activity with the highest activity observed at pH 6.013. Even if we compare the FTO activity towards m6A at pH 7.0 to that of m3U at pH 6.0 in ssRNA, we still observe an over 50-fold preference of FTO for m6A. To further validate this conclusion, we synthesized and tested ssRNA containing m3U with the same sequence as that used for m6A at 20 °C (Supplementary Fig. 7). The demethylation activity of FTO towards m6A is indeed significantly faster than towards m3U at neutral pH in vitro. Under physiological conditions, m6A is the best substrate discovered so far for FTO.
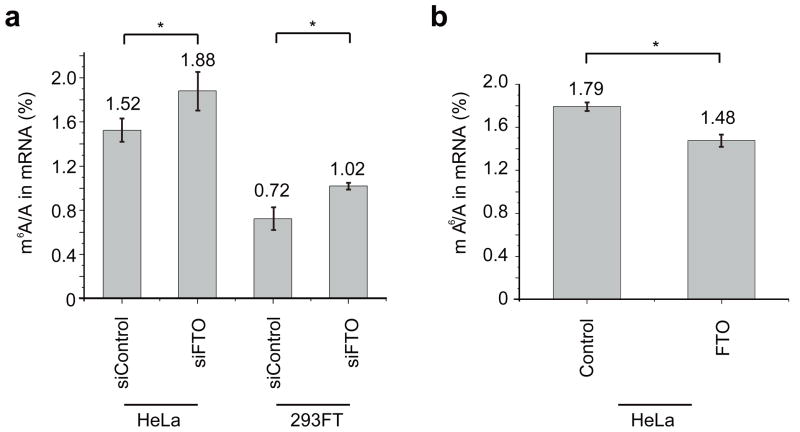
While m6A has not been identified in genomic DNA in higher eukaryotes, it is present in easily detectable levels in cellular mRNAs isolated from all higher eukaryotes and viral RNA16–19. We explored whether m6A in mRNA is a physiological substrate of FTO in vivo. We transfected HeLa and 293FT cells with FTO siRNA (see Supplementary Methods), and western blotting of the total cell lysates from transfected cells showed close to 90% knockdown of FTO 48 h post-transfection (Supplementary Fig. 8a). We then isolated total mRNA from cells treated with control siRNA and FTO siRNA using a biotin-poly (dT) probe for separation followed by a rRNA depletion step to ensure depletion of rRNA (see Supplementary Methods and Supplementary Fig. 9). We then digested the total mRNA into nucleosides, and measured the relative amount of m6A levels by LC-MS/MS. The total contents of m6A and A were quantified based on a standard curve generated using pure standards, from which the m6A/A ratio was calculated (see Supplementary Methods and Supplementary Fig. 10). Our results indicated that knockdown of the cellular FTO increased m6A in mRNA by 23% in HeLa cells and 42% in 293FT cells (Fig. 2a). We also overexpressed wild-type FTO using a mammalian expression vector. Western blotting of the total cell lysates from transfected cells confirmed the overexpression of FTO by 6–8 folds after 24 h (Supplementary Fig. 8b). Total mRNA isolated from HeLa cells overexpressing the wild-type FTO showed a notable decrease of m6A (~18%) (Fig. 2b).
Figure 2. FTO regulates m6A content in mRNA in a FTO-activity dependent manner.
(a) Quantification of the m6A/A ratio in mRNA by LC-MS/MS. Significant increase of the m6A level was observed in both FTO siRNA-treated HeLa and 293FT cells after 48 h. *, P-value < 0.05, Student’s t-test; means ± s.e.m. for n = 8 experiments. (b) The m6A/A ratio of mRNA samples isolated from cells with overexpressed FTO. The overexpression of wild-type FTO decreased the m6A content significantly. * P-value < 0.05, Student’s t-test; means ± s.e.m. for n = 8 experiments.
Since a large portion of m6A is after G in the consensus sequence (Pu[G>A]m6AC[A/C/U])19, we also employed a two-dimensional thin layer chromatography (2D-TLC) method that can detect the m6A content after G in mRNA (see Supplementary Methods). The results supported the conclusion that the m6A content in mRNA is affected by the cellular activity of FTO (Supplementary Fig. 11). A dot blot assay using an anti-m6A antibody was also used to qualitatively monitor the change of m6A level with the same conclusion obtained (Supplementary Fig. 12). We performed additional western blotting experiments for MT-A70 (AdoMet-binding subunit of mRNA N6-adenosine-methyltransferase) to confirm that the observed change of the m6A level was caused by FTO, but not from the potential alteration of the m6A mRNA methyltransferase expression20 (Supplementary Fig. 13). Knockdown of FTO in HeLa cells led to a decrease of MT-A70 expression of around 15%, and overexpression of FTO led to an increase of MT-A70 expression of about 15%. Furthermore, knockdown of ABH3 in HeLa cells did not noticeably affect the total level of m6A in mRNA as a negative control (Supplementary Fig. 14). We conclude that the increase of the m6A level in the FTO knockdown sample is not due to an increased level of methyltransferase, but is a result of the depletion of cellular FTO.
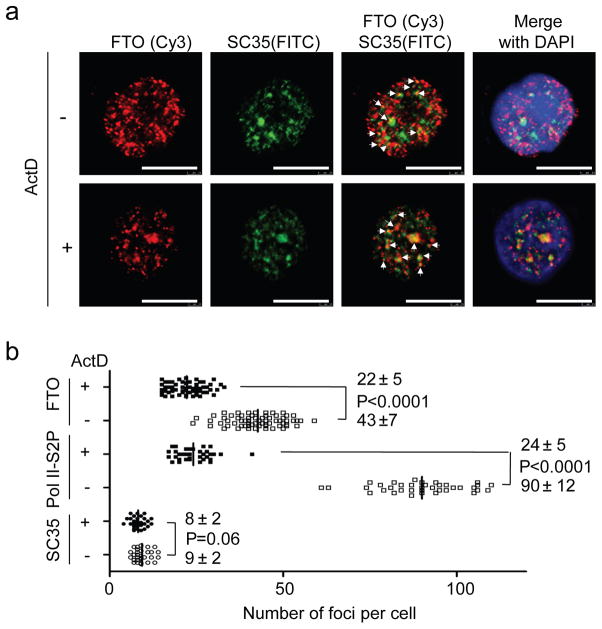
Lastly, indirect immunofluorescence analysis was performed to visualize the localization of the FTO protein in mammalian cells. FTO is known to be entirely localized to cell nuclei7. We observed that the cellular FTO protein is present in a dot-like manner in nucleoplasm, and partially colocalizes with splicing or splicing-related speckle factors SART1(U4/U6.U5 tri-snRNP-associated protein 1) and SC35 (serine/arginine-rich splicing factor 2), and RNA polymerase II phosphorylated at Ser2 (Pol II-S2P) (Fig. 3a and Supplementary Fig. 15a,b), but does not colocalize with telomere marker TRF1, replication site PCNA, Cajal body marker Coilin, cleavage body marker CstF64, or P-body marker DCP1A (Supplementary Fig. 16a and 17). To investigate whether FTO is related to mRNA processing, we performed the same immunostaining experiment upon inhibition of Pol II-S2P transcription with actinomycin D (ActD). Quantitative foci analysis showed that the FTO foci number per cell was significantly decreased from 43±7 (−ActD) to 22 ± 5 (+ActD) with P-value less than 0.0001, similar to that observed for Pol–II S2P from 90 ± 12 (−ActD) to 24 ± 5 (+ActD), but not SC3521 (Fig. 3b), upon transcription inhibition (Supplementary Fig. 18). Intriguingly, the decreased FTO nucleoplasm foci became more concentrated into discrete speckle foci after transcription inhibition by ActD (Fig. 3a and Supplementary Fig. 15a), resembling that of Pol II-S2P21 (Supplementary Fig. 15a). Moreover, the transcription inhibition induced a more pronounced colocalization of FTO with both SC35 and Pol II-S2P (Fig. 3a and Supplementary Fig. 15a). Splicing speckles are major nuclear domains enriched for components of the splicing machinery such as SC35, polyA+ RNA, and numerous mRNA metabolic factors21–24; genes associated with splicing speckles have been shown to function in spliceosome assembly or post-transcriptional splicing of pre-mRNAs23,24. Knockdown of FTO does not affect the assembly of spliceosome (Supplementary Fig. 16b,c); however, FTO shows a distribution pattern in nuclear speckles similar to other splicing factors. FTO may be recruited to the speckles by its interacting partners in the assembled nuclear speckles. In conclusion, the partial colocalization of FTO with nuclear speckles further supports the concept that m6A in nuclear RNA is a substrate of FTO, and the enzymatic alteration may be linked to processing of recently transcribed mRNA.
Figure 3. Indirect immunofluorescence analysis of endogenous FTO shows FTO partially colocalizes with nuclear speckles.
(a) HeLa cells were co-stained with antibodies against FTO and speckle marker SC35 in the presence or absence of transcription inhibitor actinomycin D (ActD). Yellow dots indicated by white arrows correspond to the colocalized spots of FTO (Red) with SC35 (Green) (N = 150). Upon transcription inhibition, the FTO loci number decreased in the nucleoplasm, but its speckle distribution became more pronounced. (b) Quantitative foci analysis of SC35, Pol II-S2P, and FTO with (+), or without (−) ActD treatment. 30, 37 and 67 cells were randomly selected for foci analysis of SC35, Pol II-S2P, and FTO, respectively. The t-test using two-way ANOVA in Grouped Analyses of Prism5 software was applied for statistical analysis. P-value less than 0.01 was considered a significant difference. The mean numbers of foci per cell for each sample with and without ActD treatment were shown above, and below the corresponding P-value, respectively. The foci number of Pol II-S2P and FTO decreased significantly upon transcription inhibition.
In summary, we show that FTO efficiently demethylates m6A at neutral pH in vitro, and the level of m6A in cellular mRNA is affected by the oxidation activity of FTO in vivo. FTO partially localizes with nuclear speckles. The pronounced concentration of FTO in speckles upon Pol II-S2P transcription inhibition suggests a dynamic interaction of FTO with the nuclear speckles. In fact, MT-A70, a critical subunit of m6A-methyltransferase that introduces m6A into mRNA, has also been shown to localize in nuclear speckles, and probably associates with nuclear pre-mRNA splicing components20. All these observations indicate that m6A in nuclear RNA is the physiological substrate of FTO, and that the function of FTO likely affects the processing of pre-mRNA and/or other nuclear RNAs.
Despite past research on m6A in mRNA, the biological function of this ubiquitous post-transcriptional modification remains unclear, particularly in mRNA from higher eukaryotes. Our cellular data suggest that a portion or subclass of m6A in mRNAs appeared to be affected by the activity of FTO in vivo. How the sequence and methylation status of m6A in mRNA affect the output of RNA still requires detailed investigation; the fact that the function of the major obesity factor FTO is to demethylate m6A in mRNA clearly indicates a novel, reversible regulatory mechanism present in mammalian cells. Methylation of DNA and histones contributes largely to epigenetic regulation in mammalian cells. The discovery of the FTO-mediated oxidative demethylation of m6A in nuclear RNA may initiate further investigations on biological regulation based on reversible chemical modification of RNA25.
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institute of Health (GM071440 to C.H.), an NIH EUREKA award (GM088599 to C.H. and T.P.), and the CAS “100-talents” Professor Program to Y.G.Y. Q.D. was supported by the Chicago Biomedical Consortium (CBC). X.Z. and Y.Y. supported by Graduate Student Program of Beijing Institute of Genomics, Chinese Academy of Sciences (CAS). T.L. supported by the CAS Senior Foreign Research Fellow Award (2009S2-1). We thank CBC/UIC RRC Proteomics and Informatics Services Facility for performing the LC-MS/MS analysis, L.A. Godley for the suggestion, and P. Jin for providing the pcDNA3-FTO plasmid. We thank Sarah Frank Reichard, M.A. for editing the manuscript.
Footnotes
Author Contributions
G.J., Y.F., and C.H. conceived the original idea. G.J., Y.F., X.Z., Y.G.Y and C.H. designed the experiments with help from T.P. Biochemistry assays and cellular analysis were performed by G.J., Y.F., and X. Z. with the help of G.Z., C.Y., and Y.Y.; Q.D. carried out the chemical synthesis; T.L. provided advice and the anti-FTO antibody; G.J., Y.F., Y.G.Y. and C.H. wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Supplementary information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Laura LJ, et al. Science. 2007;316:1341–1345. [Google Scholar]
- 2.Frayling TM, et al. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dina C, et al. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 4.Thorleifsson G, et al. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 5.Fischer J, et al. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 6.Church C, et al. Nat Genet. 2010;42:1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerken T, et al. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westbye MP, et al. J Biol Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aas PA, et al. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 11.Fu Y, et al. Angew Chem Int Ed Engl. 2010;49:8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Born E, et al. Nat Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 13.Jia G, et al. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee DH, et al. J Biol Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 15.Han Z, et al. Nature. 2010;464:1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- 16.Wei CM, Gershowitz A, Moss B. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 17.Narayan P, Rottman FM. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- 18.Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Proc Natl Acad Sci USA. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harper JE, Miceli SM, Roberts RJ, Manley JL. Nucleic Acids Res. 1990;18:5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 21.Xie SQ, et al. Mol Biol Cell. 2006;17:1723–1733. doi: 10.1091/mbc.E05-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall LL, et al. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence JB, Clemson CM. J Cell Biol. 2008;182:1035–1038. doi: 10.1083/jcb.200808121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamond AI, Spector DL. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 25.He C. Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.