Abstract
Femtosecond vibrational coherence spectroscopy was used to investigate the low frequency vibrational dynamics of the heme in the carbon monoxide oxidation activator protein (CooA) from the thermophilic anaerobic bacterium Carboxydothermus hydrogenoformans (Ch-CooA). Low frequency vibrational modes are important because they are excited by the ambient thermal bath (kBT = 200 cm-1) and participate in thermally activated barrier crossing events. However, such modes are nearly impossible to detect in the aqueous phase using traditional spectroscopic methods. Here we present the low frequency coherence spectra of the ferric, ferrous, and CO-bound forms of Ch-CooA in order to compare the protein-induced heme distortions in its active and inactive states. Distortions take place predominantly along the coordinates of low-frequency modes because of their weak force constants and such distortions are reflected in the intensity of the vibrational coherence signals. A strong mode near ~90 cm-1 in the ferrous form of Ch-CooA is suggested to contain a large component of heme ruffling, consistent with the imidazole bound ferrous heme crystal structure, which shows a significant protein-induced heme distortion along this coordinate. A mode observed at ~228 cm-1 in the six-coordinate ferrous state is proposed to be the ν(Fe-His) stretching vibration. The observation of the Fe-His mode indicates that photolysis of the N-terminal α-amino axial ligand takes place. This is followed by a rapid (~8.5 ps) transient absorption recovery, analogous to methionine rebinding in photolyzed ferrous cytochrome c. We have also studied CO photolysis in CooA, which revealed very strong photoproduct state coherent oscillations. The observation of heme-CO photoproduct oscillations is unusual because most other heme systems have CO rebinding kinetics that are too slow to make the measurement possible. The low frequency coherence spectrum of the CO-bound form of Ch-CooA shows a strong vibration at ~230 cm-1 that is broadened and up-shifted compared to the ν(Fe-His) of Rr-CooA (216 cm-1). We propose that the stronger Fe-His bond is related to the enhanced thermal stability of Ch-CooA and that there is a smaller (time dependent) tilt of the histidine ring with respect to the heme plane in Ch-CooA. The appearance of strong modes at ~48 cm-1 in both the ferrous and CO-bound forms of Ch-CooA is consistent with coupling of the heme doming distortion to the photolysis reaction in both samples. Upon CO binding and protein activation, a heme mode near 112 ± 5 cm-1 disappears, probably indicating a decreased heme saddling distortion. This reflects changes in the heme environment and geometry that must be associated with the conformational transition activating the DNA-binding domain. Protein-specific DNA binding to the CO-bound form of Ch-CooA was also investigated and, although the CO rebinding kinetics are significantly perturbed, there are negligible changes in the low-frequency vibrational spectrum of the heme.
Keywords: Ch-CooA, DNA-Protein interaction, low frequency modes, heme distortion, Fe-His stretching vibration
Introduction
The metalloproteins, especially heme proteins, exhibit a wide variety of functions such as oxygen storage/transport, electron transfer, and redox reactions of various substrates. In addition to these functions the heme is used to sense the diatomic molecules oxygen (O2), nitric oxide (NO), or carbon monoxide (CO) to regulate and control important biological processes in mammals, plants and bacteria. These heme-based sensor proteins1 include NO-sensing soluble guanylate cyclase (sGC)2, oxygen-sensing FixL3, DOS4;5, HemAT6, and CO-sensing CooA7-11.
The heme-based carbon monoxide oxidation activator (CooA) protein from Rhodospirillum rubrum (Rr-CooA) has been the paradigm for the CooA family. It senses carbon monoxide and activates genes that allow the bacterium to grow anaerobically on CO as its sole energy source. CooA is a member of the adenosine cyclic 3’,5’-adenosine phosphoate receptor protein (CRP) and fumarate/nitrate reductase regulator super family of transcriptional regulators. These family members have common sets of structural elements such as a coiled-coil dimer interface and a “hinge” region existing between the dimer interface and the DNA-binding domain. CooA regulates the expression of proteins encoded by the coo genes (CO oxidation), which are associated with CO-oxidative growth. CooA is a homodimer that contains one b-type heme per approximately 25 kDa monomer. CooA consists of two heme containing regulatory domains where the CO effector binds, plus a set of C-terminal domains that bind to DNA. The transmission of the CO signal from the heme to the DNA-binding domain is believed to involve the two long α-helices (C) at the dimer interface which interact utilizing a leucine-zipper motif.
Youn et al.12 reported the CooA homologues from the thermophile Carboxydothermus hydrogenoformans (Ch-CooA) serve as a CO-dependent transcriptional regulator analogous to Rr-CooA. Inagaki et al. 13 compared the spectroscopic and redox properties of Rr-CooA and Ch-CooA and found that the heme is 6-coordinate in the ferric, ferrous, and ferrous CO bound forms. Rr-CooA undergoes a ligand switch from Cys75 to His77 upon reduction, but both systems are 6-coordinate in the ferrous form with a histidine and an N-terminal amino acid as axial ligands (Pro2 and Ala2 as seen in Fig. 1). The N-terminal amino acids are displaced upon CO binding13. Clark et al. 8 characterized Ch-CooA spectroscopically and functionally and found that the NO bound form is 6-coordinate and also binds to DNA in vivo and in vitro at room temperature. In contrast, the NO bound form of Rr-CooA is 5-coordinate and is inactive to DNA binding. These studies support the hypothesis that binding of a small, neutral distal ligand is necessary in order to activate the CooA mechanism8.
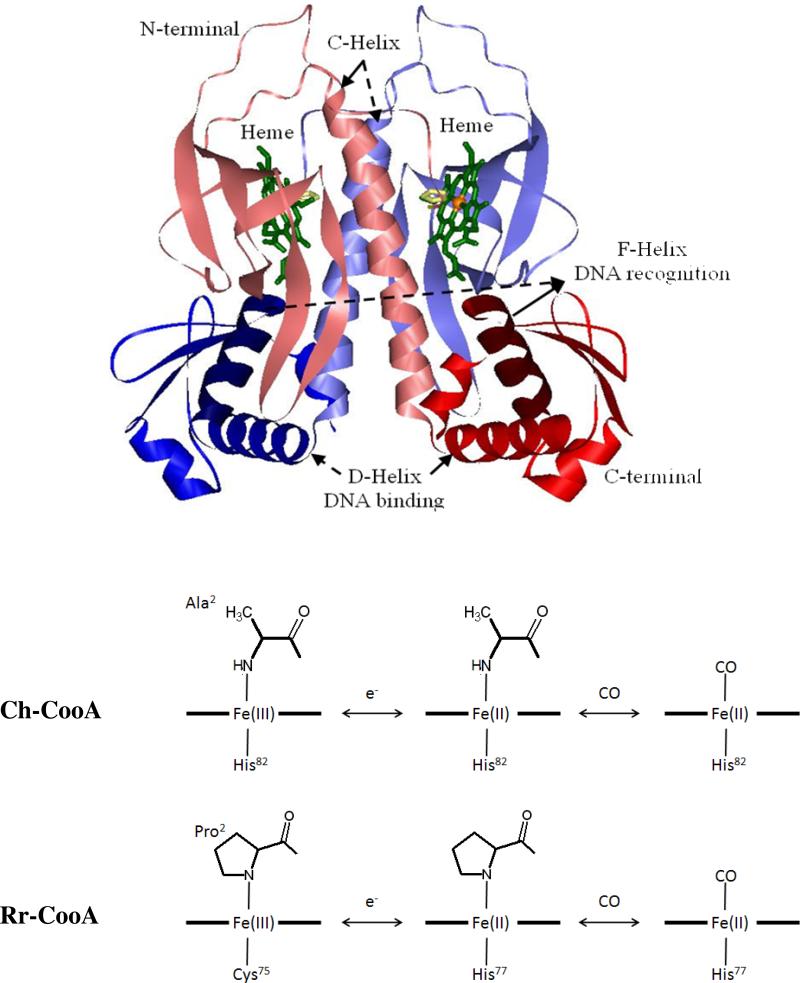
Figure 1.
The crystal structure of imidazole bound ferrous Ch-CooA obtained from PDB: 2FMY14. The Ch-CooA consists of two symmetric A and B monomers, colored red and blue, respectively. Each monomer consists of the lightly colored N-terminal heme domains and the C-terminal DNA-binding domains are colored in normal tones. The recognition F-helix is shown in a darker color. The heme and imidazole are shown in green and yellow respectively. The coordination structures of Rr-CooA and Ch-CooA are shown in the lower panel for the ferric, ferrous and CO bound forms.
The ferric heme of Ch-CooA is low-spin and 6-coordinate with an N-terminal α-amino group and the His82 as the axial ligands. Upon reduction, those ligands are retained, whereas when CO binds, the N-terminal α-amino group is replaced. Komori et al. 14 reported the crystal structure of an imidazole-bound form of Ch-CooA, but they were unable to crystallize the original protein with the natural N-terminal α−amino heme ligand. The crystal structure reveals that imidazole replaces the N-terminal α−amino heme ligand, as does CO, when it binds to the heme. The imidazole-bound structure shows two monomers that are symmetric and adopting a bent conformation that has been suggested to be in-between the active (CO bound) and inactive (ferrous) form. An intermediate conformation is also consistent with previously measured small-angle X-ray scattering experiments15. However, whatever the details of the intermediate conformation(s) may be, the replacement of the N-terminal amino acid heme ligand by imidazole does not activate CooA for DNA binding16.
The reduction and oxidation potentials of Ch-CooA have been found to be +150 mV and +230 mV, respectively13. These values are approximately +500 mV higher than for Rr-CooA, which has reduction and oxidation potentials of -320 mV and -260 mV respectively17. The differences in the redox potentials have been explained on the basis of the electronic nature of the axial ligands and on the difference in pheonotype of their original host strains (the Rr-CooA shuts down in an oxidative environment). The first “activated” state structure (analogous to CO bound) of Ch-CooA was reported by Borjigin et al.18 using the mutant designated as LL-Ch-CooA, where both Asn127 and Ser128 were replaced by leucine. The DNA-binding domain of monomer A that contains the heme is in an “active” conformation, analogous to the DNA complex of the E. coli CRP19. Upon CO binding, the N-terminal region is displaced away from the heme by ~20Å and positioned between the heme binding and DNA binding domains. This acts as bridge between the two domains and evidently provides the free energy for the large reorientation of the DNA-binding domain. Kubo et al. 20 suggested the displacement of the C-helix and a heme sliding motion upon CO activation by comparing the UV resonance Raman Trp110 bands of the active and inactive forms of Rr-CooA. They also found a change of the Fe−CO and CO stretching frequencies, but no effect on the Fe−His mode upon addition of DNA20.
This paper investigates the low frequency vibrational modes of the inactive ferric and ferrous forms of Ch-CooA, along with the active CO-bound form with and without the presence of the CooA-specific DNA. These modes are revealed by using vibrational coherence spectroscopy (VCS). The theoretical and experimental details of this nonlinear optical technique can be found elsewhere21-25. VCS has an advantage over frequency domain techniques such as infrared and Raman spectroscopy, which have difficulty probing the spectrum below ~ 200 cm-1 in aqueous solution. At room temperature, kBT~200 cm-1, these low frequency modes are thermally excited and are likely involved as reaction coordinates for protein function. For example, the doming mode near 40 cm-1 has been shown to play an important role in diatomic ligand binding to heme systems26;27. This study reveals protein-induced distortions of the heme that vary dependent upon the activation state of the protein. It also probes low frequency modes that are coupled to the photodissociation of the heme axial ligands in the Ch-CooA system.
The paper is organized as follows: The coherence and Raman spectra of the ferric form of Ch-CooA are compared and the new low-frequency modes in the region below 200 cm-1 are revealed. The vibrational coherence spectra of ferrous Ch-CooA are presented, along with evidence that indicates photodissociation and rebinding of the N-terminal α-amino acid. In the “active” CO bound state, we observe robust vibrational coherence spectra of the Ch-CooA photoproduct. The VCS measurements display a strong mode near 230 cm-1 that is assigned to a rapidly relaxing and unusually strong ν(Fe-His) bond. The strong Fe-His bond is suggested to be related to the thermal stability of the Ch-CooA. Finally, we also present results that probe perturbations of the Ch-CooA-CO heme structure following the binding of CooA-specific DNA. Although the CO rebinding kinetics are unambiguously perturbed by DNA binding, the heme structure remains largely unaffected, suggesting that the DNA binding primarily affects the configuration of the heme distal pocket but not the heme itself.
Experimental
Sample preparation
Ferric C-terminal Histidine tagged Ch-CooA from Carboxydothermus hydrogenoformans was prepared as discussed previously8;28. The ferric sample was generated by diluting the stock protein in Tris-HCl buffer (0.05 M, 0.5 M KCl, pH 8.0) to the required concentration. Ferrous: To prepare ferrous Ch-CooA (0.05 M Tris-HCl buffer, 0.5 M KCl, 0.006M CaCl2 and 0.005M DDT at pH 8.0), the ferric protein was degassed for 20 min under an argon atmosphere and subsequently, the samples were transferred into a glovebox and 3 μL of 1 M sodium dithionite (Na2S2O4) was added to 250 μL of buffered sample. Ch-CooA−CO: CO gas (Med-Tech, Medford, MA) was flushed slowly over the surface of the ferrous sample for about 40 min so that the CO concentration is saturated (~1mM). Ch-CooA−CO/DNA complex: Two strands of 26-mer oligonucleotides containing PcooF (5’−ATAACTGTCATCTGGCCGACAGACGG−3’ and 5’−CCGTCTGTCGGCCAGATGACAGTTAT−3’) were purchased from Eurofins MWG Operon, Atlanta. These strands were heated at 80 °C for 5 min and then annealed at room temperature for ~3hr to form double stranded DNA. Double strand DNA is added to Ch-CooACO, which is prepared by above procedure. The final concentrations of protein and target DNA were 50 μM and 250 μM respectively. Since Ch-CooA−CO has a Kd of 25 nM8 for the specific DNA, the concentration of specific DNA used was enough to allow 99 % of the protein to bind to the target DNA. For the vibrational coherence spectroscopy measurements, the concentration of the protein samples was adjusted so that the optical density of the sample at the pump wavelength was between 0.7 and 1.0 O.D. in a 1-mm path length spinning sample cell (50-100 μM). The average pump and probe laser power at the sample was ~8 and ~4 mW respectively. The absorption spectra of all samples were recorded after the preparation procedure to confirm that the desired chemical modifications were achieved and that they were complete. Absorption spectra were also taken after the experiments to check for possible photochemical damage and none was observed.
Optical System
The experimental optical system has been described elsewhere and the details are given in the supporting information. Two different detection geometries, each providing for optimal enhancement of a different range of frequencies, are used to obtain the vibrational coherence spectra. In an open-band detection scheme, a photodiode is used to measure the entire spectral bandwidth of the probe pulse. This optimizes the fidelity of the low frequency response in the range 20–100 cm-1. The detuned detection scheme selectively enhances the higher frequency components of the third order polarization response, which generates a coherent signal with improved reliability in the ~200–400 cm-1 region. The detuned detection is accomplished using a monochomator that disperses the probe pulse and detects 5 or 6 nm away from the carrier wavelength with a bandwidth of 0.5 nm. The results obtained from the detuned configuration allow comparison with frequency domain techniques like resonance Raman spectroscopy, which probe modes above 200 cm-1.
Data Analysis
The experimental data have components arising from ultrafast population transfer as well as vibrational coherence. To obtain the residual coherence signal, the monotonic population decay components must be removed. Digitization of the experimental signal is done using a lock-in amplifier (LIA) on a 24 bit scale, allowing for a sufficient dynamic range to detect the low amplitude coherence signals. The fractional change of transmittance, ΔT/T, for the open band oscillatory signals is on the order of 10-4-10-5. We used linear predictive singular value decomposition (LPSVD) to analyze the data, which simultaneously fits the exponential decay for population dynamics and the damped cosine functions that describe the vibrational oscillations. A maximum entropy method (MEM) algorithm is also used to remove assumptions regarding the number of exponential decay processes and retrieves the oscillatory signal with a second independent method. When the two methods agree, we are assured that the extracted low frequency oscillations are contained within the signal-to-noise of the experimental measurement.
Resonance Raman spectra
Resonance Raman spectra were obtained using a standard setup with a 90° light-collection geometry and a single grating monochromator (Acton SP2500 with 1800g/mm holographic UV optimized grating; Princeton Instruments, Princeton, NJ). An optical polarization scrambler was inserted in front of the monochromator to obtain the intensity of the scattered light without bias from the polarization-sensitive grating. The monochromator output was coupled to a liquid nitrogen-cooled CCD (SPEX 10:400B, Princeton Instruments, Princeton, NJ). In order to improve detection in the low-frequency domain of Raman shifts, an interferometric notch filter (Kaiser Optical Systems, Ann Arbor, MI) was used to extinguish the elastically and quasi-elastically scattered laser light. Samples were excited with a 413.1 nm laser line generated by a krypton laser (Innova 300, Coherent) with power of ~ 6 mW. A standard quartz cuvette (NSG Precision Cells, Farmingdale, NY) was used for the experimental Raman measurements. All Raman spectra are frequency calibrated using neat fenchone with ± 1 cm-1 spectral resolution.
Results
The x-ray crystal structure of the ferrous form of imidazole bound Ch-CooA14, reduced by xray irradiation, is shown in Fig. 1 (from PDB-2FMY with resolution of 2.2 Å). The ligation states of both Rr-CooA and Ch-CooA are shown in the lower panels. The two monomers are shown in blue and red colors. The effector-binding N-terminal (B- and C-helix and eight β-barrel) and DNA-binding C-terminal (a helix-turn-helix, E-, and F-helix) are shown in lighter and darker colors, respectively. The recognition F-helix, heme, and histidine axial ligand are shown in dark blue/red, green, and yellow colors, respectively.
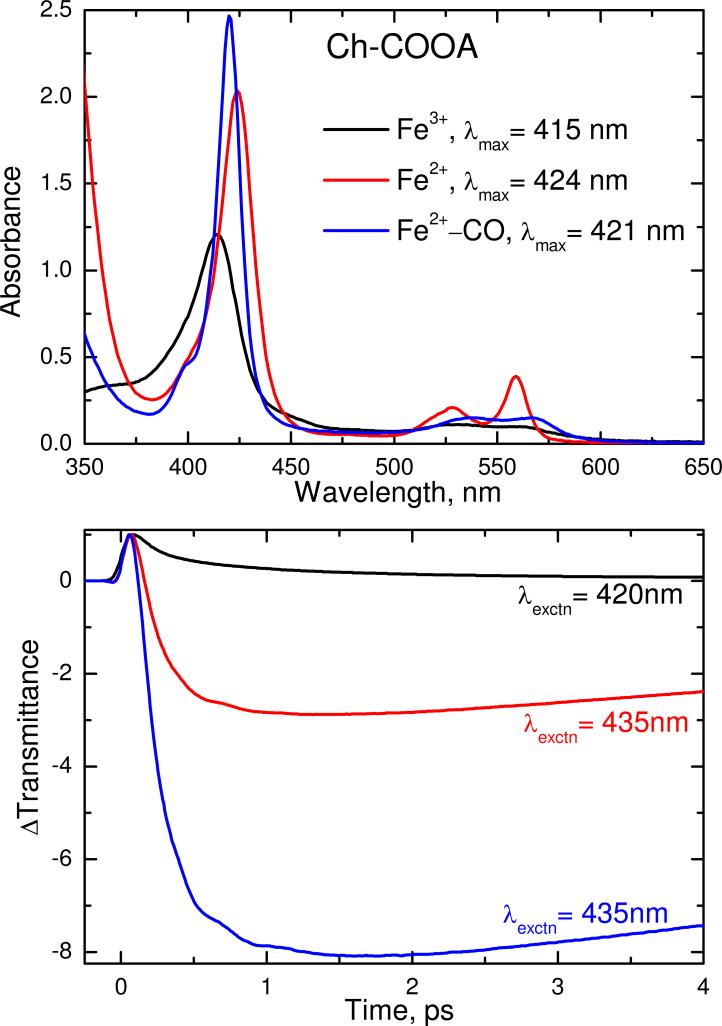
Figure 2 shows the electronic absorption spectra of the ferric, ferrous and CO bound forms of Ch-CooA. The Soret absorption maxima appeared at 415, 424 and 421 nm for the ferric, ferrous and CO-bound forms respectively. All these forms of Ch-CooA are typical of six coordinate, low spin heme proteins. The femtosecond time resolved optical transmittance of all three forms were obtained by exciting at 420 nm for ferric and at 435 nm for the ferrous and CO bound forms. The kinetic traces out to 4 ps are shown in Fig. 2B and the longer time kinetic decays for ferrous and CO bound form are given in the supporting information, Fig. S1. The ferric form has a prompt bleach (ΔT > 0) and coherence coupling signal, followed by a recovery to thermal equilibrium. The time constants (amplitudes) for the ferric sample are 162 fs (0.61) and 1.04 ps (0.34), with an offset of 0.05. In contrast, the ferrous and CO-bound forms show a strong antibleach (ΔT < 0). The primary time constant for the ferrous material is 8.46 ps. The geminate recombination of CO occurs on sub-nanosecond time scales with a high geminate yield (> 98%), and a non-exponential behavior. The details of the CO rebinding analysis will be published elsewhere (Benabbas, et al., in preparation).
Figure 2.
The upper panel compares the electronic absorption spectra of the ferric, ferrous and ferrous CO-bound forms of Ch-CooA at pH 8. The Soret absorption maxima are located at 415, 424 and 421 nm for the respective states. The lower panel displays the normalized time resolved transmittance (ΔT) of the above complexes obtained with pump/probe excitation at 420 nm for ferric and 435 nm for the ferrous and CO bound complexes. The ferric transient response shows a bleaching recovery signal (ΔT > 0). In contrast, the ferrous and CO bound forms show a strong transient absorption signal (ΔT < 0) at 435 nm that indicates ligand photolysis in both cases. The time constants are reported in the text.
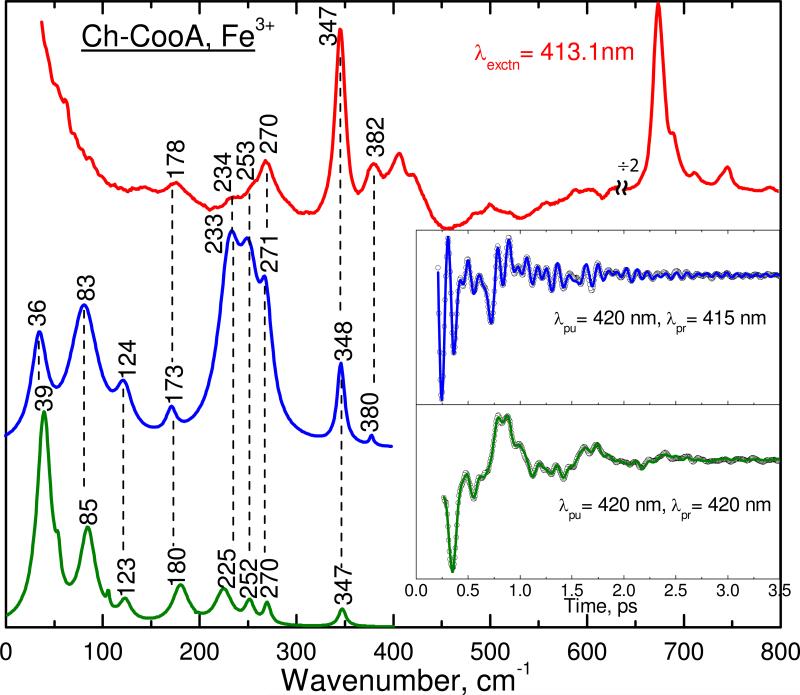
Figure 3 shows the correlation between the Raman and coherence spectra of the ferric form of Ch-CooA. The inset shows the oscillatory components of the experimental coherence data (detuned, blue upper curve and open-band, green lower curve). The spontaneous resonance Raman spectrum (red) measured at 413.1 nm is included at the top of the figure. Typically, the Raman spectrum is unable to detect vibrational frequencies below 150 cm-1 because of the strong Rayleigh and quasielastic light scattering and the nonlinearities in the low-frequency region of the cut-off filters used to attenuate the large flux at the laser excitation. There is a very good correlation between the coherence spectra and the Raman spectrum. For example, the strongest modes such as ν8 (347), 270 and 252 cm-1 in the Raman spectrum are correlated very well with the vibrational modes seen in the open band (347, 270 and 252 cm-1) and detuned detection (348, 271 and 253 cm-1). A mode at 380 cm-1 in the detuned spectrum is correlated to the 382 cm-1 mode observed in the Raman spectrum. The modes at 234 and 178 cm-1 in the Raman spectrum are correlated to vibrational activity seen at 233 and 173 cm-1 in the detuned detection and at 225 and 180 cm-1 in the open band measurements, respectively. There is also good correlation of the low-frequency modes between the open band (at 39, 85, and 123 cm-1) and the detuned detection (at 36, 83 and 124 cm-1). Overall the vibrational modes seen in the coherence spectra are correlated well to the Raman spectra; hence, the low frequency modes are revealed with confidence.
Figure 3.
The correlation between the Raman and coherence spectra for ferric Ch-CooA. The Raman spectrum (upper red) was measured with excitation at 413.1 nm, whereas the open band (lower green) and detuned (middle blue) coherence spectra were measured at a carrier wavelength of 420 nm. The detuned coherence data were collected with a 0.5 nm spectral window, located 5 nm to the blue of the carrier wavelength (415 nm). The insets show time domain oscillation data (circles) and LPSVD fits (lines) corresponding to the open band (green) and detuned (blue) detection methods. There is a very good correlation between the Raman and coherence spectral frequencies within accuracy of ± 5 cm-1
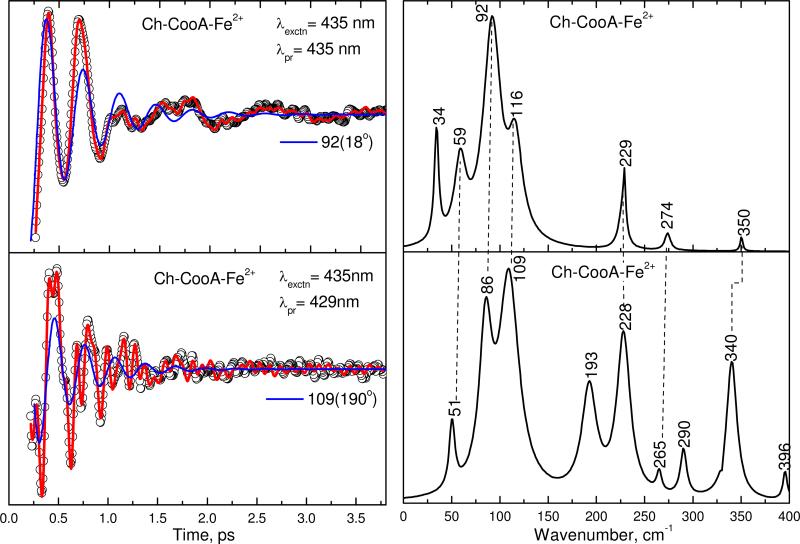
Figure 4 shows the open-band (top panel) and detuned (bottom panel) VCS spectra of the ferrous Ch-CooA at pH 8 obtained by exciting at 435 nm. The probe wavelength for the detuned detection is 429 nm. The experimental data were analyzed using LPSVD. The left panels show the oscillatory data (small circles) and the LPSVD fits (solid red line). The LPSVD components corresponding to the dominant modes at 92 and 109 cm-1 are shown as the blue lines and the LPSVD determined phases are noted in parentheses. The right panels show the corresponding amplitudes of the power spectra. There is a strong mode ~92 cm-1 along with peaks at 59 and 116 cm-1 in the open band spectra. An additional strong mode at 229 cm-1 is also seen. Within the experimental error of ±5 cm-1, these modes are also seen in the detuned spectrum along with some higher frequency modes. The difference between the vibrational modes obtained from the open band and detuned detection is partially due to the fact that detuned detection selects for, and gives, a better measurement of the higher frequencies29. Varying experimental signal-to-noise, as well as creation and build up of photoproducts can also lead to such differences.
Figure 4.
The open-band and detuned coherence spectra of ferrous Ch-CooA at pH 8. The pump and probe wavelength are given in the inset. The left panels show the oscillatory components (circles) and LPSVD fits (solid red lines). The LPSVD components corresponding to the dominant mode are also shown (blue solid lines), along with the extracted phases. The right panels show the corresponding power spectrum amplitudes. The correlated modes are shown by dotted lines.
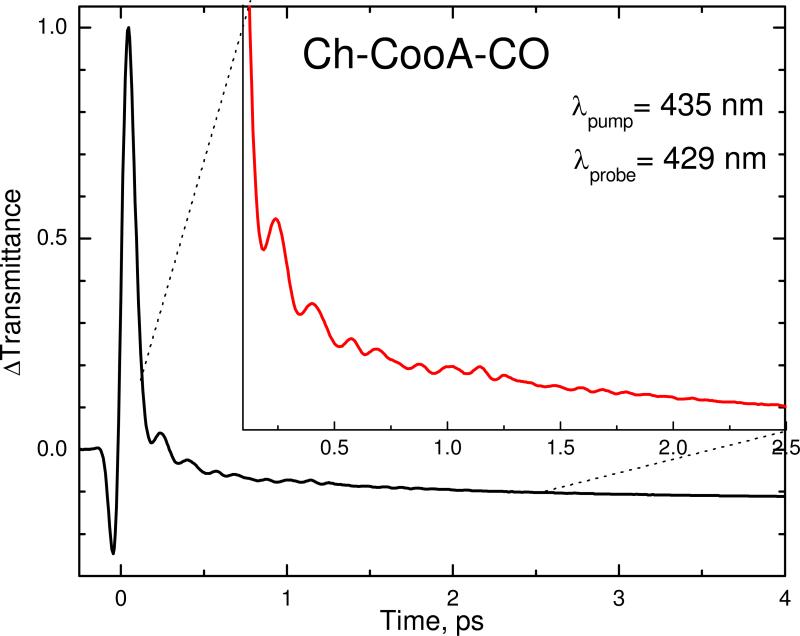
Since CO binding to Ch-CooA activates the expression of the coo operons by initiating a large conformational change of the DNA binding domains, it is important to measure the vibrational coherence spectra of the CO-bound form. Figure 5 shows the raw transient decay dynamics of the CO bound form of Ch-CooA pumped at 435 nm and probed, using detuned detection, at 429 nm. The oscillatory components of the signal are clearly seen in the expanded inset and they correspond to the coherent nuclear motion on the electronic potential surface of the newly formed 5-coordinate ferrous heme, after the photodissociation of CO. Because of its rapid CO rebinding time and large geminate yield, the CooA system offers a rare opportunity to monitor the vibrational coherence spectrum of a heme protein subsequent to CO dissociation. These measurements are possible using a high repetition rate (76 MHz) laser system only because of the ultrafast CO rebinding that takes place in CooA.
Figure 5.
The femtosecond coherence signal of CO bound Ch-CooA obtained using detuned detection. The pump wavelength is 435 nm and the probe is at 429 nm (detuned 6 nm to the blue of the carrier wavelength). The inset shows the signal from 120 fs to 2.5 ps. The oscillatory components of the signal are clearly apparent in the inset. They correspond to the strongly coupled coherent nuclear motion following CO photolysis.
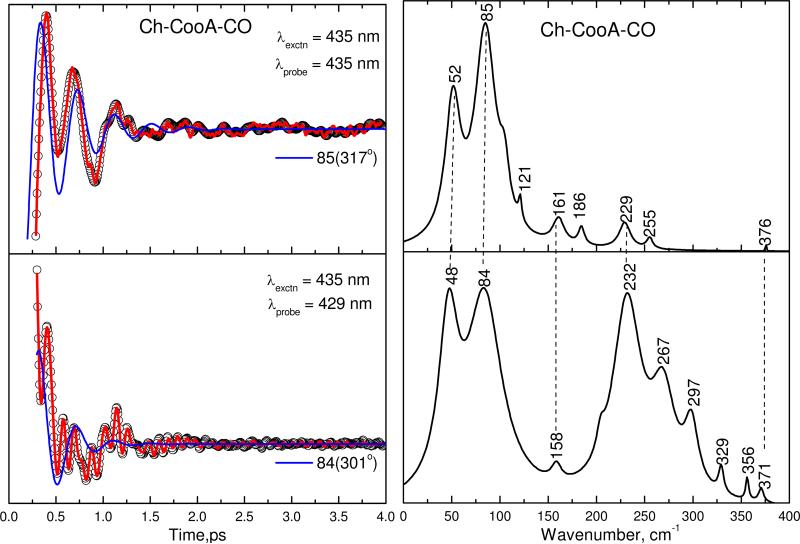
Figure 6 shows the open-band (top panel) and detuned (bottom panel) VCS spectra of Ch-CooA following CO dissociation. The left panels show the oscillatory data (small circles) and the LPSVD fits (solid red line). The LPSVD component corresponding to the dominant mode near 85 cm-1 and its phase are also shown as the blue line. The right panels show the corresponding amplitudes of the power spectra. The top panel, showing the coherence spectrum of Ch-CooA-CO using open band detection, has strong modes at 52 and 85 cm-1 that are correlated well with the detuned measurement in the lower panel, which shows modes at 48 and 83 cm-1. We also observe a strong feature near 230 cm-1 and this is assigned to the Fe−His stretching vibration of a transiently formed five-coordinated heme species.
Figure 6.
The open-band and detuned coherence spectra of the CO bound form of Ch-CooA at pH 8. The pump and probe wavelengths are given in the inset. The left panel shows the oscillatory components (circles) and the LPSVD fits (solid red lines). The LPSVD components corresponding to the dominant mode are also shown (blue solid lines), along with the extracted phases. The right panels show the corresponding power spectrum amplitudes. The correlated modes are shown by dotted lines.
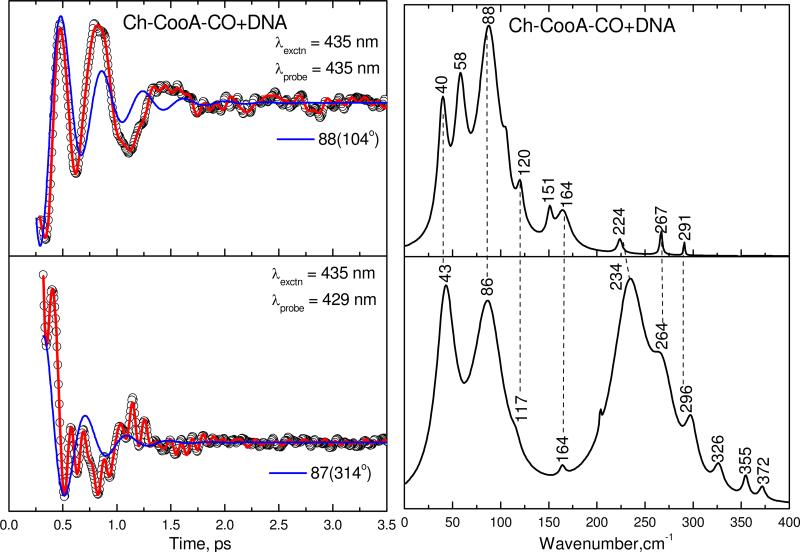
To understand the potential effect of specific DNA binding on the heme geometry and its environment, we have also measured the coherence spectra of the Ch-CooA-CO/DNA complex. Figure 7 shows the coherence spectra of the Ch-CooA-CO/DNA complex obtained using the open band (top panel) and detuned (bottom panel) methods, analogous to Fig. 6. The two strong modes at 40 and 88 cm-1 are again correlated well and are similar in the two samples (with and without the DNA). There is also a new mode appearing at 58 cm-1 that is only seen in the open band DNA bound spectra. There are some other weak modes appearing in the open band spectrum (such as 164, 224 and 267 and 291 cm-1) that can also be correlated with modes (at 164, 234, 264 and 296 cm-1) appearing in the detuned spectrum. The proposed Fe−His vibration at 234 cm-1 in the Ch-CooA-CO/DNA complex is shifted from 224 cm-1 in the open band spectrum, but this may be due to errors associated with its relatively weak intensity in the open band measurement.
Figure 7.
The open-band and detuned coherence spectra of Ch-CooA-CO that has bound to specific DNA at pH 8. The components of the figure are analogous to Fig.6.
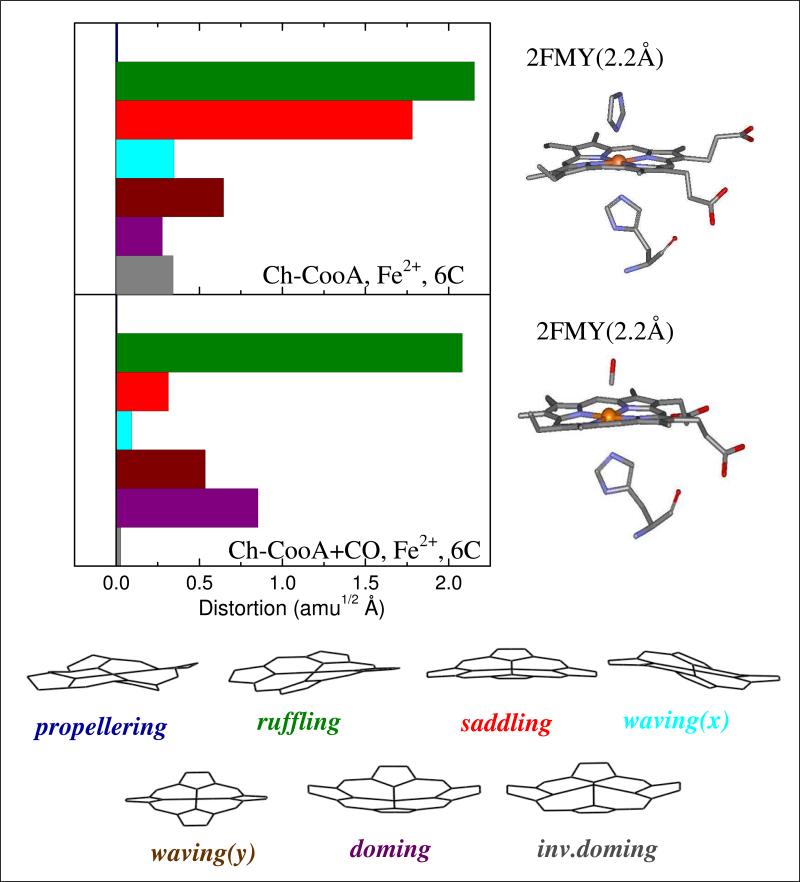
The normal coordinate structural decomposition (NSD) analysis for ferrous imidazole-bound Ch-CooA and CO-bound LL-ChCooA are shown in Fig. 8. NSD analysis determines the geometric distortion of the heme moiety in different protein environments by extracting the displacements along low-frequency normal modes that are referenced to the planar protoporphyrin IX core (porphine). The details of the NSD analysis are given in the supporting information. Basically, the x-ray structure is mapped onto the low-frequency, out-of-plane (OOP), modes of different symmetry (such as propellering, ruffling, saddling, waving(x), waving(y), doming and inverse doming). We have previously suggested that the symmetry forbidden low-frequency modes have relative intensities that are correlated with heme out-of-plane distortions, so long as the ratio of the ground and excited state force constants remains roughly independent of such distortions 23.
Figure 8.
The crystal structures and NSD analysis of the heme in imidazole-bound ferrous Ch-CooA and the CO bound form of LL-Ch-CooA. The displacement along each of the low frequency normal mode unit vectors of Fe porphine is given in mass weighted coordinates (amu1/2Å). The color coding for the modes is propellering (blue), ruffling (green), saddling (red), waving-x (light blue), waving-y (brown), doming (purple), and inverse doming (gray). The crystal structures are extracted from the protein data bank 2FMY14 and 2HKX18 for imidazole-bound ferrous Ch-CooA and CO bound LL-Ch-CooA, respectively. The NSD analysis of ferrous Ch-CooA shows strong ruffling and saddling distortions, whereas the CO bound form shows the ruffling distortion is maintained but the saddling distortion is strongly diminished.
We detect a significant amount of both the heme ruffling and heme saddling distortions to be present in ferrous Ch-CooA. In contrast, the CO-bound LL-Ch-CooA shows a significant decrease of the saddling distortion when compared to ferrous form. In addition, we observed a small increase of the doming distortion in the CO-bound form.
Discussion
Low frequency modes in the range of 10-200 cm-1 can be selectively investigated by use of femtosecond vibrational coherence spectroscopy. Specific heme modes having doming, ruffling and saddling content are found in this region and can be strongly excited by the thermal fluctuations (kBT ~ 200 cm-1 at 300K). Such modes are likely candidates for reaction coordinates that are associated with essential protein functions such as ligand binding (e.g., doming26;27) and electron transfer (e.g., ruffling30-33). In this paper we have studied the vibrational coherence spectra of Ch-CooA to investigate the following. (i) What different heme distortions are observed when the “active” and “inactive” forms of Ch-CooA are compared? (ii) What heme vibrational modes are coupled to the CO photo-dissociation reaction? (iii) Does the binding of DNA, specific to Ch-CooA, influence the heme environment? To answer these questions, we have carried out systematic experiments that compare the vibrational coherence spectra of the inactive forms of ferric and ferrous Ch-CooA, as well as the active form of Ch-CooA (i.e., the CO-bound form) with and without its specific target DNA sequence.
Ferric Ch-CooA
We have used VCS to reveal the low-frequency modes of ferric Ch-CooA and correlate the high frequency components with the steady-state resonance Raman spectrum. The mode near 35-40 cm-1 is thought to contain significant heme doming motions, while the mode near 80-85 cm-1 likely contains contributions from heme ruffling. In the case of cytochrome c, it is known that the axial methionine bond is photolabile in the ferrous form (with a rebinding time of ~6ps), but stable in the ferric form34-36. Thus, the ultrafast population decay dynamics (~1ps) seen in Fig. 2B for ferric Ch-CooA probably arise from cooling of the hot six-coordinated heme system. In further analogy with cytochrome c 34-36 the heme in ferrous Ch-CooA appears to undergo photolysis of its distal N-terminal α-amino acid ligand residue (vide infra).
Ferrous Ch-CooA
The kinetics of ferrous Ch-CooA shows a strong transient absorption at 435 nm, where 5-coordinate histidine-bound heme proteins have their absorption maxima. The time constants for ferrous Ch-CooA, measured using another laser system37 that spans time delays beyond 10 ns, are 8.7 ps (85%) and ~38 ps (15%). The longer timescale optical response is shown in the supporting information, Fig S1. The primary time constant, 8.7 ps, is assigned to the rebinding of the dissociated N-terminal α−amino acid ligand. The time constants for dissociated axial ligand rebinding to the ferrous histidine bound heme in different proteins are in this range38. For example, the time constant for rebinding of dissociated proline to ferrous Rr-CooA is 6.5 ps39 and methionine rebinding to ferrous cytochrome c takes place in 6.2 ps35. Since the rebinding can depend on the heme type, distal axial ligand, and protein environment, we suggest that the 8.7 ps phase is due to the rebinding of the N-terminal alanine with the protein in the 6-coordinate “closed” conformation. On the other hand, the slower time constant (38 ps) likely results from an inherent flexibility of the distal heme environment analogous to E. coli-DosH (35 ps) and human-neuroglobin (21 ps)38;40. This flexibility facilitates the replacement of the endogenous amino acid ligand by the functional CO ligand. Therefore, the slower time constant probably represents the rebinding of the α-amino acid from a more “open” 5-coordinate transient protein conformation. This latter conformation is functionally necessary in order to allow the CO to bind and initiate the transduction signal.
If there is distal ligand photolysis in the ferrous heme system, it should generate a transient five-coordinate His82-ligated heme having a stretching vibration around ~220-230 cm-1. In fact, we have observed a strong mode at ~228 cm-1 in the coherence spectrum of the ferrous system, Fig. 4, which supports the idea that ligand photolysis is taking place. When compared to the ν(Fe-His) of deoxy Mb at 220 cm-1, the ν(Fe-His) of Ch-CooA is up-shifted by ~8 cm-1. This may be due to a hydrogen bonding interaction between His82 and Asn47. The distance between these two amino acids is about 2.71 Å in the ferrous imidazole bound Ch-CooA14 (PDB:2FMYwith resolution 2.2 Å). Hence, based on the disappearance of transient absorption at 435 nm with a primary rate constant of 8.7 ps and a mode around 228 cm-1 in the ferrous Ch-CooA, we suggest that the photolysis and rebinding of the N-terminal alanine is taking place. The quantum yield of axial ligand photolysis in ferrous cytochrome c was found to be ~80%35 and, based on the relative intensities of the doming mode discussed below, we suggest that the photolysis yield for alanine in Ch-CooA is also less than the 100% normally found for the CO state.
The open band coherence spectra of ferrous Ch-CooA show a strong mode around 92 cm-1 and weaker modes around 59 and 116 cm-1. From earlier studies, specific heme distortion-induced low frequency modes have been revealed. For example, modes near ~40 cm-1 are suggested to involve heme doming26;27, modes near ~60 cm-1 may involve ruffling23;41 and those near ~90 cm-1 could involve saddling24;42. In our earlier study of ruffling in nitrophorin, we found that the “ruffling” mode shifted by ~20 cm-1 in the various ligand complexes23, presumabably due to the variable amounts of mode-mixing that take place at low frequency for different symmetry breaking steric situations. If the heme in the Ch-CooA thermophile is structured so that there is a general strengthening of macrocycle bonds and there is a systematic upshift in the Ch-CooA mode frequencies, the strong mode near 90 cm-1 in ferrous Ch-CooA could have a primary contribution from heme ruffling as suggested by the NSD analysis shown in Fig. 8. The modes near 109-116 cm-1 and 51-59 cm-1 would then correspond to modes having strong contributions from saddling and doming, respectively. There are two other strong modes appearing in the detuned spectra near 193 and 340 cm-1. The 340 cm-1 mode in the ferrous form is assigned to ν8, which is down-shifted somewhat when compared to the ν8 of the ferric form (347 cm-1) shown in Fig.3. The appearance of the mode at 193 cm-1 in the detuned spectrum, and its absence in the open band spectrum, is unexplained and requires further investigation.
CO bound Ch-CooA
Upon CO dissociation from Ch-CooA, the heme finds itself evolving on a product state potential energy surface, far from equilibrium. The resulting oscillations are much stronger than a ground state Raman coherence in the absence of a photochemical process (see Figs. 5 and 6). Assuming a 100% quantum yield of CO photolysis, as in the MbCO system43, all the vibrational modes observed in this experiment are due to the CO photo-dissociated ferrous heme system. There are two strong modes at 52 and 85 cm-1 that appear in the open band experiment.44 The 52 cm-1 mode is probably due to heme doming associated with the photo-dissociation44. The 85 cm-1 mode should involve heme ruffling in order to be consistent with the strong ruffling distortion revealed in the NSD analysis of CO-bound LL-CooA (see Fig. 8). Additional evidence of CO photolysis involves the appearance of a broad Fe-His stretching vibration near ~230 cm-1 in the coherence spectra (given in supporting information, Fig. S2 and S3) and a very strong transient absorption signal appearing at 435 nm (Fig. 2). The broadening in the range ~230-300 cm-1 seen, most dramatically, in the detuned measurement could arise from inhomogeneities in the protein ensemble or it could reflect relaxation of the system after the photolysis reaction. When we compare the steady-state absorption spectra of the CO-bound forms of cystathionine β-synthase, Ch-CooA and myoglobin, as shown in the supporting information, Fig, S4, we find the Soret bandwidth of Ch-CooA-CO is sharper than in the other heme systems. All of these proteins are six-coordinate, low spin, and have histidine as the proximal axial ligand. Since Ch-CooA is a thermophile, its conformation is less flexible and probably less heterogeneous. Hence, the broadening seen in the coherence spectra is probably not due to sample inhomogeneity.
To test for relaxation, the detuned data of the Ch-CooA-CO system (Fig.5) were analyzed by changing the time domain used for analysis. For example, the time domain was changed by varying the initial time, starting at 200, 300, 500 and 760 fs and the corresponding LPSVD fitted results are shown in the supporting information Fig S2. The power spectrum obtained using an initial time of 200 fs shows a broader peak at around 219 cm-1. With an initial time of 300 fs, the mode shifted to 232 cm-1 and narrows. The mode becomes weaker when time window is started at 760 fs. Alternatively, we also analyzed the data by selecting different 750 fs time windows. The period of oscillation for 220 cm-1 is 150 fs and the 750 fs window will contain several cycles in the data. We selected three 750 fs time windows beginning at 150 fs, 450 fs and 900 fs. The coherence spectra for these limited time domains contain fewer modes and are given in the supporting information, Fig. S3. It is observed that the Fe-His mode appears at 221 (broader), 233 (narrow) and 225 (narrow) cm-1, for the respective time frames. Hence the broadening of the feature and appearance of multiple modes could be due to relaxation of the hot transient 5C product, where the Fe−His mode is changing its frequency with time over the 150 fs to 4 ps time scale. Anharmonic coupling of the Fe-His mode to lower frequency modes such as heme doming45-47 as well as time dependent resonance effects associated with spectral diffusion as the system cools could contribute to the observed signals. Such effects may be analogous to time dependent evolution of the transient Fe-His mode observed following NO dissociation in myoglobin48.
The ν(Fe-His) frequency of photodissociated CooA reflects the structure of the heme environment. From the literature49, it is known that the ν(Fe-His) frequency ranges from 200-250 cm-1 for various 5-coordinate ferrous heme proteins (as can be seen in the supporting information, Table S1). The major cause of these variations is the H-bonding interaction of the histidine ligand with neighboring amino acids as well as strain and/or tilting of the Fe-His linkage with respect to the heme plane. It is interesting to compare the Fe-His mode of Ch-CooA with that of Rr-CooA, even though they have different thermal stability. The up-shift of the Fe-His vibration in Ch-CooA (~229 cm-1) compared to that of Rr-CooA (216 cm-1) could be associated with the thermal stability of Ch-CooA and the formation of a more stable binding geometry with a smaller tilt (less strain) of the His82 ring with respect to the heme plane.
It has been inferred from studies of LL-Ch-CooA that the His82-Asn47 hydrogen bond is broken upon CO binding and activation of the DNA binding domain18 . Since loss of the H-bond should lower the Fe-His frequency, the transient up-shift observed in the photoproduct of Ch-CooA-CO must be due to strengthening of the Fe-His bond on a short time scale probably by a reduction of the histidine tilt in the CO bound state. The complex pattern of the ν(Fe-His) stretching vibration seen in the Ch-CooA-CO photoproduct probably reflects time-dependent changes in the protein structure, including the orientation of the proximal histidine during rapid relaxation of the photo-dissociated system.
Comparisons to the Ch-CooA-CO/DNA complex
We investigated the possibility of heme environmental changes upon binding of specific DNA to the CO activated heme. Figs. 6 and 7 reveal a relatively small overall effect of DNA binding. However, there are some differences when the open band and detuned spectra are compared. The Fe-His mode located in the region 224-229 cm-1 upshifts and broadens in both samples. In addition, it should be noted that the open band measurement of the Ch-CooACO/DNA system shows a splitting of the mode at 52 cm-1 observed in the absence of DNA into modes at 40 and 58 cm-1. The mode at 58 cm-1 is a likely indicator of changes in the heme geometry that take place upon DNA binding. The absence of the 58 cm-1 mode in the detuned spectrum is unexpected, as are the upshift and broadening of the Fe-His mode. However, as mentioned above, such effects might arise from vibrational relaxation and rapid spectral diffusion that alters the resonance conditions when the different spectral acquisition methods are compared.
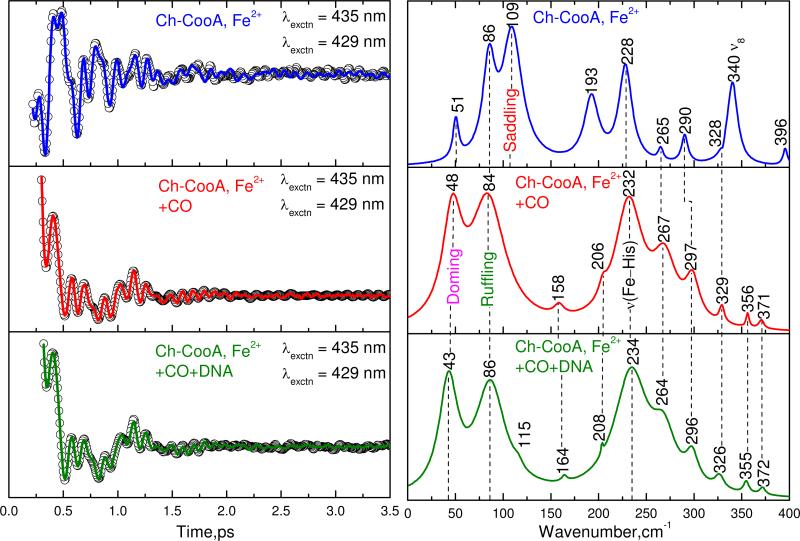
A direct comparison of the detuned coherence spectra of ferrous Ch-CooA and Ch-CooACO with and without DNA bound is shown in Fig. 9. The feature near 43-51 cm-1 is assigned to a mode with large heme doming content that arises from photodissociation of the distal CO or the N-terminal axial ligand. This mode is strongest in the CO-bound forms with and without DNA and weaker in the ferrous form. This is consistent with a large (~100%) photolysis of the CO ligand and a smaller photolysis quantum yield of the N-terminal alanine in the ferrous form..
Figure 9.
The detuned coherence spectra of ferrous, and Ch-CooA-CO with and without DNA bound. The pump and probe wavelengths are 435 and 429 nm respectively for all the three forms. The correlated modes are shown by dotted lines. The spectra of Ch-CooA-CO with and without bound DNA are similar and indicate that no significant difference in heme distortion when DNA is bound. This suggests that kinetic effects (to be published) are likely associated with changes in the distal pocket environment when DNA binds.
The mode near 86 cm-1 is correlated well in all three forms and we suggest that this mode includes significant heme ruffling motion23. This suggestion is based on the ferrous Ch-CooA and CO bound LL-Ch-CooA x-ray structures, which show a strong ruffling distortion in the NSD analysis (Fig.8). Ruffling should be correlated with one of the largest amplitude low frequency modes in the coherence spectrum. The mode near 109 cm-1 in the ferrous form is suggested to have a large contribution from heme saddling. This is based on the decreased saddling distortion of the heme in the CO bound form of LL-Ch-CooA as seen in Fig. 8. This mode disappears in both of the Ch-CooA-CO samples (with and without DNA) as seen in Fig. 9. Thus, the saddling distortion, evidently present in the resting ferrous form, is removed upon CO binding and this change in heme geometry is probably associated with the conformation transition that takes place when CO binds to the heme. The change in the heme conformation could arise directly from the exchange of the sixth ligand from alanine to CO, or more indirectly from the structural rearrangements of the N-terminal amino acids (or both). If replacement of the alanine ligand by CO independently alters the heme saddling geometry, this could presumably help to trigger protein conformational changes that activate the DNA binding domain.
The use of the 6-coordinate equilibrium crystal structure to analyze the coherence spectra of a photolabile species is useful, but can be potentially problematic if the protein relaxation is fast enough to reflect the heme surroundings of the newly formed pentacoordinate state. In the case of the ferrous protein, the relaxation in the 0-4 ps time window of the coherence experiment appears to be slow enough that we obtain a valid imprint of the initial state protein surroundings. The open band coherence spectra of the ferrous form (Fig. 4, upper panel) is consistent with the strong ruffling and saddling heme distortions obtained from the NSD analysis of the 6-coordinate crystal structure as shown in Fig. 8. The strong 92 and 116 cm-1 modes in the open band experiment are correlated with the 86 and 109 cm-1 modes seen in the detuned measurement. The disappearance of the 116 cm-1 mode in the open band measurement of the CO bound state (Fig. 6, upper panel) is therefore consistent with the decreased saddling distortion seen in the NSD analysis when CO replaces the N-terminal alanine as the sixth ligand.
Another strong mode that appears in all the three forms is found near ~230 cm-1. This mode is proposed to be the ν(Fe-His) stretching vibration of the five-coordinate high-spin ferrous heme system. Even though the difference of the mode frequencies is close to the measurement error, it is conceivable that the proximal histidine binding geometry is changing systematically as CO and DNA bind to the system and the VCS measurement is sensing these effects as well as transient relaxation following photolysis.
More generally, when we compare the vibrational frequencies of the Ch-CooA−CO and Ch-CooA−CO/DNA complexes, all mode frequencies are matching well with each other within the error of ± 5 cm-1. Since there is no major difference outside experimental error, this indicates that there is no major heme environmental change upon DNA binding. The results on Ch-CooA are consistent with prior observations of the heme environment in Rr-CooA20. Kubo et al 20 used resonance Raman spectroscopy to detect the ν(Fe-His) frequency of photodissociated Rr-CooA−CO at 216 cm-1 and observed no change in frequency upon binding the target DNA. This strongly suggests that binding of target DNA to CooA does not significantly alter the proximal side of the heme20. Finally, the mode at 340 cm-1, ν8, due to the Fe-N(pyrrole) stretching and pyrrole substituent bending, in the ferrous form of Ch-CooA is seen to disappear in the active CO bound form either with or without DNA. Similar to the disappearance of the 109 cm-1 “saddling” mode, this could reflect heme distortions present in the ferrous form that are removed upon CO binding. Once again, if these heme geometry changes are a direct result of the ligand exchange, they become candidates for helping to trigger the large conformational changes that activate the DNA binding domain when CO binds.
Summary
We have used vibrational coherence spectroscopy to investigate the low frequency vibrational modes of the heme in Ch-CooA. These modes are excited by the thermal bath and can assist the heme group in reaching thermally activated transition states. The ferric protein displays low-frequency heme oscillations that involve both heme doming and ruffling motions. There is also an excellent correlation between the VCS spectra and the higher frequency Raman modes. Ferric Ch-CooA does not appear to undergo photolysis and only rapid cooling processes and ground state coherences were observed.
In contrast, the ferrous sample is subject to photodissociation of the distal N-terminal α-amino group. This conclusion is based on the appearance of the Fe-His mode at ~228 cm-1, which is uniquely observed in the 5-coordinate ferrous state. The rebinding of the distal N-terminal α-amino group (alanine) was monitored using the transient optical response at 435 nm and the time constant for ligand rebinding of the majority “closed” population was found to be 8.5 ps. The functionally important “open” population was found to rebind in 38 ps. A strong mode observed at ~85-90 cm-1 in the ferrous sample is suggested to contain a significant contribution from heme ruffling, consistent with the protein-induced heme distortion along this coordinate derived from the NSD analysis.
When CO binds, the N-terminal α-amino group is replaced by CO, which activates the CooA for specific DNA binding by inducing a large conformational change in the DNA binding domain. VCS allowed us to study the accompanying differences in the heme structure between the inactive (ferrous) and active (CO bound) states. The low frequency spectra of both the CO bound and ferrous forms show a strong vibration at ~230 cm-1 that is assigned to the Fe-His stretching mode of the respective photoproducts. The appearance of strong heme “doming” modes in the vicinity of 40-50 cm-1 in Fig. 9 confirms the strong coupling of this mode to the CO photolysis reaction. Insofar as similar nuclear motions (e.g., heme doming) take place on the ground electronic state surface, this mode must also be an important reaction coordinate for CO rebinding to the photolyzed 5-cordinate state26. On the other hand, the details of how the CO ligand exchange in the 6-coordinate ferrous species takes place are not fully determined. One likely possibility is that an “open” form of the protein distal pocket slows the geminate rebinding of the alanine just enough for bimolecular CO binding to be competitive.
The change of the heme environment by adding CooA-specific DNA to Ch-CooA−CO was also investigated and no significant changes were observed in the low-frequency spectra. The VCS activity of vibrational modes at 109 cm-1 and 340 cm-1, which appear to involve heme saddling and Fe pyrrole motions, differentiate the state of the heme in the active and inactive forms of Ch-CooA. The coherence measurements indicate that significant heme perturbations, involving these coordinates take place upon CO binding.
Supplementary Material
Acknowledgment
We thank Gary Roberts Robert L. Kerby, Jose Serate and Nicholas D. Lanz for helpful comments and the expression and purification of Ch-CooA. This work is supported by grants from the NIH (DK35090) and the NSF (MCB-0744738).
Footnotes
Vibrational dynamics of Ch-CooA
Supporting Information Available
The details of the experimental set up, procedure for NSD analysis, the long time kinetic decays of the ferrous and CO bound forms of Ch-CooA with and without specific DNA, absorption spectra of the CO bound forms of cystathionine β-synthase, Ch-CooA and myoglobin, the LPSVD analysis of the CO bound form of Ch-CooA over different time domains, and the ν(Fe-His) stretching frequencies of different ferrous 5C heme proteins. This material is available free of charge via the internet at http://pubs.acs.org
Reference List
- 1.Vos MH. Biochim.Biophys.Acta. 2008;1777:15–31. doi: 10.1016/j.bbabio.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Cary SPL, Winger JA, Derbyshire ER, Marletta MA. Trends in Biochem.Sci. 2006;31:231–239. doi: 10.1016/j.tibs.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Rodgers KR, Lukat-Rodgers GS. J.Inorg.Biochem. 2005;99:963–977. doi: 10.1016/j.jinorgbio.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- 5.Liebl U, Bouzhir-Sima L, Negrerie M, Martin JL, Vos MH. Proc.Natl.Acad.Sci.U.S A. 2002;99:12771–12776. doi: 10.1073/pnas.192311699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimura H, Yoshioka S, Kobayashi K, Ohta T, Uchida T, Kubo M, Kitagawa T, Aono S. Biochemistry. 2006;45:8301–8307. doi: 10.1021/bi060315c. [DOI] [PubMed] [Google Scholar]
- 7.Aono S. Dalton Trans. 2008:3137–3146. doi: 10.1039/b802070c. [DOI] [PubMed] [Google Scholar]
- 8.Clark RW, Lanz ND, Lee AJ, Kerby RL, Roberts GP, Burstyn JN. Proc.Natl.Acad.Sci.U.S A. 2006;103:891–896. doi: 10.1073/pnas.0505919103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim M, Kuchinskas M, Youn H, Kerby RL, Roberts GP, Poulos TL, Spiro TG. J.Inorg.Biochem. 2007;101:1776–1785. doi: 10.1016/j.jinorgbio.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts GP, Youn H, Kerby RL. Microbiol.Mol.Biol.Rev. 2004;68:453–473. doi: 10.1128/MMBR.68.3.453-473.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puranik M, Nielsen SB, Youn H, Hvitved AN, Bourassa JL, Case MA, Tengroth C, Balakrishnan G, Thorsteinsson MV, Groves JT, McLendon GL, Roberts GP, Olson JS, Spiro TG. J.Biol.Chem. 2004;279:21096–21108. doi: 10.1074/jbc.M400613200. [DOI] [PubMed] [Google Scholar]
- 12.Youn H, Kerby RL, Conrad M, Roberts GP. J.Bacteriol. 2004;186:1320–1329. doi: 10.1128/JB.186.5.1320-1329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inagaki S, Masuda C, Akaishi T, Nakajima H, Yoshioka S, Ohta T, Pal B, Kitagawa T, Aono S. J.Biol.Chem. 2005;280:3269–3274. doi: 10.1074/jbc.M409884200. [DOI] [PubMed] [Google Scholar]
- 14.Komori H, Inagaki S, Yoshioka S, Aono S, Higuchi Y. J.Mol.Biol. 2007;367:864–871. doi: 10.1016/j.jmb.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 15.Akiyama S, Fujisawa T, Ishimori K, Morishima I, Aono S. J.Mol.Biol. 2004;341:651–668. doi: 10.1016/j.jmb.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 16.Youn H, Kerby RL, Roberts GP. J.Biol.Chem. 2004;279:45744–45752. doi: 10.1074/jbc.M404336200. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima H, Honma Y, Tawara T, Kato T, Park SY, Miyatake H, Shiro Y, Aono S. J.Biol.Chem. 2001;276:7055–7061. doi: 10.1074/jbc.M003972200. [DOI] [PubMed] [Google Scholar]
- 18.Borjigin M, Li H, Lanz ND, Kerby RL, Roberts GP, Poulos TL. Acta Crystallogr.D Biol.Crystallogr. 2007;63:282–287. doi: 10.1107/S0907444906051638. [DOI] [PubMed] [Google Scholar]
- 19.Parkinson G, Wilson C, Gunasekera A, Ebright YW, Ebright RE, Berman HM. J.Mol.Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 20.Kubo M, Inagaki S, Yoshioka S, Uchida T, Mizutani Y, Aono S, Kitagawa T. J.Biol.Chem. 2006;281:11271–11278. doi: 10.1074/jbc.M513261200. [DOI] [PubMed] [Google Scholar]
- 21.Kumar ATN, Rosca F, Widom A, Champion PM. J.Chem.Phys. 2001;114:701–724. [Google Scholar]
- 22.Kumar ATN, Rosca F, Widom A, Champion PM. J.Chem.Phys. 2001;114:6795–6815. [Google Scholar]
- 23.Kubo M, Gruia F, Benabbas A, Barabanschikov A, Montfort WR, Maes EM, Champion PM. J.Am.Chem.Soc. 2008;130:9800–9811. doi: 10.1021/ja800916d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruia F, Kubo M, Ye X, Ionascu D, Lu C, Poole RK, Yeh SR, Champion PM. J.Am.Chem.Soc. 2008;130:5231–5244. doi: 10.1021/ja7104027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruia F, Ionascu D, Kubo M, Ye X, Dawson J, Osborne RL, Sligar SG, Denisov I, Das A, Poulos TL, Terner J, Champion PM. Biochemistry. 2008;47:5156–5167. doi: 10.1021/bi7025485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye X, Ionascu D, Gruia F, Yu A, Benabbas A, Champion PM. Proc.Natl.Acad.Sci.U.S A. 2007;104:14682–14687. doi: 10.1073/pnas.0702622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srajer V, Reinisch L, Champion PM. J.Am.Chem.Soc. 1988;110:6656–6670. [Google Scholar]
- 28.Kerby RL, Youn H, Thorsteinsson MV, Roberts GP. J.Mol.Biol. 2003;325:809–823. doi: 10.1016/s0022-2836(02)01203-2. [DOI] [PubMed] [Google Scholar]
- 29.Rosca F, Kumar ATN, Ye X, Sjodin T, Demidov AA, Champion PM. J.Phys.Chem A. 2000;104:4280–4290. [Google Scholar]
- 30.Liptak MD, Wen X, Bren KL. J.Am.Chem.Soc. 2010;132:9753–9763. doi: 10.1021/ja102098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Chem.Soc.Rev. 1998;27:31–41. [Google Scholar]
- 32.Walker FA. Coord.Chem.Rev. 1999;186:471–534. [Google Scholar]
- 33.Ma JG, Zhang J, Franco R, Jia SL, Moura I, Moura JJ, Kroneck PM, Shelnutt JA. Biochemistry. 1998;37:12431–12442. doi: 10.1021/bi981189i. [DOI] [PubMed] [Google Scholar]
- 34.Negrerie M, Cianetti S, Vos MH, Martin JL, Kruglik SG. J.Phys.Chem.B. 2006;110:12766–12781. doi: 10.1021/jp0559377. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Ye X, Demidov AA, Rosca F, Sjodin T, Cao W, Sheeran M, Champion PM. J.Phys.Chem.B. 2000;104:10789–10801. [Google Scholar]
- 36.Zang C, Stevens JA, Link JJ, Guo L, Wang L, Zhong D. J.Am.Chem Soc. 2009;131:2846–2852. doi: 10.1021/ja8057293. [DOI] [PubMed] [Google Scholar]
- 37.Yu AC, Ye X, Ionascu D, Cao WX, Champion PM. Rev.Sci.Inst. 2005;76 [Google Scholar]
- 38.Vos MH, Battistoni A, Lechauve C, Marden MC, Kiger L, Desbois A, Pilet E, de Rosny E, Liebl U. Biochemistry. 2008;47:5718–5723. doi: 10.1021/bi800288z. [DOI] [PubMed] [Google Scholar]
- 39.Kumazaki S, Nakajima H, Sakaguchi T, Nakagawa E, Shinohara H, Yoshihara K, Aono S. J.Biol.Chem. 2000;275:38378–38383. doi: 10.1074/jbc.M005533200. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita T, Bouzhir-Sima L, Lambry JC, Liebl U, Vos MH. Journal of Biological Chemistry. 2008;283:2344–2352. doi: 10.1074/jbc.M708123200. [DOI] [PubMed] [Google Scholar]
- 41.Karunakaran V, Benabbas A, Sun Y, Zhang Z, Singh S, Banerjee R, Champion PM. J.Phys.Chem B. 2010;114:3294–3306. doi: 10.1021/jp909700r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karunakaran V, Denisov I, Sligar SG, Champion PM. J.Phys.Chem B. 2011;115:5665–5677. doi: 10.1021/jp112298y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuresko DD, Webb WW. Biophys.J. 1978;24:382–383. doi: 10.1016/S0006-3495(78)85390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosca F, Kumar ATN, Ionascu D, Ye X, Demidov AA, Sjodin T, Wharton D, Barrick D, Sligar SG, Yonetani T, Champion PM. J.Phys.Chem A. 2002;106:3540–3552. [Google Scholar]
- 45.Korostishevsky M, Zaslavsky Z, Stavrov SS. Biophys.J. 2004;86:656–659. doi: 10.1016/S0006-3495(04)74143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bitler A, Stavrov SS. Biophys.J. 1999;77:2764–2776. doi: 10.1016/S0006-3495(99)77109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schott J, Dreybrodt W, Schweitzer-Stenner R. Biophys.J. 2001;81:1624–1631. doi: 10.1016/S0006-3495(01)75816-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruglik SG, Yoo BK, Franzen S, Vos MH, Martin JL, Negrerie M. Proc.Natl.Acad.Sci.U.S A. 2010;107:13678–13683. doi: 10.1073/pnas.0912938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitagawa T. Biological Applications of Raman Spectroscopy. John Wiley & Sons; New York: 1988. pp. 97–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.