Abstract
Cell death in bacteria can be triggered by activation of self-inflicted molecular mechanisms. Pathogenic bacteria often make use of suicide mechanisms in which the death of individual cells benefits survival of the population. Important elements for programmed cell death in bacteria are proteinaceous toxin–antitoxin systems. While the toxin generally resides dormant in the bacterial cytosol in complex with its antitoxin, conditions such as impaired de novo synthesis of the antitoxin or nutritional stress lead to antitoxin degradation and toxin activation. A widespread toxin–antitoxin family consists of the ε/ζ systems, which are distributed over plasmids and chromosomes of various pathogenic bacteria. In its inactive state, the bacteriotoxic ζ toxin protein is inhibited by its cognate antitoxin ε. Upon degradation of ε, the ζ toxin is released allowing this enzyme to poison bacterial cell wall synthesis, which eventually triggers autolysis. ε/ζ systems ensure stable plasmid inheritance by inducing death in plasmid-deprived offspring cells. In contrast, chromosomally encoded ε/ζ systems were reported to contribute to virulence of pathogenic bacteria, possibly by inducing autolysis in individual cells under stressful conditions. The capability of toxin–antitoxin systems to kill bacteria has made them potential targets for new therapeutic compounds. Toxin activation could be hijacked to induce suicide of bacteria. Likewise, the unique mechanism of ζ toxins could serve as template for new drugs. Contrarily, inhibition of virulence-associated ζ toxins might attenuate infections. Here we provide an overview of ε/ζ toxin–antitoxin family and its potential role in the development of new therapeutic approaches in microbial defense.
Keywords: Antimicrobial peptides, Biochemistry, Cell death, Drug development, Drug resistance, Infectiology
Introduction
The genomes of bacteria contain many mobile genetic elements (MGEs) such as transposons, plasmids, or bacteriophages that have been acquired by horizontal gene transfer (HGT). Many MGEs serve as shuttles for genes that are beneficial to bacteria during proliferation in a host environment or in soil. Several supportive MGEs were found in the genome of pathogenic bacteria and encode for genes conferring antibiotic resistance and virulence factors such as exotoxins, adhesins, and secretion systems [1–3]. However, this additional genetic material imposes a metabolic burden to the bacterium. Thus, genes acquired by HGT are prone to elimination from bacterial genomes in absence of selective pressure [4]. To prevent removal, MGEs contain maintenance systems that maximize their retention within a bacterial population [5–7]. The most abundant group of such stabilization systems are proteinaceous type II toxin–antitoxin (TA) systems. Initially, these systems were identified as maintenance elements of conjugable low-copy number plasmids insuring stable inheritance by a simple but effective mechanism [8–10]. Once a plasmid encoding a TA system has been acquired, the host bacterium starts to express two gene products from the TA operon: a “toxin” protein that threatens to poison the host from within and a cognate “antitoxin” protein that inactivates the toxin by complex formation. When sufficient antitoxin is present, the bacterium is little affected by the TA system. However, the antitoxin protein is prone to faster degradation by intracellular proteases than the toxin. If offspring loose the plasmid encoding for the TA system during cell division, the continuous depletion of inherited antitoxin protein causes release of the residual cytosolic toxin. Eventually, the toxin triggers post-segregational killing (PSK) of the host, since such a plasmid-free bacterium would otherwise be favored when competing with the metabolically handicapped plasmid-bearing cells. A similar genome maintenance function was also reported for chromosomally encoded type II TA systems [7, 11]. Apart from that, it is emerging that several bacterial species use TA systems for their own benefit [12–14] and some TA systems were reported to be involved in bacterial stress response [15, 16], protection against PSK [17], phage exclusion [18, 19], differentiation [20], and virulence [21, 22]. Yet, some of these proposed functions such as a role of TA systems in bacterial stress response are still a matter of ongoing debate due to a lack of sufficient supportive data [23].
A TA element commonly found in multiresistance plasmids and chromosomes of various human pathogens are the ε/ζ systems. Whereas plasmid-encoded ε/ζ systems ensure stable inheritance of plasmids, chromosomally encoded homologues were reported to contribute to virulence of pathogens. The recently uncovered mechanism of ζ toxins sheds new light on these different functions of the ε/ζ systems. Moreover, the unique enzymatic mechanism of the ζ toxins provides new targets for the development of novel antibiotics.
Cell killing by ζ toxins
As in other type II TA systems, the bacteriotoxic ζ protein normally resides dormant in the cytosol of bacteria in complex with ε. However, once the levels of ε decrease, for instance when de novo expression is impaired, inhibition of ζ fails and the toxin exerts its poisonous activity [24, 25]. Until recently, the bacteriotoxic mechanism of ζ toxins has remained elusive. Structural and mutational studies of the ε/ζ system from Streptococcus pyogenes and pneumococcal ε/ζ antitoxin–toxin (PezAT) from Streptococcus pneumoniae suggested that ζ toxins might be kinases that phosphorylate an enigmatic target under consumption of ATP [26–28]. This hypothesis was based on the structure of the ζ toxins, since ζ resembles nucleoside monophosphate kinases and polynucleotide kinases and mutations of putative catalytically important residues rendered the toxins inactive [28, 29]. Moreover, antitoxins occlude the proposed ATP-binding site of ζ toxins and thereby inactivate the toxins within the complexes.
First hints for deciphering the functional mechanism of ζ toxins came from an in vivo study by Zielenkiewicz and Ceglowski who showed that ζ is bactericidal in the gram-positive Bacillus subtilis and causes growth arrest and filamentation in the gram-negative Escherichia coli [25]. Furthermore, an attenuated ζ variant was shown to inhibit cell proliferation, causing cell lysis of a subpopulation in B. subtilis [24]. In addition, Lioy et al. found that elevated expression of ζ in E. coli causes a pleiotropic inhibition of DNA replication, transcription, and translation without specifically targeting any of these cellular events.
Recently, we deciphered the molecular mechanism of ζ-induced cell death using the chromosomally encoded ζ toxin PezT [30]. Expression of an attenuated PezT variant in E. coli caused a phenotype like that observed during treatment with β-lactam antibiotics. Cytosolic levels of PezT inhibited binary fission and caused formation of mid-cell positioned bulges, which eventually burst and lead to cell death (Fig. 1a). We showed that ζ toxins specifically phosphorylate the peptidoglycan precursor UDP-N-acetylglucosamine (UNAG) at the 3′-OH group of the N-acetylglucosamine moiety and thus define a new family of small-molecule kinases (Fig. 1b). The toxic reaction product, UDP-N-acetylglucosamine-3-phosphate (UNAG-3P), accumulates in the cytosol and competitively inhibits MurA. Since MurA is the essential enzyme that catalyzes the first committed step of peptidoglycan synthesis [31], UNAG-3P thereby impairs the entire pathway (Fig. 1c). The key residues for UNAG phosphorylation are well conserved in ζ homologues from other bacteria (Fig. 2), and we could indeed verify the UNAG kinase activity for plasmid-borne ζ toxin. Thus, formation of UNAG-3P is also the cause for PSK induced by plasmid-stabilizing ε/ζ systems.
Fig. 1.

Bacteriotoxic mechanism of ζ toxins. a Cytosolically expressed ζ toxins in E. coli cause inhibition of binary fission, membrane bulge formation, and eventually autolysis. b ζ toxins catalyze the ATP-dependent phosphorylation of the nucleotide sugar UDP-N-acetylglucosamine (UNAG) at the 3′-OH of the N-acetylglucosamine moiety producing UDP-N-acetylglucosamine-3-phosphate (UNAG-3P). c This reaction introduces a detrimental side branch to bacterial peptidoglycan synthesis. UNAG is the universal precursor of the sugar backbone of the peptidoglycan macromolecule, and normally, a large fraction of the nucleotide sugar is converted into UDP-N-acetyl muramic acid (UDP-MurNAc) through modification at the 3′-end of UNAG. The processing of the UNAG 3′-OH involves an enolpyruvyl transfer from phopsphoenolpyruvate (PEP) catalyzed by MurA, which is followed by an NADH-dependent reduction catalyzed by MurB. This step prepares UNAG for the subsequent addition of the muropeptide. However, the phosphorylation of UNAG by ζ converts the former product of MurA into a competitive inhibitor that blocks cell wall synthesis. In the presence of the antitoxin ε, however, the kinase activity of ζ is inhibited
Fig. 2.

Sequence alignment of representative ζ toxins from gram-positive and gram-negative bacteria. The NCBI accession numbers are given in brackets. S. pneumoniae (NP_345525), S. agalactiae (NP_735780), Oribacterium sinus (ZP_03991854), Eggerthella lenta (YP_003180798), E. faecalis (ZP_07107801), S. pyogenes (YP_232762), Lactobacillus antri (ZP_05746663), Bacillus fusiformis (ZP_07050953), E. coli O103:H2 (NC_013353), Neisseria cinerea (ZP_05983406), Bacteroides intestinalis (ZP_03012857), and Capnocytophaga ochracea (YP_003140484). Identical residues are colored in dark green and according to the decreasing similarity from light green through orange to yellow. Secondary structure elements of ζ from S. pyogenes are shown above the sequence. Note that 71 N-terminal residues of the E. coli sequence were omitted from the alignment, and some sequences were truncated at their C-terminus. Squares indicate those residues involved in the interaction with UNAG in the ζ from S. pyogenes. Diamonds indicate those residues whose mutation by site-directed mutagenesis in PezT and ζ from S. pyogenes were shown to abolish toxicity. The conserved Walker A motif that is crucial for ATP binding is marked in pink, and the loop region forming contacts to the UNAG phosphates is marked in blue
Plasmid-borne ε/ζ systems
ε/ζ systems encoded on plasmids of gram-positive pathogens
The plasmid-borne ε/ζ system was originally discovered on the low-copy number plasmid pSM19035 from a clinical isolate of the gram-positive pathogen S. pyogenes [32]. This plasmid confers resistances to high levels of erythromycin and lincomycins [33] and is member of the broad host family of Inc18-like plasmids. Although pSM19035 has a low-copy number (two to five plasmids per cell [34]), it is maintained in absence of any selective antibiotic. This stable inheritance is conferred by plasmid-encoded segregation (seg) loci, whose gene products control the plasmid copy number, improve plasmid partitioning after replication, ensure a better-than-random segregation during cell division, and trigger PSK (see Lioy et al. [6]). Among those different maintenance mechanisms, PSK leading to suicide of plasmid-devoid offspring primarily contributes to stable inheritance of pSM19035 [25]. PSK is evoked by the segB1 locus that encodes three proteins: ω, a global transcription repressor of pSM19035, the ε antitoxin, and the ζ toxin [32, 35] (Fig. 3a). The proteins ε and ζ are translated from a bicistronic ε–ζ mRNA, whose transcription is regulated by the ω repressor [36]. Upon plasmid loss, the pool of proteolytically unstable ε antitoxin is eliminated by ATP-dependent proteases and the released ζ prevents further proliferation of plasmid-free cells [24, 25, 37].
Fig. 3.

Operon architecture of the plasmid-borne ε/ζ systems and the chromosomal PezAT system. a Architecture of the ω/ε/ζ operon that stabilizes multiresistance plasmids in a broad range of bacteria. The ω/ε/ζ operon contains the two discrete promoters Pω and Pε. Pω produces transcripts containing either the ω, the ω/ε, or the ω/ε/ζ ORFs and is auto-repressed by ω (yellow). Pε is a weak constitutive promoter that produces transcripts containing the ORFs of ε alone or ε/ζ. The antitoxin ε (blue) and the toxin ζ (red) proteins form an inactive complex after translation. In case the encoding plasmid is lost, ζ is released from the ε/ζ complex through continuous degradation of ε by constitutive proteases. b Organization of the pezAT operon from S. pneumoniae. In contrast to the ω/ε/ζ system, no ω operon is present and autoregulation of the operon is achieved by the antitoxin PezA (blue) with the toxin PezT (red) acting as corepressor. Similar to ε/ζ, PezA and PezT form a stable complex. When and how the toxin PezT is released from the PezAT complex is unknown
More recently, a plethora of homologous ω–ε–ζ loci were identified on multiple resistance plasmids of other gram-positive pathogens such as Enterococcus faecalis and Enterococcus faecium [38–43] or Streptococcus agalactiae [44]. Some of these ω–ε–ζ loci evoke stabilization of conjugable Inc18-like plasmids that encode for the vancomycin and teicoplanin resistance determinant vanA in enterococci. Importantly, such Inc18-like plasmids were not exclusively found in hospital strains but also in common human community and animal-derived vancomycin-resistant enterococci (VRE) strains [45, 46]. This suggests that these glycopeptide-resistant enterococci propagate within the food chain. VRE strains are capable of transferring vanA to other pathogens, such as the methicillin-resistant Staphylococcus aureus (MRSA) [47–50], and it is conceivable that ε/ζ systems also participate in the stable persistence of vanA in the resulting vancomycin-resistant S. aureus strains, which has evolved to a major public health concern.
A thorough sequence analysis divided ε/ζ systems into two distinct phylogenetic groups that differed in their efficiency to stabilize plasmids in E. faecalis [42, 43]. Notably, these two different groups show sequence variation in the ε and ζ protein interface [28, 43] but are conserved in the active site of ζ. This suggests that fine-tuning of the ε–ζ affinity is crucial for the efficiency of plasmid stabilization and is an evolved, host-adaptive feature.
ε/ζ systems encoded on plasmids of gram-negative bacteria
Originally, ε/ζ systems were presumed to be confined to gram-positive bacteria but were recently identified on IncP1 plasmids of Neisseria gonorrhoeae isolates from American and Dutch patients [14, 51]. Within the tetracycline determinant tetM of these plasmids, two different ε/ζ loci were identified, but no open reading frame (ORF) for an ω homologue was found. This suggests that the ε/ζ system from IncP1 plasmids is regulated differently when compared with those found in Inc18-like plasmids. ORFs encoding for ε/ζ systems were further identified on resistance plasmids in Bacteroides fragilis [52]. A recent phylogenetic analysis of type II TA systems suggested that homologues of ε/ζ systems are widespread in all bacterial kingdoms [53]. This study also showed that most ζ toxins reside on bacterial chromosomes rather than plasmids.
Chromosomal ε/ζ systems
The PezAT system from S. pneumoniae
The human pathogen S. pneumoniae is one of the leading causes for bacterial pneumonia, otitis media, and invasive disease such as bacteremia and meningitis. More than 90 different pneumococcal serotypes have been classified [54] which differ strongly in their virulence. However, only a limited number of ∼16 serotypes are found worldwide to cause the bulk of invasive diseases [55, 56]. Since a strong variation in virulence was reported for strains belonging to identical serotypes, the content of the polysaccharide capsule is not the main and exclusive determinant for pathogenicity [57–59]. Differences in virulence result rather from genomic heterogeneity caused by variable regions in the genome of S. pneumoniae. Genes in these accessory regions encode important virulence factors such as adhesion factors, transporters, and metabolic enzymes [58, 60–62]. The chromosomally encoded ζ homologue PezT (Fig. 2) is expressed from the bicistronic pezAT operon (locus tag: SP1050/SP1051) located on the pneumococcal pathogenicity island PPI-1 [21, 59, 63] and, most importantly, was shown to be required to establish full virulence in mice [21]. However, unlike the otherwise stably integrated and conserved PPI region, the pezAT operon resides in the recombination hot-spot PPI-1v that is highly diverse in its gene content [59]. Similar to the plasmid-borne ε/ζ systems, the bacteriotoxic UNAG kinase activity of PezT was shown to be inhibited by high-affinity binding of the antitoxin PezA [28, 64]. In contrast to plasmid-borne ε/ζ systems whose expression relies on the ω repressor, PezA harbors a transcription repressor domain in addition to its ε homology domain (Fig. 3b).
The contribution of PezT to pneumococcal virulence
A major step in connecting PezT with virulence in S. pneumoniae came from the work by Brown et al. who compared clinical serotype 3 isolates with PezT knockout strains in mixed systemic and respiratory infections of mice [21]. The authors showed that disruption of the PezT ORF resulted in a variant that was outcompeted by wild-type strains in mouse challenge models. Only a single isolate out of 500 that were recovered from mice either 24 h after systemic infections or 72 h after respiratory infections contained a disrupted PezT ORF. Furthermore, a considerably attenuated infection progress in mice was observed when strains deprived of the PezT ORF were compared with wild-type strains. However, PezT-negative strains had no growth defect in laboratory broth, serum, or blood. This demonstrated that the phenotype of PezT disruption is virulence specific and not a general growth defect. Along this line, a comparison of the gene content of serotype 1 isolates from indigenous Australian patients revealed that the pezAT ORF was exclusively contained in the recombination hot-spot PPI-1v of hypervirulent isolates, whereas it was absent in the PPI-1v of noninvasive and intermediately virulent strains [59]. Similar to as found by Brown et al., these hypervirulent strains caused rapid lethal infection in intranasally challenged mice, whereas the infection progress of the pezAT-negative, intermediately virulent isolates was strongly attenuated. Furthermore, when the pezAT-positive PPI-1v element of the virulent laboratory S. pneumoniae D39 strain was replaced by a pezAT-negative PPI-1v element from a noninvasive isolate, this hybrid strain was readily outcompeted by wild-type strains in nasal lavage fluid, lungs, and blood models [59]. In contrast, replacing the D39 PPI-1v with one from the hypervirulent serotype 1 isolates resulted in a hybrid strain that was as effective in infection as the wild-type strain. It is noteworthy that the occurrence of pezAT locus is not the only difference within the gene content of these PPI-1v regions [59]. Thus, the increased pathogenicity of pezAT-positive PPI-v regions does not unambiguously demonstrate that pezAT is the exclusive virulence factor within PPI-1v. Nonetheless, its exclusive occurrence in hypervirulent strains is striking.
The strong correlation of pezAT loci in PPI-1v and occurrence of hypervirulence suggests that the bacteria have taken advantage of the TA system to increase the speed of infection. It is rather unlikely that PezAT is a simple maintenance system of PPI-1v, since despite the pezAT locus, the other gene content of PPI-1v is highly variable. More likely, the originally as being detrimental perceived UNAG kinase activity of PezT could support pneumococcal virulence directly. PezT might be released from PezA during environmental stresses such as lack of nutrients, presence of antibiotics, or the immune response, as observed for other TA systems [16, 65–67]. In this scenario, UNAG-3P production would result in inhibition of cell wall synthesis and lysis of a pneumococcal subpopulation. The ability to undergo lysis under stress conditions is a peculiar feature of pneumococci. It is evoked either by self-induced autolysis [68, 69] or by fratricide, in which competent cells induce lysis of their noncompetent sister cells [70]. Generally, lysis of gram-positive bacteria has great impact on virulence, since it leads to accelerated release of cellular components that are detrimental to the infected host. Components which are well established as virulence factors are teichoic acids, lipoteichoic acids, and bacterial DNA [71]. In addition, cell fragments from lysed pneumococci inhibit phagocytosis and impair phagocyte-mediated defense against living pneumococci [72]. Most importantly, pneumococcal lysis leads to triggered release of pneumolysin (Ply), which has multiple detrimental effects for the host organism (see [73]). Low Ply concentrations are already sufficient to trigger apoptosis [74], to activate host complement [75], and to induce proinflammatory reactions [76]. At elevated concentrations, Ply causes general cellular damage in the host organism by its cholesterol-dependent ability to form large membrane pores [77]. However, unlike other bacterial exotoxins, Ply is not directly secreted into the bacteria environment and its release relies on pneumococcal lysis after peptidoglycan degradation as, for example, caused by the major pneumococcal autolysin LytA [78], and similar to PezT, impairment of Ply and LytA function also causes attenuated virulence in infection models [79–82]. Thus, it is likely that PezT indeed supports pneumococcal lysis causing cellular fragmentation and by triggered release of virulence factors.
A remaining conundrum is how activation of PezT is accomplished during infection, since PezA and PezT establish an extremely tight complex and house-keeping proteases of E. coli and B. subtilis were unable to disassemble this complex [64]. This implies that a specific pneumococcal protease or protease adaptor protein is required to remove and degrade PezA from PezT. More experimental data are required to decipher the molecular mechanisms of PezAT's contribution to pneumococcal virulence. Likewise, the role of PezAT homologues in other streptococcal pathogens such as S. agalactiae and Streptococcus suis [63] remains completely unexplored. Moreover, the presence of ζ homologues in the chromosomes of pathogenic E. coli and Capnocytophaga strains (Fig. 3) raises interesting questions about a general implication of the UNAG kinases family in virulence of gram-negative pathogens.
ζ toxins in drug development
Specific activation of ζ toxins
Drugs that disrupt normal operation of TA systems (Fig. 4a) are considered an attractive target for development of new therapeutic agents [83–85]. Pathogenic species could be specifically affected, without eliminating the bulk of the mutualistic bacterial flora. On the one hand, compounds could be developed that directly interfere with toxin inhibition by the antitoxin such as for instance disrupting the TA interaction [85] (Fig. 4b). On the other hand, specific compounds that impair expression of a TA operon could trigger toxin release indirectly (Fig. 4c). In fact, interference with antitoxin inhibition seems to be explored by bacteria themselves. The quorum-sensing pentapeptide extracellular death factor (EDF) has been shown to stimulate the MazF endoribonuclease of the chromosomal MazEF system [86]. Although the antitoxin MazE is present, the endoribunclease activity is enhanced by EDF and this is thought to trigger stress-induced cell death in E. coli [86].
Fig. 4.
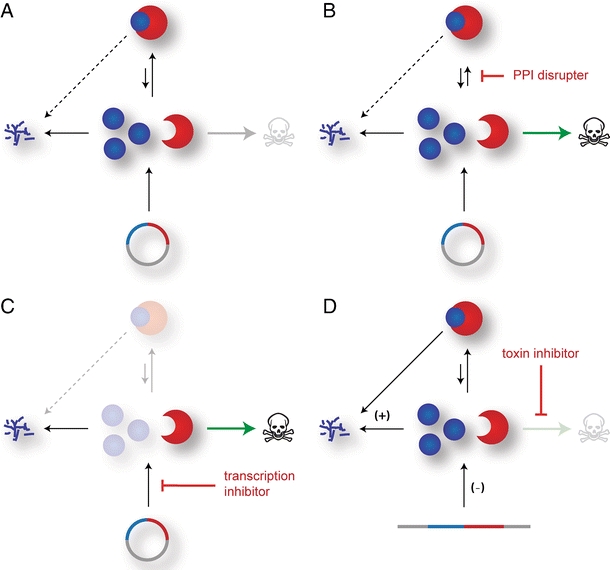
Different possibilities to target TA systems. a In a genome maintenance TA system, the excess production of the labile antitoxin and its high affinity to the toxin prevent cell death. b An agent that would shift the toxin–antitoxin equilibrium towards an active toxin species, for example by masking the toxin binding site of the antitoxin, could trigger bacterial cell death. c Likewise, any suppression of de novo synthesis from the TA operon as for example by an mRNA antisense mechanism would result in toxin release as consequence of antitoxin degradation. d Chromosomal TA systems whose function is beneficial for the fitness of the bacterial population are often also activated by downregulation of de novo synthesis, by upregulation of antitoxin degradation, or both mechanisms in parallel. However, a cellular activation of such a TA system could be prevented by specific inhibition of the toxin activity
Similarly, an artificial disruption of the toxin–antitoxin interaction of the ε/ζ TA system from S. pyogenes has recently been described [87]. In a high-throughput approach, a mixture of oligopeptide libraries was screened for their ability to impair the ε–ζ interaction. Thereby, sub-libraries containing 17 amino acids long peptides were shown to interfere with the ε–ζ interaction. However, further subfractionation of this mixed oligopeptide libraries lead to loss of this capability and the authors suggested that avidity effects from several weak binders could have caused disruption of the protein interaction [87]. It remains to be shown whether those peptide inhibitors can also restore UNAG kinase activity of ζ, as they could block the ATP binding similarly as the antitoxin ε does. Whether dismantling of the ε–ζ interaction is always feasible remains questionable, since this relies heavily on the dissociation kinetics of the complex. For instance, dissociation of the ultra-tight PezAT complex is extremely slow, rendering it difficult to disrupt the complex by any peptide inhibitor [64].
Another very promising approach to achieve ζ activation and toxins of other TA systems directly is interference with de novo synthesis by use of antisense RNA mechanisms (Fig. 4c). This would provoke specific degradation of the TA mRNA and thereby impair de novo synthesis of the TA proteins. For instance, such successful knockdowns of genes essential for bacterial viability were achieved by liposome-delivered antisense RNA [88] or peptide nucleic acids [89–92].
It remains to be shown whether ζ or PezT activation is indeed always a beneficial approach for microbial defense. In cases where the TA system is associated with virulence, activation of the toxin could increase infectiousness by enhanced cell lysis and boosted virulence factor release from the pathogenic bacteria. In this case, an as-yet-unknown but specific inhibitor of ζ toxin itself would be expected to eliminate the toxic effects of the TA system (Fig. 4d).
UNAG-3P in drug development
The enzymatic product of this kinase family, UNAG-3P, is a promising lead compound for the development of a new broad-spectrum antibiotic. UNAG-3P inhibits MurA [30] and most likely also its paralogue MurZ, which are conserved enzymes in peptidoglycan synthesis in all gram-positive and gram-negative bacteria. This family of enzymes is an attractive drug target for microbial defense, since no eukaryotic homolog exists [93]. However, apart from the naturally occurring, broad-spectrum antibiotic fosfomycin, no specific MurA inhibitors have been applied so far [94]. Because an increasing number of bacteria have developed resistance, fosfomycin has turned into a relatively inefficient drug [94]. Thus, the discovery of UNAG-3P discloses a novel and attractive compound that has the potential to evolve into a drug inhibiting MurA.
Moreover, UNAG-3P most likely interferes with bacterial biosynthetic pathways of other glycoconjugates as well. For instance, formation of UNAG-3P might interfere with lipopolysaccharide (LP) production in gram-negative bacteria. LP biosynthesis requires the unique and distinctive building block lipid A. This phosphoglycolipid is highly conserved [95], and its formation is initiated by condensation of the 3′-OH group of the N-acetylglucosamine moiety of UNAG with a lipid anchor [96]. It remains to be shown for this pathway whether UNAG-3P is an inhibitor similar as it is for peptidoglycan synthesis or is just a dead-end nucleotide sugar. LPs are part of the outer membrane of gram-negative bacteria and protect bacterial cells from cell-damaging agents. Moreover, LPs act as endotoxins and can cause septic shocks due to stimulation of excessive levels of proinflammatory cytokines [97]. Thus, UNAG-3P could not only impair bacterial growth but could also reduce LP-induced toxic shock responses during infection.
Moreover, teichoic acids, which are diverse multifunctional glycopolymers that reside in the cell wall of gram-positive bacteria, require UNAG as precursor. Similar to inhibition of teichoic acid synthesis leading to bacteriostasis [98] and impaired host colonization and tissue invasion in MRSA [99], UNAG-3P might impair these bacterial mechanisms as well.
The discovery of UNAG-3P as a new natural product has uncovered the basic principles for the development of a new broad-spectrum antibiotic, and new challenges need to be managed to turn it into an effective drug. An obvious and required improvement is to render this compound permeable for cell membranes. UNAG-3P is a highly charged anionic molecule and thus most likely will not penetrate through membranes. Further synthetic modifications of UNAG-3P should aim at creating membrane-permeable derivates of UNAG-3P and probe their activity against gram-positive and gram-negative bacteria. For instance, reversible masking of the phosphate groups should help to reduce the permeability barrier [100, 101].
Furthermore, it remains unclear whether UNAG-3P has any detrimental effect on eukaryotes, since UNAG is also an essential metabolite for these organisms. Previous studies have shown that ectopic expression of ζ from S. pyogenes in Saccharomyces cerevisiae caused growth retardation, albeit the general toxicity of ζ was much weaker when compared with the bacteria [102]. This growth retardation in yeast might be explained by a toxic effect of UNAG-3P. However, a more likely explanation is an artificial depletion of cells from UNAG by ζ's enzymatic activity. Cytosolic concentrations of UNAG are approximately 10 times lower in S. cerevisiae (∼30 μM) than in E. coli [103], and a rapid depletion of UNAG by ζ will most probably impair chitin synthesis in S. cerevisiae [104]. Although UNAG-3P is a very promising lead compound for drug development, it might also have undesired adverse reactions similar to those of other lytic antibiotics when detrimental effects upon endotoxin release have to be expected [105].
Inhibition of PezT
Both triggered induction of cell death upon treatment with UNAG-3P-derived drugs or upon activation of PezT toxins will boost the release of virulence factors from pathogenic bacteria into their environment. The involvement of PezT toxins in virulence makes it conceivable that an attenuation of fulminant pneumococcal infections could be achieved by the inhibition of PezT's lytic activity. If the increase of the survival time during fulminant infections in humans is similar to what was observed after PezT inactivation in mice, such an inhibitor could help to increase the time window for therapeutic intervention and allow a retarded kill-off of pathogens.
Another interesting, although still speculative, aspect of PezT's contribution to virulence is a possible involvement as endotoxin during infection. Similar to the cytolysin-mediated translocation of the toxic S. pyogenes NAD glycohydrolase into host cells [106], PezT could enter the cytosol of the infected host cells. In this scenario, PezT would aggravate eukaryotic cells by its fatal enzymatic mechanism similarly as observed during ectopic expression in S. cerevisiae [102]. A PezT inhibitor that enters eukaryotic cells as well would have dual function and could not only counteract pneumococcal autolysis but additionally abolish the toxin's activity in the infected host.
Conclusion
The recent discovery of the UNAG kinase activity of ζ toxins and its implementation in programmed cell death has expanded the repertoire of cellular structures targeted by TA systems to include corruption of cell wall synthesis. Whereas all other type II TA systems either interfere with replication or translation [13, 14], ζ toxins are the first reported inhibitors of peptidoglycan synthesis and most probably of other biosynthetic pathways involved in cell wall synthesis as well. Thus, the list of cellular structures targeted by TA systems becomes more complete and TA systems seem to impair the same cellular mechanisms that commonly applied antibiotics target as well. Exploring especially ε/ζ TA systems in our microbial defense is a promising approach, since this system provides several important points of attack that can be targeted by drugs. Furthermore, any compound interfering with this element will be of general impact, since loci encoding for ε/ζ systems are widely distributed among pathogenic bacteria.
Acknowledgment
We want to apologize to all colleagues whose scientific contributions we could not mention because of space restraints. We are very grateful to RL Shoeman, M Cryle, and J Reinstein the for comments and corrections. We thank I Schlichting for the continuous encouragement and support. Our work is financially supported by the Max Planck Society.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Palmer KL, Kos VN, Gilmore MS. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol. 2010;13:632–639. doi: 10.1016/j.mib.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly BG, Vespermann A, Bolton DJ. Horizontal gene transfer of virulence determinants in selected bacterial foodborne pathogens. Food Chem Toxicol. 2009;47:969–977. doi: 10.1016/j.fct.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 4.Kunin V, Ouzounis CA. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 2003;13:1589–1594. doi: 10.1101/gr.1092603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 6.Lioy VS, Pratto F, de la Hoz AB, Ayora S, Alonso JC. Plasmid pSM19035, a model to study stable maintenance in Firmicutes. Plasmid. 2010;64:1–17. doi: 10.1016/j.plasmid.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Wozniak RA, Waldor MK. A toxin–antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5:e1000439. doi: 10.1371/journal.pgen.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen RB, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 9.Zielenkiewicz U, Ceglowski P. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim Pol. 2001;48:1003–1023. [PubMed] [Google Scholar]
- 10.Hayes F. Toxins–antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 11.Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. Chromosomal toxin–antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol Microbiol. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- 12.Hayes CS, Sauer RT. Toxin–antitoxin pairs in bacteria: killers or stress regulators? Cell. 2003;112:2–4. doi: 10.1016/s0092-8674(02)01282-5. [DOI] [PubMed] [Google Scholar]
- 13.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 14.Van Melderen L, Saavedra De Bast M. Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin–antitoxin loci as stress-response elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 17.Saavedra De Bast M, Mine N, Van Melderen L. Chromosomal toxin–antitoxin systems may act as antiaddiction modules. J Bacteriol. 2008;190:4603–4609. doi: 10.1128/JB.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazan R, Sat B, Reches M, Engelberg-Kulka H. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J Bacteriol. 2001;183:2046–2050. doi: 10.1128/JB.183.6.2046-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin–antitoxin pair. Proc Natl Acad Sci U S A. 2009;106:894–899. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 21.Brown JS, Gilliland SM, Spratt BG, Holden DW. A locus contained within a variable region of pneumococcal pathogenicity island 1 contributes to virulence in mice. Infect Immun. 2004;72:1587–1593. doi: 10.1128/IAI.72.3.1587-1593.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daines DA, Jarisch J, Smith AL. Identification and characterization of a nontypeable Haemophilus influenzae putative toxin–antitoxin locus. BMC Microbiol. 2004;4:30. doi: 10.1186/1471-2180-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L. What is the benefit to Escherichia coli of having multiple toxin–antitoxin systems in its genome? J Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lioy VS, Martin MT, Camacho AG, Lurz R, Antelmann H, Hecker M, Hitchin E, Ridge Y, Wells JM, Alonso JC. pSM19035-encoded zeta toxin induces stasis followed by death in a subpopulation of cells. Microbiology. 2006;152:2365–2379. doi: 10.1099/mic.0.28950-0. [DOI] [PubMed] [Google Scholar]
- 25.Zielenkiewicz U, Ceglowski P. The toxin–antitoxin system of the streptococcal plasmid pSM19035. J Bacteriol. 2005;187:6094–6105. doi: 10.1128/JB.187.17.6094-6105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meinhart A, Alonso JC, Strater N, Saenger W. Crystal structure of the plasmid maintenance system ε/ζ: functional mechanism of toxin ζ and inactivation by ε2/ζ2 complex formation. Proc Natl Acad Sci U S A. 2003;100:1661–1666. doi: 10.1073/pnas.0434325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowakowska B, Kern-Zdanowicz I, Zielenkiewicz U, Ceglowski P. Characterization of Bacillus subtilis clones surviving overproduction of Zeta, a pSM19035 plasmid-encoded toxin. Acta Biochim Pol. 2005;52:99–107. [PubMed] [Google Scholar]
- 28.Khoo SK, Loll B, Chan WT, Shoeman RL, Ngoo L, Yeo CC, Meinhart A. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J Biol Chem. 2007;282:19606–19618. doi: 10.1074/jbc.M701703200. [DOI] [PubMed] [Google Scholar]
- 29.Meinhart A, Alings C, Strater N, Camacho AG, Alonso JC, Saenger W. Crystallization and preliminary X-ray diffraction studies of the epsilonzeta addiction system encoded by Streptococcus pyogenes plasmid pSM19035. Acta Crystallogr D Biol Crystallogr. 2001;57:745–747. doi: 10.1107/s0907444901004176. [DOI] [PubMed] [Google Scholar]
- 30.Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. A novel mechanism of programmed cell death in bacteria by toxin–antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011;9:e1001033. doi: 10.1371/journal.pbio.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:168–207. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 32.Ceglowski P, Boitsov A, Karamyan N, Chai S, Alonso JC. Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035-derived plasmids in Bacillus subtilis. Mol Genet Genomics. 1993;241:579–585. doi: 10.1007/BF00279900. [DOI] [PubMed] [Google Scholar]
- 33.Dixon JM, Lipinski AE. Resistance of group A beta-hemolytic streptococci to lincomycin and erythromycin. Antimicrob Agents Chemother. 1972;1:333–339. doi: 10.1128/aac.1.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clewell DB. Plasmids, drug resistance, and gene transfer in the genus Streptococcus. Microbiol Rev. 1981;45:409–436. doi: 10.1128/mr.45.3.409-436.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceglowski P, Boitsov A, Chai S, Alonso JC. Analysis of the stabilization system of pSM19035-derived plasmid pBT233 in Bacillus subtilis. Gene. 1993;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- 36.de la Hoz AB, Ayora S, Sitkiewicz I, Fernandez S, Pankiewicz R, Alonso JC, Ceglowski P. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc Natl Acad Sci U S A. 2000;97:728–733. doi: 10.1073/pnas.97.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camacho AG, Misselwitz R, Behlke J, Ayora S, Welfle K, Meinhart A, Lara B, Saenger W, Welfle H, Alonso JC. In vitro and in vivo stability of the ε2ζ2 protein complex of the broad host-range Streptococcus pyogenes pSM19035 addiction system. Biol Chem. 2002;383:1701–1713. doi: 10.1515/BC.2002.191. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz FV, Perreten V, Teuber M. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid. 2001;46:170–187. doi: 10.1006/plas.2001.1544. [DOI] [PubMed] [Google Scholar]
- 39.Dahl KH, Mater DD, Flores MJ, Johnsen PJ, Midtvedt T, Corthier G, Sundsfjord A. Transfer of plasmid and chromosomal glycopeptide resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist stably in vivo in the absence of glycopeptide selection. J Antimicrob Chemother. 2007;59:478–486. doi: 10.1093/jac/dkl530. [DOI] [PubMed] [Google Scholar]
- 40.Moritz EM, Hergenrother PJ. Toxin–antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci U S A. 2007;104:311–316. doi: 10.1073/pnas.0601168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sletvold H, Johnsen PJ, Simonsen GS, Aasnaes B, Sundsfjord A, Nielsen KM. Comparative DNA analysis of two vanA plasmids from Enterococcus faecium strains isolated from poultry and a poultry farmer in Norway. Antimicrob Agents Chemother. 2007;51:736–739. doi: 10.1128/AAC.00557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sletvold H, Johnsen PJ, Hamre I, Simonsen GS, Sundsfjord A, Nielsen KM. Complete sequence of Enterococcus faecium pVEF3 and the detection of an omega-epsilon-zeta toxin–antitoxin module and an ABC transporter. Plasmid. 2008;60:75–85. doi: 10.1016/j.plasmid.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Rosvoll TC, Pedersen T, Sletvold H, Johnsen PJ, Sollid JE, Simonsen GS, Jensen LB, Nielsen KM, Sundsfjord A. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol Med Microbiol. 2010;58:254–268. doi: 10.1111/j.1574-695X.2009.00633.x. [DOI] [PubMed] [Google Scholar]
- 44.Brantl S, Behnke D, Alonso JC. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAM beta 1 and pSM19035. Nucleic Acids Res. 1990;18:4783–4790. doi: 10.1093/nar/18.16.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klare I, Heier H, Claus H, Bohme G, Marin S, Seltmann G, Hakenbeck R, Antanassova V, Witte W. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb Drug Resist. 1995;1:265–272. doi: 10.1089/mdr.1995.1.265. [DOI] [PubMed] [Google Scholar]
- 46.Kuhn I, Iversen A, Finn M, Greko C, Burman LG, Blanch AR, Vilanova X, Manero A, Taylor H, Caplin J, et al. Occurrence and relatedness of vancomycin-resistant enterococci in animals, humans, and the environment in different European regions. Appl Environ Microbiol. 2005;71:5383–5390. doi: 10.1128/AEM.71.9.5383-5390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perichon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:4580–4587. doi: 10.1128/AAC.00346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu W, Clark NC, McDougal LK, Hageman J, McDonald LC, Patel JB. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob Agents Chemother. 2008;52:452–457. doi: 10.1128/AAC.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, Clark NC, Anderson KF, McDougal LK, Hageman JC, Olsen-Rasmussen M, et al. Dissemination of an Enterococcus Inc18-Like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54:4314–4320. doi: 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loomba PS, Taneja J, Mishra B. Methicillin and vancomycin resistant S. aureus in hospitalized patients. J Glob Infect Dis. 2010;2:275–283. doi: 10.4103/0974-777X.68535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pachulec E, van der Does C. Conjugative plasmids of Neisseria gonorrhoeae. PLoS One. 2010;5:e9962. doi: 10.1371/journal.pone.0009962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 53.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, Van Melderen L (2011) Diversity of bacterial type II toxin–antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. doi:10.1093/nar/gkr131 [DOI] [PMC free article] [PubMed]
- 54.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott JA, Hall AJ, Dagan R, Dixon JM, Eykyn SJ, Fenoll A, Hortal M, Jette LP, Jorgensen JH, Lamothe F, et al. Serogroup-specific epidemiology of Streptococcus pneumoniae: associations with age, sex, and geography in 7,000 episodes of invasive disease. Clin Infect Dis. 1996;22:973–981. doi: 10.1093/clinids/22.6.973. [DOI] [PubMed] [Google Scholar]
- 56.Johnson HL, Deloria-Knoll M, Levine OS, Stoszek SK, Freimanis Hance L, Reithinger R, Muenz LR, O'Brien KL. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. PLoS Med. 2010;7(10):pii:e1000348. doi: 10.1371/journal.pmed.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Forbes ML, Horsey E, Hiller NL, Buchinsky FJ, Hayes JD, Compliment JM, Hillman T, Ezzo S, Shen K, Keefe R, et al. Strain-specific virulence phenotypes of Streptococcus pneumoniae assessed using the Chinchilla laniger model of otitis media. PLoS One. 2008;3:e1969. doi: 10.1371/journal.pone.0001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–4777. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harvey RM, Stroeher UH, Ogunniyi AD, Smith-Vaughan HC, Leach AJ, Paton JC. A variable region within the genome of Streptococcus pneumoniae contributes to strain–strain variation in virulence. PLoS One. 2011;6:e19650. doi: 10.1371/journal.pone.0019650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 61.Silva NA, McCluskey J, Jefferies JM, Hinds J, Smith A, Clarke SC, Mitchell TJ, Paterson GK. Genomic diversity between strains of the same serotype and multilocus sequence type among pneumococcal clinical isolates. Infect Immun. 2006;74:3513–3518. doi: 10.1128/IAI.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blomberg C, Dagerhamn J, Dahlberg S, Browall S, Fernebro J, Albiger B, Morfeldt E, Normark S, Henriques-Normark B. Pattern of accessory regions and invasive disease potential in Streptococcus pneumoniae. J Infect Dis. 2009;199:1032–1042. doi: 10.1086/597205. [DOI] [PubMed] [Google Scholar]
- 63.Croucher NJ, Walker D, Romero P, Lennard N, Paterson GK, Bason NC, Mitchell AM, Quail MA, Andrew PW, Parkhill J, et al. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniaeSpain23F ST81. J Bacteriol. 2009;191:1480–1489. doi: 10.1128/JB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mutschler H, Reinstein J, Meinhart A. Assembly dynamics and stability of the pneumococcal epsilon zeta antitoxin toxin (PezAT) system from Streptococcus pneumoniae. J Biol Chem. 2010;285:21797–21806. doi: 10.1074/jbc.M110.126250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, Tarone AM, Benedik MJ, Peti W, Page R, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bordes P, Cirinesi AM, Ummels R, Sala A, Sakr S, Bitter W, Genevaux P. SecB-like chaperone controls a toxin-antitoxin stress-responsive system in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;108:8438–8443. doi: 10.1073/pnas.1101189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin–antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5:e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinas GE, Cortes PR, Orio AG, Echenique J. Acidic stress induces autolysis by a CSP-independent ComE pathway in Streptococcus pneumoniae. Microbiology. 2008;154:1300–1308. doi: 10.1099/mic.0.2007/015925-0. [DOI] [PubMed] [Google Scholar]
- 69.Regev-Yochay G, Trzcinski K, Thompson CM, Lipsitch M, Malley R. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J Bacteriol. 2007;189:6532–6539. doi: 10.1128/JB.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nau R, Eiffert H. Modulation of release of proinflammatory bacterial compounds by antibacterials: potential impact on course of inflammation and outcome in sepsis and meningitis. Clin Microbiol Rev. 2002;15:95–110. doi: 10.1128/CMR.15.1.95-110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martner A, Skovbjerg S, Paton JC, Wold AE. Streptococcus pneumoniae autolysis prevents phagocytosis and production of phagocyte-activating cytokines. Infect Immun. 2009;77:3826–3837. doi: 10.1128/IAI.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirst RA, Kadioglu A, O'Callaghan C, Andrew PW. The role of pneumolysin in pneumococcal pneumonia and meningitis. Clin Exp Immunol. 2004;138:195–201. doi: 10.1111/j.1365-2249.2004.02611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Braun JS, Sublett JE, Freyer D, Mitchell TJ, Cleveland JL, Tuomanen EI, Weber JR. Pneumococcal pneumolysin and H(2)O(2) mediate brain cell apoptosis during meningitis. J Clin Invest. 2002;109:19–27. doi: 10.1172/JCI12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mitchell TJ, Andrew PW, Saunders FK, Smith AN, Boulnois GJ. Complement activation and antibody binding by pneumolysin via a region of the toxin homologous to a human acute-phase protein. Mol Microbiol. 1991;5:1883–1888. doi: 10.1111/j.1365-2958.1991.tb00812.x. [DOI] [PubMed] [Google Scholar]
- 76.Cockeran R, Theron AJ, Steel HC, Matlola NM, Mitchell TJ, Feldman C, Anderson R. Proinflammatory interactions of pneumolysin with human neutrophils. J Infect Dis. 2001;183:604–611. doi: 10.1086/318536. [DOI] [PubMed] [Google Scholar]
- 77.Gilbert RJ, Jimenez JL, Chen S, Tickle IJ, Rossjohn J, Parker M, Andrew PW, Saibil HR. Two structural transitions in membrane pore formation by pneumolysin, the pore-forming toxin of Streptococcus pneumoniae. Cell. 1999;97:647–655. doi: 10.1016/s0092-8674(00)80775-8. [DOI] [PubMed] [Google Scholar]
- 78.Martner A, Dahlgren C, Paton JC, Wold AE. Pneumolysin released during Streptococcus pneumoniae autolysis is a potent activator of intracellular oxygen radical production in neutrophils. Infect Immun. 2008;76:4079–4087. doi: 10.1128/IAI.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berry AM, Lock RA, Hansman D, Paton JC. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989;57:2324–2330. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berry AM, Yother J, Briles DE, Hansman D, Paton JC. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect Immun. 1989;57:2037–2042. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lock RA, Hansman D, Paton JC. Comparative efficacy of autolysin and pneumolysin as immunogens protecting mice against infection by Streptococcus pneumoniae. Microb Pathog. 1992;12:137–143. doi: 10.1016/0882-4010(92)90116-6. [DOI] [PubMed] [Google Scholar]
- 82.Berry AM, Paton JC. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect Immun. 2000;68:133–140. doi: 10.1128/iai.68.1.133-140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12:66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 84.DeNap JC, Hergenrother PJ. Bacterial death comes full circle: targeting plasmid replication in drug-resistant bacteria. Org Biomol Chem. 2005;3:959–966. doi: 10.1039/b500182j. [DOI] [PubMed] [Google Scholar]
- 85.Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles' heel of drug-resistant bacteria. Curr Opin Chem Biol. 2008;12:389–399. doi: 10.1016/j.cbpa.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Belitsky M, Avshalom H, Erental A, Yelin I, Kumar S, London N, Sperber M, Schueler-Furman O, Engelberg-Kulka H. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol Cell. 2011;41:625–635. doi: 10.1016/j.molcel.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 87.Lioy VS, Rey O, Balsa D, Pellicer T, Alonso JC. A toxin–antitoxin module as a target for antimicrobial development. Plasmid. 2010;63:31–39. doi: 10.1016/j.plasmid.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 88.Meng J, Wang H, Hou Z, Chen T, Fu J, Ma X, He G, Xue X, Jia M, Luo X. Novel anion liposome-encapsulated antisense oligonucleotide restores susceptibility of methicillin-resistant Staphylococcus aureus and rescues mice from lethal sepsis by targeting mecA. Antimicrob Agents Chemother. 2009;53:2871–2878. doi: 10.1128/AAC.01542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol. 2001;19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- 90.Kurupati P, Tan KS, Kumarasinghe G, Poh CL. Inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant beta-lactamase-producing Klebsiella pneumoniae strain. Antimicrob Agents Chemother. 2007;51:805–811. doi: 10.1128/AAC.00709-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nekhotiaeva N, Awasthi SK, Nielsen PE, Good L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol Ther. 2004;10:652–659. doi: 10.1016/j.ymthe.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 92.Tan XX, Actor JK, Chen Y. Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob Agents Chemother. 2005;49:3203–3207. doi: 10.1128/AAC.49.8.3203-3207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Du W, Brown JR, Sylvester DR, Huang J, Chalker AF, So CY, Holmes DJ, Payne DJ, Wallis NG. Two active forms of UDP-N-acetylglucosamine enolpyruvyl transferase in gram-positive bacteria. J Bacteriol. 2000;182:4146–4152. doi: 10.1128/jb.182.15.4146-4152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Popovic M, Steinort D, Pillai S, Joukhadar C. Fosfomycin: an old, new friend? Eur J Clin Microbiol Infect Dis. 2009;29:127–142. doi: 10.1007/s10096-009-0833-2. [DOI] [PubMed] [Google Scholar]
- 95.Kabanov DS, Prokhorenko IR. Structural analysis of lipopolysaccharides from Gram-negative bacteria. Biochemistry (Mosc) 2010;75:383–404. doi: 10.1134/s0006297910040012. [DOI] [PubMed] [Google Scholar]
- 96.Trent MS. Biosynthesis, transport, and modification of lipid A. Biochem Cell Biol. 2004;82:71–86. doi: 10.1139/o03-070. [DOI] [PubMed] [Google Scholar]
- 97.Fujihara M, Muroi M, Tanamoto K, Suzuki T, Azuma H, Ikeda H. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: roles of the receptor complex. Pharmacol Ther. 2003;100:171–194. doi: 10.1016/j.pharmthera.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 98.Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem Biol. 2009;4:875–883. doi: 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suzuki T, Swoboda JG, Campbell J, Walker S, Gilmore MS. In vitro antimicrobial activity of wall teichoic acid biosynthesis inhibitors against Staphylococcus aureus isolates. Antimicrob Agents Chemother. 2011;55:767–774. doi: 10.1128/AAC.00879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schultz C, Vajanaphanich M, Harootunian AT, Sammak PJ, Barrett KE, Tsien RY. Acetoxymethyl esters of phosphates, enhancement of the permeability and potency of cAMP. J Biol Chem. 1993;268:6316–6322. [PubMed] [Google Scholar]
- 101.Jiang T, Sweeney G, Rudolf MT, Klip A, Traynor-Kaplan A, Tsien RY. Membrane-permeant esters of phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:11017–11024. doi: 10.1074/jbc.273.18.11017. [DOI] [PubMed] [Google Scholar]
- 102.Zielenkiewicz U, Kowalewska M, Kaczor C, Ceglowski P. In vivo interactions between toxin-antitoxin proteins epsilon and zeta of streptococcal plasmid pSM19035 in Saccharomyces cerevisiae. J Bacteriol. 2009;191:3677–3684. doi: 10.1128/JB.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Namboori SC, Graham DE. Enzymatic analysis of uridine diphosphate N-acetyl-D-glucosamine. Anal Biochem. 2008;381:94–100. doi: 10.1016/j.ab.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmidt M. Survival and cytokinesis of Saccharomyces cerevisiae in the absence of chitin. Microbiology. 2004;150:3253–3260. doi: 10.1099/mic.0.27197-0. [DOI] [PubMed] [Google Scholar]
- 105.Holzheimer RG. Antibiotic induced endotoxin release and clinical sepsis: a review. J Chemother. 2001;13 Spec No 1(1):159–172. doi: 10.1179/joc.2001.13.Supplement-2.159. [DOI] [PubMed] [Google Scholar]
- 106.Madden JC, Ruiz N, Caparon M. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 2001;104:143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]


