Abstract
The binding sites of Hoechst 33258, netropsin and distamycin on three DNA restriction fragments from plasmid pBR322 were compared by footprinting with methidiumpropyl-EDTA X Fe(II) [MPE X Fe(II)]. Hoechst, netropsin and distamycin share common binding sites that are five +/- one bp in size and rich in A X T DNA base pairs. The five base pair protection patterns for Hoechst may result from a central three base pair recognition site bound by two bisbenzimidazole NHs forming a bridge on the floor of the minor groove between adjacent adenine N3 and thymine O2 atoms on opposite helix strands. Hydrophobic interaction of the flanking phenol and N-methylpiperazine rings would afford a steric blockade of one additional base pair on each side.
Full text
PDF

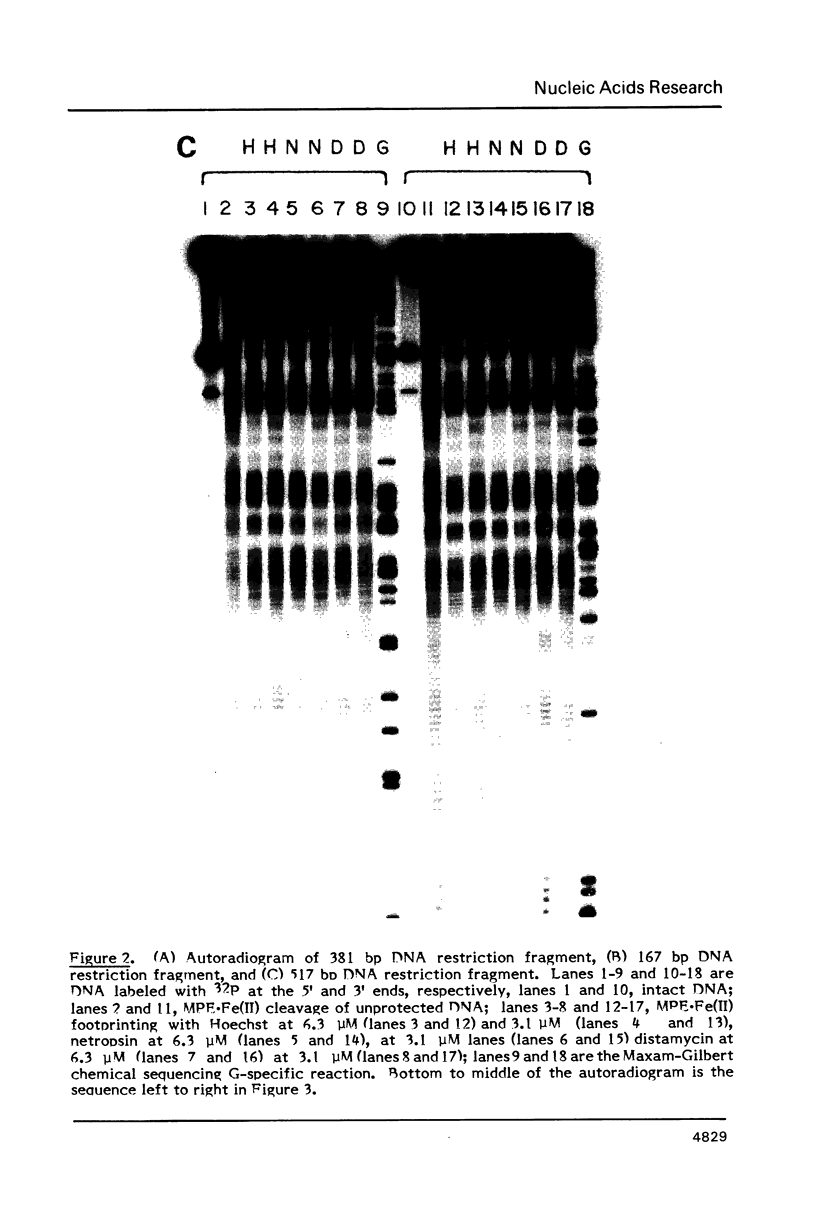






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman H. M., Neidle S., Zimmer C., Thrum H. Netropsin, a DNA-binding oligopeptide structural and binding studies. Biochim Biophys Acta. 1979 Jan 26;561(1):124–131. doi: 10.1016/0005-2787(79)90496-9. [DOI] [PubMed] [Google Scholar]
- Gursky G. V., Zasedatelev A. S., Zhuze A. L., Khorlin A. A., Grokhovsky S. L., Streltsov S. A., Surovaya A. N., Nikitin S. M., Krylov A. S., Retchinsky V. O. Synthetic sequence-specific ligands. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):367–378. doi: 10.1101/sqb.1983.047.01.043. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov A. S., Grokhovsky S. L., Zasedatelev A. S., Zhuze A. L., Gursky G. V., Gottikh B. P. Quantitative estimation of the contribution of pyrrolcarboxamide groups of the antibiotic distamycin A into specificity of its binding to DNA AT pairs. Nucleic Acids Res. 1979 Jan;6(1):289–304. doi: 10.1093/nar/6.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck G., Zimmer C., Reinert K. E., Arcamone F. Specific interactions of distamycin A and its analogs with (A-T) rich and (G-C) rich duplex regions of DNA and deoxypolynucleotides. Nucleic Acids Res. 1977 Aug;4(8):2655–2670. doi: 10.1093/nar/4.8.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Blumenfeld K. S., Breslauer K. J. Calorimetric and spectroscopic investigation of drug-DNA interactions. I. The binding of netropsin to poly d(AT). Nucleic Acids Res. 1983 May 11;11(9):2857–2870. doi: 10.1093/nar/11.9.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. F., Holmes N. Use of an 125I-labelled DNA ligand to probe DNA structure. 1983 Mar 31-Apr 6Nature. 302(5907):452–454. doi: 10.1038/302452a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D. Theoretical calculations of the helix-coil transition of DNA in the presence of large, cooperatively binding ligands. Biopolymers. 1976 Jul;15(7):1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. L. Netropsin-poly(dA-dT) complex in solution: structure and dynamics of antibiotic-free base pair regions and those centered on bound netropsin. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5207–5211. doi: 10.1073/pnas.74.12.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. M., Dervan P. B. Echinomycin binding sites on DNA. Science. 1984 Sep 14;225(4667):1122–1127. doi: 10.1126/science.6089341. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
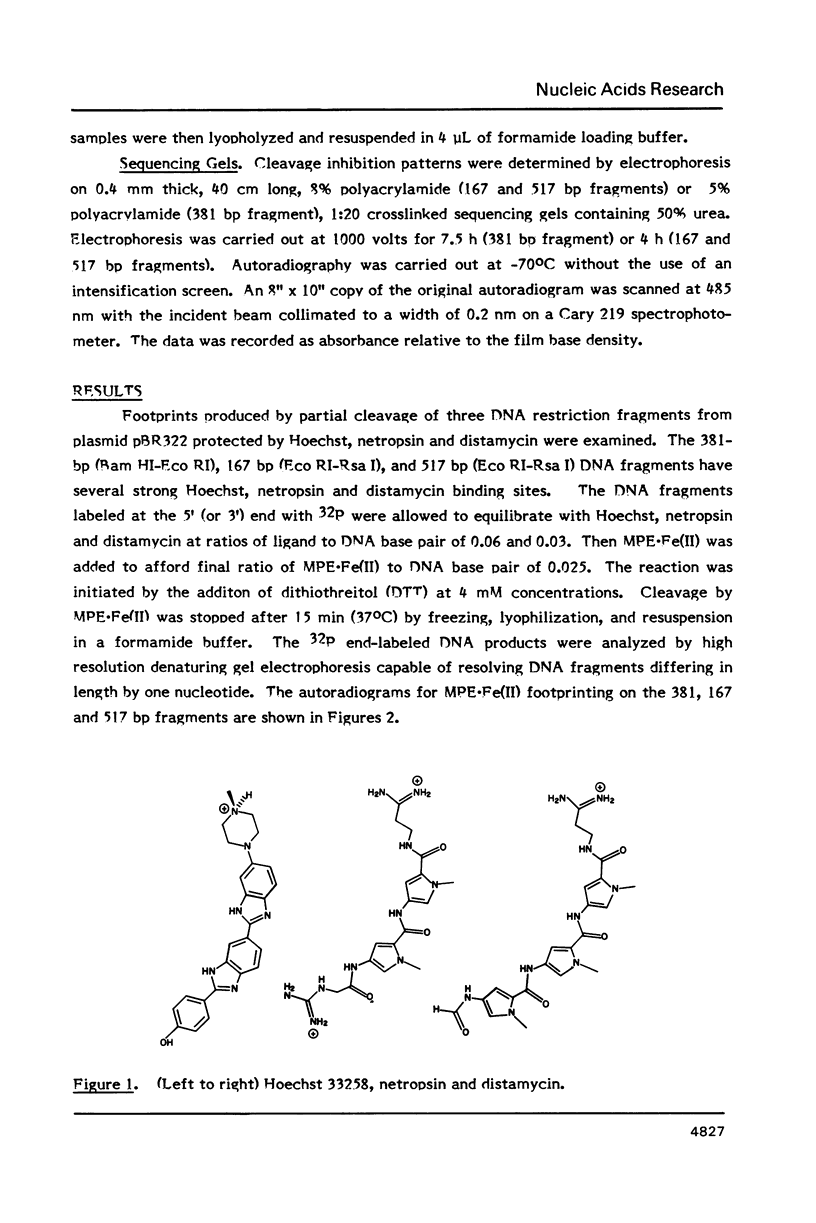
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngquist R. S., Dervan P. B. Sequence-specific recognition of B-DNA by oligo(N-methylpyrrolecarboxamide)s. Proc Natl Acad Sci U S A. 1985 May;82(9):2565–2569. doi: 10.1073/pnas.82.9.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska K., Lavery R., Pullman B. Theoretical studies of the selective binding to DNA of two non-intercalating ligands: netropsin and SN 18071. Nucleic Acids Res. 1983 Dec 20;11(24):8825–8839. doi: 10.1093/nar/11.24.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]