Abstract
Phenotypic differences in Schwann cells (SCs) may help guide axonal regeneration down motor or sensory specific pathways following peripheral nerve injury. The goal of this study was to identify phenotypic markers for SCs harvested from the cutaneous (sensory) and quadriceps (motor) branches of the rat femoral nerve and to study the effects of expansion culture on the expression patterns of these motor or sensory phenotypic markers. RNA was extracted from SCs harvested from the motor and sensory branches of the rat femoral nerve and analyzed using Affymetrix Gene Chips© (Rat Genome 230 v2.0 Array A). Genes that were upregulated in motor SCs compared to the sensory SCs or vice versa were identified, and the results were verified for a subset of genes using quantitative real time polymerase chain reaction (qRT-PCR). The expression levels of the “phenotype-specific” genes were then evaluated in SC expansion cultures at various timepoints over 30 days by qRT-PCR to determine the effect of expansion on SC phenotype. Expression levels of the phenotype-specific genes were significantly altered after expansion culture for both the motor and sensory markers compared to fresh nerve tissue. These results indicate that both motor and sensory SC gene expression patterns are disrupted during expansion in vitro and may affect the ability of SCs to express phenotype specific genes after transplantation.
Keywords: nerve regeneration, cell transplantation, peripheral nerve injury
INTRODUCTION
Peripheral nerve injury (PNI) due to a complete nerve transection results in a loss of function. Ideally, the two severed ends of the nerves can be rejoined using a direct end to end coaptation. However, in larger nerve gap injuries, a direct coaptation can introduce unnecessary tension that may impede regeneration. To prevent tension and to bridge the nerve defect, an autograft can be used to provide extracellular matrix (ECM) molecules and growth factors (GFs) to promote regeneration of axons across the nerve gap. Although the nerve autograft remains the gold standard of care, this method has limitations including donor site morbidity, lack of sufficient donor tissue, and size mismatches at the injury site (Burnett and Zager 2004; Schmidt and Leach 2003). Currently, investigators are searching for alternative therapies to bridge nerve gaps following injury, such as acellular nerve grafts (ANGs).
ANGs have been used to support the growth of regenerating axons from the proximal nerve into the distal stump (Hare et al. 1993; Hare et al. 1995; Levi et al. 1994). Compared to nerve conduits, ANGs support superior nerve regeneration because they contain an intact microstructure consisting of endoneurial tubes and ECM that supports regenerating axons (Johnson et al. 1982; Sondell et al. 1998; Whitlock et al. 2009). However, the regenerative capacity of ANGs is still inferior to autografts because they lack SCs (Whitlock et al. 2009). SCs provide GFs and ECM to promote neuronal survival and axonal regeneration (Brenner et al. 2005; Bunge 1993; Frostick et al. 1998; Nagarajan et al. 2002; Reynolds and Woolf 1993). Addition of SCs to ANGs has been proposed as a method to enhance their regenerative capacity.
When given equal access to motor and sensory pathways, the motor axon tends to regenerate down the motor pathway. This phenomenon, preferential motor reinnervation (PMR), may be influenced by trophic support from the end organs (with muscle greatly outweighing skin in PMR) or the SCs present within terminal nerve pathways (Brushart 1988; Brushart 1993; Madison et al. 1996; Madison et al. 1999; Madison et al. 2007; Madison et al. 2009). Previous studies have shown that sensory axons may also regenerate preferentially down sensory pathways (Hoke et al. 2006). In addition to guiding the regeneration of axons, SCs derived from the cutaneous branch of the rat femoral nerve and the SCs derived from ventral root of the sciatic nerve exhibit phenotypic differences that may influence the regenerating pathway (Hoke et al. 2006). After denervation, SCs de-differentiate into an immature state (Mirsky and Jessen 1996) and secrete GFs and ECM to enhance regeneration (Bunge 1993; Bunge 1994; Bunge et al. 1986). Although SCs de-differentiate, they may retain a “phenotypic memory” that allows them to re-differentiate into their original phenotype during regeneration, as evidenced by different expression profiles observed by motor and sensory SCs even after prolonged contact with axons of the opposite phenotype (Hoke et al. 2006).
Understanding how SC gene expression changes during de-differentiation and following transplantation could enable motor or sensory specific nerve regeneration using ANGs seeded with phenotype specific SCs. In this study, a set of phenotypic markers were identified for the motor and sensory SCs, harvested from the motor and sensory branches of the rat femoral nerve, using Affymetrix Gene Chips© and quantitative real time polymerase chain reaction (qRT-PCR) and to study the effects of expansion culture conditions on the SC gene expression profiles. The results indicate that both motor and sensory SCs have unique phenotypes that are disrupted when expanded in vitro.
MATERIALS AND METHODS
RNA preparation
Male Lewis rats (400–500 grams) were anesthetized to undergo bilateral harvest of the sensory and motor branches of the femoral nerve and were then euthanized. The nerves were stored at −80°C in RNAlater® solution (Ambion, Austin, TX). Total RNA was extracted from homogenized nerves using an acid phenol extraction (TRIzol Reagent, Invitrogen, Carlsboro, CA). The aqueous layer was collected, and the samples were purified using an RNeasy Mini Kit (Qiagen, Valencia, CA). The presence of the RNA was assessed by electrophoresis using 2% agarose gels after running reverse transcriptase PCR with a β-actin primer. To verify that the mRNA extracted from the nerves met the quality standards for further experiments, the mRNA concentration (0.2 – 0.6 μg/μL) was determined by measuring the absorbance at 260 nm and the quality was verified using an absorbance ratio of A260/A280. The ratio threshold was held at 1.8, which signifies a high purity of RNA in the sample (Wilfinger et al. 1997). The extracted RNA was used for further experiments. Since the majority of the nerves are composed of SCs (~90%) (Oda et al. 1989), the harvested RNA was assumed to be representative of the SC RNA present in fresh nerve tissue.
Gene Chips
The RNA (10 nerves pooled from different male Lewis rats per sample) required for gene chip analysis was prepared according to the standard protocol provided by the Siteman Cancer Center GeneChip Facility at Washington University and run in triplicate. Purified total RNA (10 μg) was spiked with a set of four synthetic, polyadenylated, and bacterial transcripts (Lys, Phe, Thr, and Trp) diluted to defined copy numbers. Oligonucleotide probes for these transcripts were present on all Affymetrix GeneChips, thus monitoring the expression level of these internal standards provided an indication of the total technical variability associated with the experiment. Spiked RNA was converted to cDNA, purified, and then used as a template for in vitro transcription of biotin-labeled antisense RNA. All protocols were performed as recommended by the manufacturer (Affymetrix, Santa Clara, CA). Each biotinylated antisense RNA preparation (20 μg) was fragmented, assessed by gel electrophoresis, and placed in hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre). Samples were hybridized to Affymetrix Rat GeneChip® Rat Genome 230 2.0 Array A for 16 h (n = 3 chips for each condition). GeneChips were washed and stained using the instrument's standard Eukaryotic GE Wash 2 protocol, using antibody mediated signal amplification. The images from the scanned chips were processed using Affymetrix Microarray Analysis Suite 4.0. Genes that did not hybridize to the probe for any sample were excluded from the analysis.
The results obtained from the gene chips were analyzed using the exclusion criterion (Costigan et al. 2002) in combination with Spotfire DecisionSite 9.0 and Microsoft Excel. If the gene was not present in at least two of the chips in each group, then it was excluded from the gene set. The detection signal was then z-score normalized and statistical analysis (ANOVA) paired with a t-test was performed to compare the detection signals of the sensory nerves and the motor nerves, and the gene was excluded if the p-value was greater than 0.05, which is the threshold of significance. The remaining data was imported to Microsoft Excel. Genes that had a mean value varying by less than two-fold between sensory and motor or with a standard deviation between replicates in the same group that was greater than 35% of the mean were excluded from the analysis.
Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
cDNA was synthesized from the isolated RNA using the QuantiTect® Reverse Trascription Kit (Qiagen). Using the QuantiTect® SYBR® Green PCR mastermix (Qiagen) in combination with gene specific QuantiTect® primer assays, qRT-PCR was performed using an Applied Biosystems 7000 Real-Time PCR thermocycler. The genes studied included vascular endothelial cell growth factor (VEGF), nerve growth factor (NGF), brain derived neurotrophic factor (BDNF), pleiotrophin (PTN), glial-derived neurotrophic factor (GDNF), myelin basic protein (MBP), protein kinase C iota (PRKCi), neuroligin 1 (NLGN1), and neurofilament (NEFL). The primers for those mentioned proteins were added to the cDNA for each sample present for the motor and sensory nerves. The qRT-PCR was conducted using the following conditions: (1) 50°C for 2 min (2) 95°C for 15 min, and (3) forty cycles of 95°C for 15 seconds, 55°C for 30 seconds, and 72°C for 30 seconds (Gaumond et al. 2006). Target genes were normalized to an internal control (β-actin) to account for the variation in cDNA concentration between samples, and appropriate negative control samples were present (no template control). The QuantiTect® primer assays are validated to have a PCR efficiency of 100%. To estimate the mRNA concentrations, the differences in gene expression levels between two different samples were calculated using the comparative delta crossover threshold (Ct) method (Livak and Schmittgen 2001).
SC Culture Preparation for Time Study
SC cultures were prepared as previously described (Pruss 1982; Raff et al. 1978). Briefly, the sensory and motor branches of the rat femoral nerve were harvested and placed in Leibovitz's L-15 medium (Invitrogen, Carlsbad, CA). Collagenase I (1%) (Fisher, Pittsburgh, PA) and trypsin (2.5%) (Invitrogen) were added to the fascicles and incubated for 30 min at 37°C. After centrifugation at 130 × g for 5 min, the pellet was washed with Dulbecco's modified Eagle medium (DMEM, Invitrogen) supplemented with 10% heat-activated fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO) and 1% antibiotic antimycotic (ABAM, Invitrogen). The cells were then seeded on 24 well plates coated with poly-L-lysine (pLL) (Sigma-Aldrich). Tissue culture plates were prepared by coating with 1 mL 0.01% pLL in sterile water and washing twice with sterile water. On day 2 of culture, 10 μM Cytosine-beta-arabino furanoside hydrochloride (Ara-C) (Sigma-Aldrich), was added to cultures along with the media containing DMEM, FBS and ABAM. On day 6, the fibroblasts were complement-killed using an anti-Thy 1.1 antibody (1:40 dilution in media, Serotec, Raleigh, NC) and guinea pig complement (1:4 dilution in media, Sigma-Aldrich). On subsequent days the culture media was supplemented with 2 μM forskolin (Sigma-Aldrich), and 20 μg/mL pituitary extract (PE) (Biomedical Tech, Inc., Stoughton, MA). RNA was extracted from Days 1, 3, 7, 14 and 30 using an acid-phenol extraction and was purified using an RNeasy Mini Kit (Qiagen). qRT-PCR was performed for each gene at each time point and compared to the gene expression each gene in freshly harvested femoral motor and sensory nerves.
Statistical Analysis
Statistical analyses were performed using SigmaStat 3.0 (Systat Software, San Jose, CA), and all data were evaluated with one-way analysis of variance (ANOVA), followed by a Scheffe's F test for comparisons between groups when significance (p<0.05) was present. All results are reported as mean ± standard deviation.
RESULTS
Gene Chips
The differences in gene expression between SCs in the motor and sensory branches of the rat femoral nerve were evaluated using Affymetrix gene chips and qRT-PCR. Similar to findings in literature, we assumed that the majority of the RNA harvested from these nerves (~90%) was from SCs (Oda et al. 1989). RNA was extracted from the motor and sensory branches of fresh rat femoral nerves, and analyzed using gene chips to obtain a set of genes that were differentially upregulated in the motor SCs or sensory SCs when compared to each other. Using a stringent criteria, which has been shown to generate the lowest number of false positives (Costigan et al. 2002), ~100 genes were identified to be differentially upregulated in either the sensory (76) or motor (23) branches of the femoral nerve (data not shown).
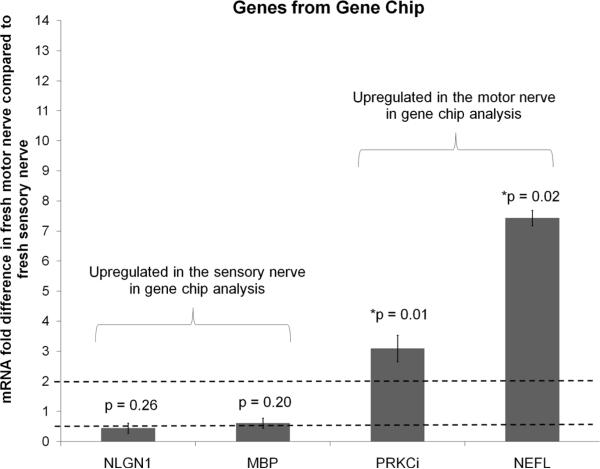
In motor SCs, a subset of the upregulated genes was identified to be involved in different functions related to motor nerve myelination and signaling (Table I). For example, NEFL has been shown to be an important factor in the myelination of the motor axons (Roxanne et. al 2003, Roberson et. al 1992), which suggests that NEFL may be a good marker for motor SCs. In SCs, PRKCi may act through the p75NTR activation pathway to promote survival or apoptosis (Mamidipudi et. al 2002). PRKCi has also been shown to interact with Rab2 to promote vesicle budding for exosome formation to facilitate intercellular signaling between SCs and axons (Tisdale 2000; van Niel et al. 2006). The increased expression of PRKCi in the motor SCs may be attributed to having more signaling between the motor SCs and axons than the sensory SCs due to the higher number of motor neurons (and thus motor axons and motor SCs) present in the peripheral nervous system (PNS).
Table I.
Genes that are upregulated in the motor branch of the femoral nerve versus sensory branch. M – average signal intensity in motor nerve group, S – average signal intensity in sensory nerve group (n = 3)
| Gene name | Gene Common name | Accession Number | Fold Difference M/S | std dev/average |
|---|---|---|---|---|
| Peripheral myelin protein 2 | Pmp2 | AW533483 | 5.62 | 0.19 |
| Four and a half LIM domains 1 | Fhl1 | BI298356 | 4.83 | 0.22 |
| Gap junction membrane channel protein beta 2 | Gjb2 | AI179953 | 3.48 | 0.21 |
| Neurofilament, light polypeptidea | Nefl | NM_031783 | 3.46 | 0.02 |
| Prostaglandin D2 synthase | Ptgds | J04488 | 2.33 | 0.1 |
| Ubiquitin carboxy-terminal hydrolase L1 | Uchl1 | NM_017237 | 2.28 | 0.03 |
| Calsequestrin 2 | Casq2 | NM_017131 | 2.07 | 0.31 |
| Amphiphysin 1 | Amph1 | NM_022217 | 2.05 | 0.28 |
| Fucosidase, alpha-L- 2, plasma | Fuca2 | BM389993 | 2.04 | 0.11 |
| Tubulin, beta 2b | Tubb2 | X03369 | 2.01 | 0.07 |
| Protein kinase C, iotaa | Prkci | AB020615 | 2.00 | 0.11 |
indicates genes used for further studies
A subset of the genes identified to be differentially upregulated in the sensory SCs has been shown to be involved in promoting sensory nerve myelination and maturation (Table II). Neuroligin 1 (NLGN1) is a component of the myelin sheath made by SCs (Jahn et al. 2009; Song et al. 1999). Because the expression of NLGN1 in SCs is increased during sensory nerve depolarization and signal conduction in the SC-associated axons, NLGN1 makes a good candidate as a sensory SC marker (Biswas et al. 2010). Although MBP is present in both motor and sensory SCs and promotes myelination (Eylar et al. 1971) of axons, increased intracellular progesterone in SCs, (Guennoun et al. 2001; Robert et al. 2001), increases the expression of MBP in sensory SCs, thus making MBP a good marker for sensory SCs.
Table II.
Genes that are upregulated in the sensory branch of the femoral nerve versus motor branch. M – average signal intensity in motor nerve group, S – average signal intensity in sensory nerve group (n = 3)
| Gene name | Gene common name | Accession Numbers | Fold Difference S/M | std dev/average |
|---|---|---|---|---|
| Neuroligin 1a | Nlgn1 | BF400127 | 3.36 | 0.34 |
| Ankyrin 3, epithelial | Ank3 | AJ428573 | 2.52 | 0.03 |
| S100 calcium binding protein A9 (calgranulin B) | S100a9 | NM_053587 | 2.23 | 0.14 |
| Microtubule-associated protein tau | Mapt | BE107978 | 2.22 | 0.33 |
| Scavenger receptor class B, member 1 | Scarb1 | NM_031541 | 2.21 | 0.07 |
| L1 cell adhesion molecule | L1cam | NM_017345 | 2.19 | 0.09 |
| Neurotrophic tyrosine kinase, receptor, type 2 | Ntrk2 | BE102996 | 2.17 | 0.24 |
| Myelin basic proteina | Mbp | BE109730 | 2.14 | 0.12 |
| Platelet derived growth factor receptor, beta polypeptide | Pdgfrb | AI071374 | 2.13 | 0.06 |
| Neural cell adhesion molecule 1 | Ncam1 | AI576209 | 2.05 | 0.06 |
| Nuclear receptor subfamily 4, group A, member 2 | Nr4a2 | U72345 | 2.04 | 0.23 |
indicates genes used for further studies
Verification of genes using qRT-PCR
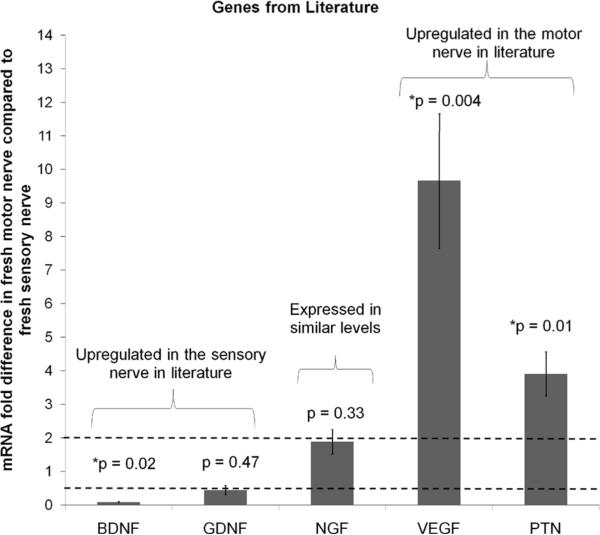
For further analysis, a set of genes was selected from the gene chip analysis that have been shown in previous studies to be involved in either sensory or motor function development and growth: neuroligin 1 (NLGN1, sensory), myelin basic protein (MBP, sensory), protein kinase C iota (PRKCi, motor), and neurofilament (NEFL, motor) (Biswas et al. 2010; Roberson et al. 1992; Robert et al. 2001; Tisdale 2000). Along with these markers, genes that were previously shown to be differentially expressed in motor and sensory SCs were also used for analysis: VEGF (motor), PTN (motor), GDNF (sensory), BDNF (sensory), and NGF (similar expression in motor and sensory) (Hoke et al. 2006). The expression levels in fresh nerve tissue were evaluated for the selected genes using qRT-PCR to verify that the chosen genes agreed with the trends found in the gene chip analysis and in literature. PRKCi and NEFL were upregulated in motor SCs compared to sensory SCs (Figure 1), which agreed with the trends observed with the gene chip analysis. Previously identified markers from literature were also analyzed by qRT-PCR (Figure 2), and those results showed similar trends to those found in the literature. The genes VEGF and PTN were upregulated in the motor SCs compared to the sensory SCs, BDNF was downregulated in motor SCs compared to the sensory SCs, and NGF was expressed in similar levels in both types of SCs. These results suggest that we can use these gene markers to identify the phenotype of the SCs because they correlated well with results from the gene chips and literature.
Figure 1. Verification of gene chip trend with qRT-PCR.
The expression level of genes that were identified to be differentially expressed in the motor and sensory branches of the rat femoral nerve was determined by qRT-PCR. The values were normalized to β-actin, and the fold difference versus sensory nerve was calculated. PRKCi and NEFL are upregulated in the motor branch similar to the gene chip results. Error bars represent the standard deviation (n=3). The dotted line at 2 is the threshold value for up regulation, and the dotted line at 0.5 is the threshold for down regulation. * denotes p-values are the significance levels between the ΔCt (motor – β-actin) and Δ Ct (sensory – β-actin)
Figure 2. Expression of genes previously identified as markers for sensory and motor SCs.
The expression level of genes that were reported in the literature to be differentially regulated in the motor and sensory SCs (Hoke et al. 2006) was determined by qRT-PCR. The values were normalized to β-actin, and the fold difference versus sensory nerve was calculated. BDNF is upregulated in the sensory branch, NGF is similarly expressed in both branches, and VEGF and PTN are upregulated the motor branch. Error bars represent the standard deviation (n=3). The dotted line at 2 is the threshold value for up regulation, and the dotted line at 0.5 is the threshold for down regulation. * denotes p-values are the significance levels between the ΔCt (motor − β-actin) and Δ Ct (sensory − β-actin)
Gene expression in SCs in vitro
To evaluate the effect of expansion culture on differential gene expression, SCs from the motor and sensory branches of the femoral nerve were harvested and cultured for 30 days. Fibroblasts were eliminated from the culture using Ara-C to inhibit the proliferation of the cells (Ogbomo et al. 2008) and the cells were allowed to recover for 4 days before the remaining fibroblasts were killed using complement. SC RNA was collected at days 0, 1, 3, 7, 14, and 30 (Table III), and the expression levels of the genes (Table IV) were analyzed using qRT-PCR compared to expression in fresh nerve tissue (day 0). In these studies, a value of two or greater was selected as the minimum criteria for a significant difference in expression levels between groups (Hoke et al. 2006).
Table III.
The different conditions at which the RNA was extracted for qRT-PCR
| Day | RNA Condition |
|---|---|
| 0 | From freshly harvested nerve tissue |
| 1 | From cells plated for 24 hours on pLL coated plates |
| 3 | From cells recovering from the Ara-C treatment |
| 7 | From cells recovering from complement killing of fibroblasts |
| 14 | From cells at ~30% confluence |
| 30 | From cells at ~80% confluence |
Table IV.
The genes chosen from the gene chip analysis and literature for further analysis
| Gene | Gene Common Name | Differentially upregulated or similarly expressed in motor or sensory SCs | Function in the Nervous system |
|---|---|---|---|
| NEFL | Neurofilament Light Peptide | Motor (Gene Chips) | Helps with the axonal growth and myelination (Roberson et al. 1992) |
| PRKCi | Protein Kinase C iota | Motor (Gene Chips) | Regulates intercellular signaling between SCs and axons (Tisdale 2000; van Niel et al. 2006) |
| NLGN1 | Neuroligin 1 | Sensory (Gene Chips) | Associated with the localization in the postsynaptic compartment of excitatory synapses (Biswas et al. 2010; Scheiffele et al. 2000) |
| MBP | Myelin Basic Protein | Sensory (Gene Chips) | Responsible for the myelination of nerves in the nervous system (Eylar et al. 1971) |
| VEGF | Vascular Endothelial Growth Factor | Motor (Literature) | Creates new blood vessels during adult nervous system development (Rosenstein et al. 2008) |
| PTN | Pleiotrophin | Motor (Literature) | Neurite outgrowth promoting factor (Jin et al. 2009) |
| NGF | Nerve Growth Factor | Similar (Literature) | Important for the growth, maintenance and survival of certain target neurons (Chan et al. 2001) |
| GDNF | Glial Derived Neurotrophic factor | Sensory (Literature) | Promotes the survival and differentiation of dopaminergic neurons (Iwase et al. 2005) |
| BDNF | Brain-derived Neurotrophic factor | Sensory (Literature) | Helps with the support, survival, growth, differentiation of new neurons and synapses (Chan et al. 2001) |
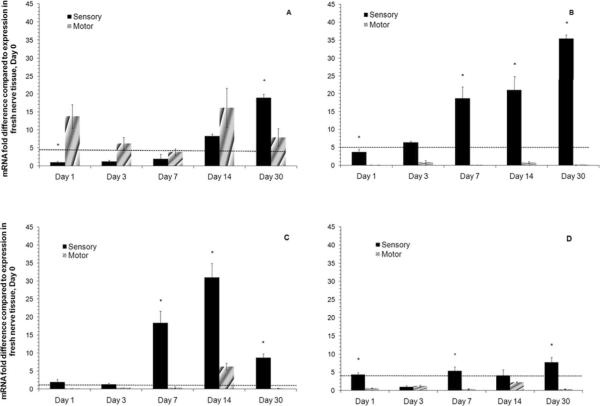
Those genes that showed significant changes in gene expression over 30 days were MBP, NEFL, PTN and PRKCi (Figure 3). MBP is differentially upregulated in the sensory branch of the femoral nerve in fresh nerve tissue. The expression in the sensory SCs increases over the course of the study as does the expression in the motor SCs. On Day 1, there is a difference in the expression of the MBP, which is greater in motor SCs versus sensory SCs. Eventually the gene is expressed at similar levels until day 30 when the expression in the sensory SCs increases and expression in motor SCs drops (Figure 3A). Of the other genes that were evaluated, NEFL, PTN and PRKCi (all motor markers) showed changes in the expression over 30 days. NEFL (Figure 3B) expression decreased in the motor SCs but in the sensory SCs there was a 40 fold increase over the fresh tissue after 30 days. PTN expression levels were upregulated in the sensory SCs and increased to a maximum expression at day 14 with a 35 fold increase when compared to the expression in the fresh tissue (Figure 3C). In the motor SCs, the PTN gene was downregulated over 30 days of expansion culture. PRKCi showed similar expression levels when compared to the fresh tissue of the motor nerve, but the expression levels in the sensory SCs increased approximately 8 fold when compared to the fresh sensory nerve tissue (Figure 3D). The gene expression for the remaining five genes, GDNF, BDNF, NGF, VEGF, and NLGN1 showed no significant increases in expression because all the mRNA fold difference values were below the value of two (data not shown). From these results, we can see that the phenotypic expression of these genes in motor or sensory SCs is significantly affected when SCs are expanded in vitro.
Figure 3. Gene expression in motor and sensory SCs over 30 days of in vitro culture by qRT-PCR.
A) MBP B) NEFL C) PTN and D) PRKCi. The gene expression for each time point was normalized to β-actin, and then the fold difference versus fresh tissue (Day 0) was calculated. The error bars represent the standard deviation (n =3). * denotes p < 0.05 when compared to the gene expression in sensory SCs to motor SCs at that time point. The dotted line at 2 is the threshold value for up regulation.
DISCUSSION
A number of studies have shown that the transplantation of SCs at the injury site improves the regeneration of peripheral nerves (Brenner et al. 2005; Fox et al. 2005). However, SCs need to be expanded in culture to transplant a sufficient number of cells at the injury site. During expansion, it is necessary to understand the effects of expansion culture on SC phenotype. In this study, we used a rat femoral nerve model to identify a set of phenotypic markers (from literature and gene chips) to monitor the expression profiles while the SCs were expanded in vitro.
There are two types of SCs, myelinating and nonmyelinating, present in healthy nerves (Mirsky and Jessen 1996). Myelinating SCs associate with axons in a 1:1 ratio and aid in saltatory conduction. Alternatively, the nonmyelinating SCs associate with several non-myelinated axons that conduct signals with wave-like impulses. From previous studies, it has been shown that the cutaneous nerve and ventral root have ~20% and ~33% myelinated axons (MAx) respectively and ~80% and ~66% nonmyelinated axons (NMAx) respectively (Castro et al. 2008; Schmalbruch 1986). These different ratios in the number of MAx versus NMAx suggests that there are different proportions of the myelinating and nonmyelinating SCs present in the nerves, which may have an effect on the differences in the expression profiles on a healthy nerve. However, in the denervated nerve, both myelinating and nonmyelinating SCs regress into a more immature phenotype (Jessen and Mirsky 2005; Mirsky and Jessen 1996). Similarly, when the SCs are harvested from the fresh tissue and expanded in vitro the SCs may be reverting to a similar immature phenotype due to the lack of cues from the environment (e.g. interaction with axons). Previous studies have shown that cutaneous nerve and ventral root SCs exhibit phenotypic differences, but also differentiate into their original phenotype during regeneration even after prolonged contact with axons of the opposite phenotype (Hoke et al. 2006). The implication that SCs having different expression profiles, which may influence preferential motor regeneration (PMR), suggests that transplanting phenotype specific-SCs may be potentially a promising therapeutic strategy for PMR.
To monitor the expression level changes in the SCs harvested from the motor and sensory branches of the rat femoral nerve, phenotypic markers were chosen from the gene chip analysis and from previously identified markers in literature. Although the gene chip analysis revealed ~100 genes that were differentially regulated in the motor or sensory SCs, the phenotypic markers chosen for this study were chosen because of the implication in SC function in the PNS. Additionally, a few genes (VEGF, GDNF, NGF, BDNF, and PTN) were selected from previously published literature (Hoke et al. 2006). These five genes may not have been identified from the gene chip analysis due to the stringent criteria (Costigan et al. 2002) used to identify the phenotypic markers. However, the markers chosen from the gene chips and literature were validated using qRT-PCR to show that they express similar trends to literature and the gene chips (Figures 1 & 2).
As mentioned earlier, the genes we chose as sensory markers (MBP, NLGN1, BDNF and GDNF) were chosen based on the role each gene plays in sensory SC function (Biswas et al. 2010; Guennoun et al. 2001; Jahn et al. 2009; Robert et al. 2001; Song et al. 1999). BDNF and GDNF were chosen from literature as markers of sensory SC expression because these growth factors are predominantly expressed in the cutaneous nerve derived from the rat femoral branch (Hoke et al. 2006). The motor markers chosen were PRKCi, NEFL, VEGF and PTN. PRKCi and NEFL have been implicated in aiding with motor SC signaling and myelination (Lariviere and Julien 2004; Mamidipudi and Wooten 2002; Roberson et al. 1992; Tisdale 2000). The remaining two genes, VEGF and PTN, were chosen from literature because they are predominantly expressed in the ventral root after denervation (Hoke et al. 2006). Due to the involvement of these genes in SC function, these markers can be used to monitor the changes in phenotypic gene expression profiles as SCs are expanded in vitro.
The expression profiles for the cells harvested from the motor and sensory branches of the rat femoral nerve were monitored by evaluating the relative mRNA levels of each gene (Table IV) compared to the expression levels in fresh tissue. As hypothesized, the expression patterns of these genes were altered as the SCs were expanded in vitro. Of the nine genes evaluated, the genes that were dysregulated in culture were MBP, NEFL, PRKCi, and PTN.
A GeneGo network pathway analysis on the results obtained through the gene chip experiment revealed pathways that may contribute to the changes in gene expression observed during SC expansion. A subset of genes from the Sox family has previously been shown to contribute to the neuronal development. One particular gene, Sox6, controls the transcription of the MBP (Stolt et al. 2006). Because MBP, a sensory marker, was upregulated in the motor SCs during culture, Sox6 alone with other transcription factors, such as SP1, Oct6, and Krox20, may be either be over- or under-expressed in the cell, thus altering the expression of MBP (Kao et al. 2009). Additionally, interaction with the sensory neurons promotes the synthesis of intracellular progesterone in SCs and thus may control the expression of Krox20, which regulates the expression of MBP (Guennoun et al. 2001; Robert et al. 2001). The absence of neurons in culture may affect the expression levels of MBP in the SCs.
NEFL interacts with the family of microtubule-associated proteins (MAP) (Frappier et al. 1991), which bind to tubulin subunits to support the assembly of microtubules in neurons. SCs also express NEFL in the healthy nerve to support the axon as well as aid in efficient signal conduction (Roberson et al. 1992). The dysregulation of NEFL may be due to the fact that the SCs are no longer supporting axons in cell culture and thus altering the expression patterns in the sensory SCs when grown in vitro in the absence of neurons. PRKCi plays a role in axonal transport, microtubule dynamics as well as SC survival and intercellular signaling (Mamidipudi and Wooten 2002; Tisdale 2000; van Niel et al. 2006). In SCs, these interactions are necessary to regulate SC apoptosis as well as assisting with vesicle budding and exosome formation for extracellular signaling to other SCs and axons. After expansion in culture, the expression of PRKCi may be dysregulated because the SCs are not in contact with axons. The PTN gene is important during neural development and promotes neurite outgrowth, cell proliferation, and cell migration. In recent studies, PTN has been shown to aid in the guidance of axonal regeneration and muscle reinnervation after injury (Deuel et al. 2002; Jin et al. 2009). The changes in the gene expression levels, especially at day 14, may be due to the fact that the SCs are missing the necessary cues from axons to keep the PTN expression levels similar to expression in fresh tissue.
Additionally, at different time points during the SC expansion culture, the SCs are treated with different chemicals and mitogens (Table III) to eliminate fibroblasts and to promote the proliferation of the cells using mitogenic supplements. Previously, it is have been shown that de-differentiation of SCs may be linked to proliferation (Guertin et al. 2005). However, recent studies have revealed that SC de-differentiation is independent of mitogenic signaling and also uncoupled to proliferation (Monje et al. 2010). De-differentiated SCs do not proliferate unless treated with mitogenic supplements, whereas differentiated post-mitotic SCs do not respond to mitogenic additions to the culture media. Since differentiation of the SCs is dependent on high levels of intracellular cyclic adenosine monophosphate (cAMP), the SCs in the present study may have de-differentiated in culture due to the decrease in intracellular cAMP levels and loss of signaling from the environment in vivo and thus proliferate in response to mitogenic additions to the media.
The de-differentiated SCs in vitro may be mimicking the de-differentiated state the SCs revert to after injury and may be awaiting cues from the environment to induce differentiation into its native phenotype. In vivo, this immature state promotes SC proliferation and secretion GFs to aid axonal regeneration (Jessen and Mirsky 2005). As the axons grow, cues in the environment, such as GFs (Chan et al. 2001; Iwase et al. 2005), neurotransmitters (Vrbova et al. 2009), or supporting cells such as fibroblasts (Parrinello et al. 2010), may guide the SCs differentiation back into their native phenotype to support the regenerating axons. To understand the differentiation and maintenance of SC phenotypes, further studies need to be conducted to evaluate the effects of culturing SCs with different environmental cues.
In conclusion, phenotype specific genes are differentially expressed in the motor branch and sensory branches of the femoral nerve. Additionally, we observed that these gene expression patterns were disrupted when motor and sensory SCs were expanded in culture. These results suggest that although motor and sensory SCs have different phenotype, future studies need to be performed to identify the environmental cues that influence and maintain the SC phenotype in a healthy nerve. GFs as well as neurotransmitters and supporting cells may help maintain the SC phenotype in vivo as well as in vitro. Understanding the cues that guide differentiation or maintenance of SC phenotype may improve SC transplantation therapies for the improvement of motor or sensory specific regeneration across nerve gaps using ANGs after injury.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the Hope Center for Neurological Disorders for use of the qRT-PCR thermocycler.
Funding sources: NIH Neuroscience Blueprint Interdisciplinary Center Core (P30 NS057105); NIH 5R01NS033406; Hope Center for Neurological Disorders, Washington University in St.Louis
REFERENCES
- Biswas S, Reinhard J, Oakeshott J, Russell R, Srinivasan MV, Claudianos C. Sensory regulation of neuroligins and neurexin I in the honeybee brain. PLoS One. 2010;5(2):e9133. doi: 10.1371/journal.pone.0009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner MJ, Lowe JB, 3rd, Fox IK, Mackinnon SE, Hunter DA, Darcy MD, Duncan JR, Wood P, Mohanakumar T. Effects of Schwann cells and donor antigen on long-nerve allograft regeneration. Microsurgery. 2005;25(1):61–70. doi: 10.1002/micr.20083. [DOI] [PubMed] [Google Scholar]
- Brushart TM. Preferential reinnervation of motor nerves by regenerating motor axons. J Neurosci. 1988;8(3):1026–1031. doi: 10.1523/JNEUROSCI.08-03-01026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brushart TM. Motor axons preferentially reinnervate motor pathways. J Neurosci. 1993;13(6):2730–2738. doi: 10.1523/JNEUROSCI.13-06-02730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge RP. Expanding roles for the Schwann cell: ensheathment, myelination, trophism and regeneration. Current opinion in neurobiology. 1993;3(5):805–809. doi: 10.1016/0959-4388(93)90157-t. [DOI] [PubMed] [Google Scholar]
- Bunge RP. The role of the Schwann cell in trophic support and regeneration. J Neurol. 1994;242(1 Suppl 1):S19–21. doi: 10.1007/BF00939235. [DOI] [PubMed] [Google Scholar]
- Bunge RP, Bunge MB, Eldridge CF. Linkage between axonal ensheathment and basal lamina production by Schwann cells. Annu Rev Neurosci. 1986;9:305–328. doi: 10.1146/annurev.ne.09.030186.001513. [DOI] [PubMed] [Google Scholar]
- Burnett MG, Zager EL. Pathophysiology of peripheral nerve injury: a brief review. Neurosurgical focus. 2004;16(5):E1. doi: 10.3171/foc.2004.16.5.2. [DOI] [PubMed] [Google Scholar]
- Castro J, Negredo P, Avendano C. Fiber composition of the rat sciatic nerve and its modification during regeneration through a sieve electrode. Brain Res. 2008;1190:65–77. doi: 10.1016/j.brainres.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Chan JR, Cosgaya JM, Wu YJ, Shooter EM. Neurotrophins are key mediators of the myelination program in the peripheral nervous system. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14661–14668. doi: 10.1073/pnas.251543398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D'Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuel TF, Zhang N, Yeh HJ, Silos-Santiago I, Wang ZY. Pleiotrophin: a cytokine with diverse functions and a novel signaling pathway. Arch Biochem Biophys. 2002;397(2):162–171. doi: 10.1006/abbi.2001.2705. [DOI] [PubMed] [Google Scholar]
- Eylar EH, Brostoff S, Hashim G, Caccam J, Burnett P. Basic A1 protein of the myelin membrane. The complete amino acid sequence. J Biol Chem. 1971;246(18):5770–5784. [PubMed] [Google Scholar]
- Fox IK, Schwetye KE, Keune JD, Brenner MJ, Yu JW, Hunter DA, Wood PM, Mackinnon SE. Schwann-cell injection of cold-preserved nerve allografts. Microsurgery. 2005;25(6):502–507. doi: 10.1002/micr.20152. [DOI] [PubMed] [Google Scholar]
- Frappier T, Stetzkowski-Marden F, Pradel LA. Interaction domains of neurofilament light chain and brain spectrin. Biochem J. 1991;275(Pt 2):521–527. doi: 10.1042/bj2750521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostick SP, Yin Q, Kemp GJ. Schwann cells, neurotrophic factors, and peripheral nerve regeneration. Microsurgery. 1998;18(7):397–405. doi: 10.1002/(sici)1098-2752(1998)18:7<397::aid-micr2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Gaumond G, Tyropolis A, Grodzicki S, Bushmich S. Comparison of direct fluorescent antibody staining and real-time polymerase chain reaction for the detection of Borrelia burgdorferi in Ixodes scapularis ticks. J Vet Diagn Invest. 2006;18(6):583–586. doi: 10.1177/104063870601800610. [DOI] [PubMed] [Google Scholar]
- Guennoun R, Benmessahel Y, Delespierre B, Gouezou M, Rajkowski KM, Baulieu EE, Schumacher M. Progesterone stimulates Krox-20 gene expression in Schwann cells. Brain Res Mol Brain Res. 2001;90(1):75–82. doi: 10.1016/s0169-328x(01)00094-8. [DOI] [PubMed] [Google Scholar]
- Guertin AD, Zhang DP, Mak KS, Alberta JA, Kim HA. Microanatomy of axon/glial signaling during Wallerian degeneration. J Neurosci. 2005;25(13):3478–3487. doi: 10.1523/JNEUROSCI.3766-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare GM, Evans PJ, Mackinnon SE, Nakao Y, Midha R, Wade JA, Hunter DA, Hay JB. Effect of cold preservation on lymphocyte migration into peripheral nerve allografts in sheep. Transplantation. 1993;56(1):154–162. doi: 10.1097/00007890-199307000-00029. [DOI] [PubMed] [Google Scholar]
- Hare GM, Evans PJ, Mackinnon SE, Wade JA, Young AJ, Hay JB. Phenotypic analysis of migrant, efferent lymphocytes after implantation of cold preserved, peripheral nerve allografts. J Neuroimmunol. 1995;56(1):9–16. doi: 10.1016/0165-5728(94)00120-d. [DOI] [PubMed] [Google Scholar]
- Hoke A, Redett R, Hameed H, Jari R, Zhou C, Li ZB, Griffin JW, Brushart TM. Schwann cells express motor and sensory phenotypes that regulate axon regeneration. J Neurosci. 2006;26(38):9646–9655. doi: 10.1523/JNEUROSCI.1620-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T, Jung CG, Bae H, Zhang M, Soliven B. Glial cell line-derived neurotrophic factor-induced signaling in Schwann cells. J Neurochem. 2005;94(6):1488–1499. doi: 10.1111/j.1471-4159.2005.03290.x. [DOI] [PubMed] [Google Scholar]
- Jahn O, Tenzer S, Werner HB. Myelin proteomics: molecular anatomy of an insulating sheath. Mol Neurobiol. 2009;40(1):55–72. doi: 10.1007/s12035-009-8071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6(9):671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Jin L, Jianghai C, Juan L, Hao K. Pleiotrophin and peripheral nerve injury. Neurosurg Rev. 2009;32(4):387–393. doi: 10.1007/s10143-009-0202-8. [DOI] [PubMed] [Google Scholar]
- Johnson PC, Duhamel RC, Meezan E, Brendel K. Preparation of cell-free extracellular matrix from human peripheral nerve. Muscle Nerve. 1982;5(4):335–344. doi: 10.1002/mus.880050410. [DOI] [PubMed] [Google Scholar]
- Kao SC, Wu H, Xie J, Chang CP, Ranish JA, Graef IA, Crabtree GR. Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science. 2009;323(5914):651–654. doi: 10.1126/science.1166562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere RC, Julien JP. Functions of intermediate filaments in neuronal development and disease. J Neurobiol. 2004;58(1):131–148. doi: 10.1002/neu.10270. [DOI] [PubMed] [Google Scholar]
- Levi AD, Evans PJ, Mackinnon SE, Bunge RP. Cold storage of peripheral nerves: an in vitro assay of cell viability and function. Glia. 1994;10(2):121–131. doi: 10.1002/glia.440100206. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Madison RD, Archibald SJ, Brushart TM. Reinnervation accuracy of the rat femoral nerve by motor and sensory neurons. J Neurosci. 1996;16(18):5698–5703. doi: 10.1523/JNEUROSCI.16-18-05698.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison RD, Archibald SJ, Lacin R, Krarup C. Factors contributing to preferential motor reinnervation in the primate peripheral nervous system. J Neurosci. 1999;19(24):11007–11016. doi: 10.1523/JNEUROSCI.19-24-11007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison RD, Robinson GA, Chadaram SR. The specificity of motor neurone regeneration (preferential reinnervation) Acta physiologica (Oxford, England) 2007;189(2):201–206. doi: 10.1111/j.1748-1716.2006.01657.x. [DOI] [PubMed] [Google Scholar]
- Madison RD, Sofroniew MV, Robinson GA. Schwann cell influence on motor neuron regeneration accuracy. Neuroscience. 2009;163(1):213–221. doi: 10.1016/j.neuroscience.2009.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidipudi V, Wooten MW. Dual role for p75(NTR) signaling in survival and cell death: can intracellular mediators provide an explanation? J Neurosci Res. 2002;68(4):373–384. doi: 10.1002/jnr.10244. [DOI] [PubMed] [Google Scholar]
- Mirsky R, Jessen KR. Schwann cell development, differentiation and myelination. Current opinion in neurobiology. 1996;6(1):89–96. doi: 10.1016/s0959-4388(96)80013-4. [DOI] [PubMed] [Google Scholar]
- Monje PV, Soto J, Bacallao K, Wood PM. Schwann cell dedifferentiation is independent of mitogenic signaling and uncoupled to proliferation: role of cAMP and JNK in the maintenance of the differentiated state. J Biol Chem. 2010;285(40):31024–31036. doi: 10.1074/jbc.M110.116970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(13):8998–9003. doi: 10.1073/pnas.132080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Okada Y, Katsuda S, Ikeda K, Nakanishi I. A simple method for the Schwann cell preparation from newborn rat sciatic nerves. Journal of Neuroscience Methods. 1989;28(3):163–169. doi: 10.1016/0165-0270(89)90032-0. [DOI] [PubMed] [Google Scholar]
- Ogbomo H, Michaelis M, Klassert D, Doerr HW, Cinatl J., Jr. Resistance to cytarabine induces the up-regulation of NKG2D ligands and enhances natural killer cell lysis of leukemic cells. Neoplasia. 2008;10(12):1402–1410. doi: 10.1593/neo.08972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Napoli I, Ribeiro S, Digby PW, Fedorova M, Parkinson DB, Doddrell RD, Nakayama M, Adams RH, Lloyd AC. EphB signaling directs peripheral nerve regeneration through Sox2-dependent Schwann cell sorting. Cell. 2010;143(1):145–155. doi: 10.1016/j.cell.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss RM. The potential use of cultured cells to detect non-neuronal targets of neuropeptide action. Peptides. 1982;3(3):231–233. doi: 10.1016/0196-9781(82)90083-3. [DOI] [PubMed] [Google Scholar]
- Raff MC, Abney E, Brockes JP, Hornby-Smith A. Schwann cell growth factors. Cell. 1978;15(3):813–822. doi: 10.1016/0092-8674(78)90266-0. [DOI] [PubMed] [Google Scholar]
- Reynolds ML, Woolf CJ. Reciprocal Schwann cell-axon interactions. Current opinion in neurobiology. 1993;3(5):683–693. doi: 10.1016/0959-4388(93)90139-p. [DOI] [PubMed] [Google Scholar]
- Roberson MD, Toews AD, Goodrum JF, Morell P. Neurofilament and tubulin mRNA expression in Schwann cells. J Neurosci Res. 1992;33(1):156–162. doi: 10.1002/jnr.490330120. [DOI] [PubMed] [Google Scholar]
- Robert F, Guennoun R, Desarnaud F, Do-Thi A, Benmessahel Y, Baulieu EE, Schumacher M. Synthesis of progesterone in Schwann cells: regulation by sensory neurons. Eur J Neurosci. 2001;13(5):916–924. doi: 10.1046/j.0953-816x.2001.01463.x. [DOI] [PubMed] [Google Scholar]
- Schmalbruch H. Fiber composition of the rat sciatic nerve. Anat Rec. 1986;215(1):71–81. doi: 10.1002/ar.1092150111. [DOI] [PubMed] [Google Scholar]
- Schmidt CE, Leach JB. Neural tissue engineering: strategies for repair and regeneration. Annu Rev Biomed Eng. 2003;5:293–347. doi: 10.1146/annurev.bioeng.5.011303.120731. [DOI] [PubMed] [Google Scholar]
- Sondell M, Lundborg G, Kanje M. Regeneration of the rat sciatic nerve into allografts made acellular through chemical extraction. Brain Res. 1998;795(1–2):44–54. doi: 10.1016/s0006-8993(98)00251-0. [DOI] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Sudhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(3):1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, Wegner M. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11(5):697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Tisdale EJ. Rab2 requires PKC iota/lambda to recruit beta-COP for vesicle formation. Traffic. 2000;1(9):702–712. doi: 10.1034/j.1600-0854.2000.010903.x. [DOI] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Vrbova G, Mehra N, Shanmuganathan H, Tyreman N, Schachner M, Gordon T. Chemical communication between regenerating motor axons and Schwann cells in the growth pathway. Eur J Neurosci. 2009;30(3):366–375. doi: 10.1111/j.1460-9568.2009.06847.x. [DOI] [PubMed] [Google Scholar]
- Whitlock EL, Tuffaha SH, Luciano JP, Yan Y, Hunter DA, Magill CK, Moore AM, Tong AY, Mackinnon SE, Borschel GH. Processed allografts and type I collagen conduits for repair of peripheral nerve gaps. Muscle Nerve. 2009;39(6):787–799. doi: 10.1002/mus.21220. [DOI] [PubMed] [Google Scholar]
- Wilfinger WW, Mackey K, Chomczynski P. Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques. 1997;22(3):474–476. 478–481. doi: 10.2144/97223st01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.