Abstract
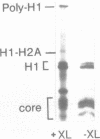
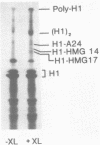
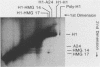
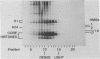
We have studied the assembly of histone H1 and the high mobility group nonhistones 14/17 by isopycnic analysis after crosslinking density labeled MSB cell nuclei or chromatin. Carbodiimide crosslinking produces dense poly-H1 and hybrid density H1-H2A histone dimers, indicating that new H1 is deposited nonrandomly, albeit nonconservatively relative to new core histones. Core histone-HMG crosslinking with succinimidyl propionate yields dense HMG 14 in uniformly dense particles and new HMG 17 crosslinked to both dense and light protein, implying that HMG 14 and 17 each deposit nonrandomly; but differently with respect to new core octamers. Propionimidate crosslinking yields dense H1-HMG 17 dimers, suggesting that the interactions of new 14/17 with H1 (new HMG 14-old H1, new HMG 17-new H1) are reciprocal to their interactions with the core histones.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama Y., Kato S. Two cell lines from lymphomas of Marek's disease. Biken J. 1974 Sep;17(3):105–116. [PubMed] [Google Scholar]
- Albright S. C., Wiseman J. M., Lange R. A., Garrard W. T. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980 Apr 25;255(8):3673–3684. [PubMed] [Google Scholar]
- Annunziato A. T., Schindler R. K., Riggs M. G., Seale R. L. Association of newly synthesized histones with replicating and nonreplicating regions of chromatin. J Biol Chem. 1982 Jul 25;257(14):8507–8515. [PubMed] [Google Scholar]
- Annunziato A. T., Schindler R. K., Thomas C. A., Jr, Seale R. L. Dual nature of newly replicated chromatin. Evidence for nucleosomal and non-nucleosomal DNA at the site of native replication forks. J Biol Chem. 1981 Nov 25;256(22):11880–11886. [PubMed] [Google Scholar]
- Ball D. J., Gross D. S., Garrard W. T. 5-methylcytosine is localized in nucleosomes that contain histone H1. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5490–5494. doi: 10.1073/pnas.80.18.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum J., Levinger L., Varshavsky A. On the chromatin structure of the amplified, transcriptionally active gene for dihydrofolate reductase in mouse cells. J Biol Chem. 1982 May 10;257(9):5274–5282. [PubMed] [Google Scholar]
- Bonner W. M., Pollard H. B. The presence of F3-F2a1 dimers and F1 oligomers in chromatin. Biochem Biophys Res Commun. 1975 May 5;64(1):282–288. doi: 10.1016/0006-291x(75)90250-8. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Stedman J. D. Histone 1 is proximal to histone 2A and to A24. Proc Natl Acad Sci U S A. 1979 May;76(5):2190–2194. doi: 10.1073/pnas.76.5.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Chalkley R., Hunter C. Histone-histone propinquity by aldehyde fixation of chromatin. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1304–1308. doi: 10.1073/pnas.72.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R. D., Lawson G. M., Hsiang M. W. H1 histone and the condensation of chromatin and DNA. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):253–263. doi: 10.1101/sqb.1978.042.01.027. [DOI] [PubMed] [Google Scholar]
- Gazit B., Cedar H. Nuclease sensitivity of active chromatin. Nucleic Acids Res. 1980 Nov 25;8(22):5143–5155. doi: 10.1093/nar/8.22.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin G. H., Sanders C., Johns E. W. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973 Sep 21;38(1):14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. The multifunctional role of histone H1, probed with the SV40 minichromosome. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):215–226. doi: 10.1101/sqb.1978.042.01.024. [DOI] [PubMed] [Google Scholar]
- Hancock R. Assembly of new nucleosomal histones and new DNA into chromatin. Proc Natl Acad Sci U S A. 1978 May;75(5):2130–2134. doi: 10.1073/pnas.75.5.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. A reevaluation of new histone deposition on replicating chromatin. J Biol Chem. 1981 May 25;256(10):5095–5103. [PubMed] [Google Scholar]
- Jackson V., Chalkley R. Separation of newly synthesized nucleohistone by equilibrium centrifugation in cesium chloride. Biochemistry. 1974 Sep 10;13(19):3952–3956. doi: 10.1021/bi00716a021. [DOI] [PubMed] [Google Scholar]
- Jackson V., Granner D., Chalkley R. Deposition of histone onto the replicating chromosome: newly synthesized histone is not found near the replication fork. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2266–2269. doi: 10.1073/pnas.73.7.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Conservative segregation of nucleosome core histones. Nature. 1984 Jan 5;307(5946):82–85. doi: 10.1038/307082a0. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Decreased protein staining after chemical crosslinking. Anal Biochem. 1983 Nov;135(1):95–101. doi: 10.1016/0003-2697(83)90735-2. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Stability of the conservative mode of nucleosome assembly. Nucleic Acids Res. 1983 May 11;11(9):2717–2732. doi: 10.1093/nar/11.9.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy W B., Wong N. C., Dixon G. H. Selective association of the trout-specific H6 protein with chromatin regions susceptible to DNase I and DNase II: possible location of HMG-T in the spacer region between core nucleosomes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2810–2814. doi: 10.1073/pnas.74.7.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littau V. C., Burdick C. J., Allfrey V. G., Mirsky S. A. The role of histones in the maintenance of chromatin structure. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1204–1212. doi: 10.1073/pnas.54.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Olins D. E., Wright E. B. Glutaraldehyde fixation of isolated eucaryotic nuclei. Evidence for histone-histone proximity. J Cell Biol. 1973 Nov;59(2 Pt 1):304–317. doi: 10.1083/jcb.59.2.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani A., Goodwin G. H., Johns E. W. High mobility group non-histone chromosomal proteins from chicken erythrocytes. Biochem Biophys Res Commun. 1978 Mar 30;81(2):351–358. doi: 10.1016/0006-291x(78)91540-1. [DOI] [PubMed] [Google Scholar]
- Reeves R., Chang D. Investigations of the possible functions for glycosylation in the high mobility group proteins. Evidence for a role in nuclear matrix association. J Biol Chem. 1983 Jan 10;258(1):679–687. [PubMed] [Google Scholar]
- Ring D., Cole R. D. Chemical cross-linking of H1 histone to the nucleosomal histones. J Biol Chem. 1979 Nov 25;254(22):11688–11695. [PubMed] [Google Scholar]
- Ring D., Cole R. D. Close contacts between H1 histone molecules in nuclei. J Biol Chem. 1983 Dec 25;258(24):15361–15364. [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale R. L., Annunziato A. T., Smith R. D. High mobility group proteins: abundance, turnover, and relationship to transcriptionally active chromatin. Biochemistry. 1983 Oct 11;22(21):5008–5015. doi: 10.1021/bi00290a020. [DOI] [PubMed] [Google Scholar]
- Seale R. L. Temporal relationships of chromatin protein synthesis, DNA synthesis, and assembly of deoxyribonucleoprotein. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2270–2274. doi: 10.1073/pnas.73.7.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senshu T., Fukuda M., Ohashi M. Preferential association of newly synthesized H3 and H4 histones with newly replicated DNA. J Biochem. 1978 Oct;84(4):985–988. doi: 10.1093/oxfordjournals.jbchem.a132213. [DOI] [PubMed] [Google Scholar]
- Smith P. A., Jackson V., Chalkley R. Two-stage maturation process for newly replicated chromatin. Biochemistry. 1984 Mar 27;23(7):1576–1581. doi: 10.1021/bi00302a036. [DOI] [PubMed] [Google Scholar]
- Stalder J., Groudine M., Dodgson J. B., Engel J. D., Weintraub H. Hb switching in chickens. Cell. 1980 Apr;19(4):973–980. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Khabaza A. J. Cross-linking of histone H1 in chromatin. Eur J Biochem. 1980 Dec;112(3):501–511. doi: 10.1111/j.1432-1033.1980.tb06113.x. [DOI] [PubMed] [Google Scholar]
- Trempe J., Leffak M. Assembly of semihistone A24. Nucleic Acids Res. 1982 Sep 25;10(18):5467–5481. doi: 10.1093/nar/10.18.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T., Singer M. F. The effect of superhelicity on the interaction of histone f1 with closed circular duplex DNA. J Biol Chem. 1976 Apr 25;251(8):2334–2338. [PubMed] [Google Scholar]
- Weintraub H. Assembly of an active chromatin structure during replication. Nucleic Acids Res. 1979 Oct 10;7(3):781–792. doi: 10.1093/nar/7.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of actively transcribed nucleosomes using immobilized HMG 14 and 17 and an analysis of alpha-globin chromatin. Cell. 1981 Feb;23(2):391–400. doi: 10.1016/0092-8674(81)90134-3. [DOI] [PubMed] [Google Scholar]
- Worcel A., Han S., Wong M. L. Assembly of newly replicated chromatin. Cell. 1978 Nov;15(3):969–977. doi: 10.1016/0092-8674(78)90280-5. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]