Abstract
A random forest method has been selected to perform both gene selection and classification of the microarray data. In this embedded method, the selection of smallest possible sets of genes with lowest error rates is the key factor in achieving highest classification accuracy. Hence, improved gene selection method using random forest has been proposed to obtain the smallest subset of genes as well as biggest subset of genes prior to classification. The option for biggest subset selection is done to assist researchers who intend to use the informative genes for further research. Enhanced random forest gene selection has performed better in terms of selecting the smallest subset as well as biggest subset of informative genes with lowest out of bag error rates through gene selection. Furthermore, the classification performed on the selected subset of genes using random forest has lead to lower prediction error rates compared to existing method and other similar available methods.
Keywords: Random forest, gene selection, classification, microarray data, cancer classification, gene expression data
Background
Through various biological experiments conducted worldwide, large datasets of information has been increasing rapidly and more analysis is conducted each day to sort out the puzzle. Since there are many separate methods available for performing gene selection as well as classification [1], finding similar approach for both, has been of interest to many researchers. Gene selection focuses at identifying a small subset of informative genes from the initial data in order to obtain high predictive accuracy for classification. Gene selection can be considered as a combinatorial search problem and therefore can be suitably handled with optimization methods. Besides that, gene selection plays an important role preceding to tissue classification [2], as only important and related genes are selected for the classification. The main reason to perform gene selection is to identify a small subset of informative genes from the initial data before classification in order to obtain higher prediction accuracy. Many researchers use single variable rankings of the gene relevance and random thresholds to select the number of genes, which can only be applied to two class problems. Random forest can be used for problems arising from more than two classes (multi class) as stated by Díaz-Uriarte R & Alvarez de Andrés (2006) [3]. Classification is carried out to correctly classify the testing samples according to the class. Therefore, performing gene selection antecedent to classification would severely improve the prediction accuracy of the microarray data. Random forest is an ensemble classifier which uses recursive partitioning to generate many trees and then combine the result. Using a bagging technique first proposed by Breiman (1996) [4], each tree is independently constructed using a bootstrap sample of the data. Classification generates gene expression profiles which can discriminate between different known cell types or conditions as described by Lee et al. (2004) [1]. A classification problem is said to be binary in the event when there are only two class labels present [5] and a classification problem is said to be a multiclass classification problem if there are at least three class labels. An enhanced version of gene selection using random forest is proposed to improve the gene selection as well as classification in order to achieve higher prediction accuracy. The proposed idea is to select the smallest subset of genes with the lowest out of bag (OOB) error rates for classification. Besides that, the selection of biggest subset of genes with the lowest OOB error rates is also available to further improve the classification accuracy. Both options are provided as the gene selection technique is designed to suit the clinical or research application and it is not restricted to any particular microarray dataset. Apart from that, the option for setting the minimum number of genes to be selected is added to further improve the functionality of the gene selection method. Therefore, the minimum number of genes required can be set for gene selection process.
Methodology
Few improvements have been made to the existing random forest gene selection, which includes automated dataset input that simplifies the task of loading and processing of the dataset to an appropriate format so that it can be used in this software. Furthermore, the gene selection technique is improved by focusing on smallest subset of genes while taking into account lowest OOB error rates as well as biggest subset of genes with lowest OOB error rates that could increase the prediction accuracy. Besides that, additional functionalities are added to suite different research outcome and clinical application such as the range of the minimum required genes to be selected as a subset. Integration of the different approaches into a single function with parameters as an option allows greater usability while maintaining the computation time required.
Selection of Smallest Subset of Genes with Lowest OOB Error Rates:
The existing method performs gene selection based on random forest to select smallest subset of genes while compromising on the out of bag (OOB) error rates. The subset of genes is usually small but the OOB error rates are not the lowest out of all the possible selection through backward elimination. Therefore, enhancement has been made to improve the prediction accuracy by selecting the smallest subset with the lowest OOB error rates. Hence, lower prediction error rates can be achieved for classification of the samples. This technique is implemented in the random forest gene selection method and shown in Figure 3 (see supplementary material) under supplementary section. During each subset selection based on backward elimination, the mean OOB error rate and standard deviation OOB error rate are tracked at every loop as the less informative genes are removed gradually. Once the loop terminates the subset with the smallest number of variables and lowest OOB error rates are selected for classification. The subset of genes is located based on the last iteration with the smallest OOB error rates. During the backward elimination process, the number of selected variables decreases as the iteration increases.
Selection of Biggest Subset of Genes with Lowest OOB Error Rates:
Another method for improving the prediction error rates is by selecting the biggest subset with the lowest OOB error rates. This is due to the fact that any two or more subsets with different number of selected variables with same lowest error rates indicates that the informative genes level are the same, but the contribution of each genes towards the prediction accuracy is not the same. So, having more informative genes can increase the classification accuracy of the sample. The technique applied for the selection of biggest subset of genes with the lowest OOB error rates are similar to the smallest subset of genes with the lowest OOB error rates, except that the selection is done by picking the first subset with the lowest OOB error rates from all the selected subset which has the lowest error rates. If there is more than one subset with lowest OOB error rates, the selection of the subset is done by selecting the one with highest number of variables for this method. The detailed process flow for this method can be seen in the Figure 4 (see supplementary material). This technique is implemented to assist researches that require filtration of genes for reducing the size of microarray dataset while making sure that the numbers of informative genes are high. This is achieved by eliminating unwanted genes as low as possible while achieving highest accuracy in prediction. Further enhancement is made to the existing random forest gene selection process by adding an extra functionality for specifying the minimum number of genes to be selected in the gene selection process that is included into the classification of the samples. This option allows flexibility of the program to suite the clinical research requirements as well as other application requirement based on the number of genes needed to be considered for classification. The selected minimum values are used during the backward elimination process which takes place in determining the best subset of genes based on out of bag (OOB) error rates.
Performance Measurement:
For gene selection using random forest, backward elimination using OOB error rates is used as the final set of genes is selected based on the lowest out of bag (OOB) error rates as random forest returns a measure of error rate based on the out-of-bag cases for each fitted tree. The classification performance of the microarray data using random forest is measured using .632 bootstrap methods. In this method, the prediction error rates obtained is used to compare the performance of the random forest in classification where lower error rates means higher prediction accuracy. In the .632 bootstrap, accuracy is estimated as followed. Given a dataset of size n, a bootstrap sample is created by sampling n instances uniformly from the data (with replacement). Since the dataset is sampled with replacement, the probability of any given instance not being chosen after n samples is given in the supplementary material.
The expected number of distinct instances from the original dataset appearing in the test set is thus 0.632. The accuracy estimate is derived by using the bootstrap sample for training and the rest of the instances for testing. Given a number b, the number of bootstrap samples, let c0i be the accuracy estimate for bootstrap sample i. The .632 bootstrap estimates are defined as given in the supplementary material. The assessment method used has been able to populate and list the overall performance of the algorithm with other similar algorithms and techniques through prediction error rates calculation comparison.
Results & Discussion
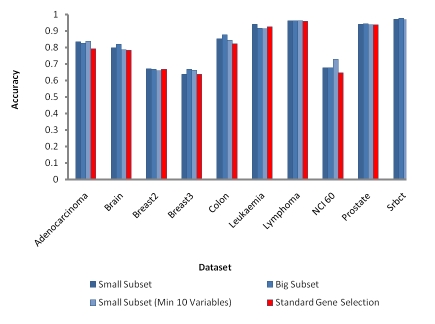
In this section, the full result of all the options used is compared. In Figure 1, the result for each dataset is plotted against the accuracy, therefore the higher the values the lower is the error rates. Based on the Figure 1, the enhanced random forest gene selection performs better compared to standard method. Though, different options have different effects to the datasets being tested. Most of the datasets tested showed larger improvement in terms of accuracy achieved for classification when the subset of genes selected is larger. The detailed information regarding the datasets has been tabulated in Table 1 (see supplementary material). However, some datasets with smaller subset of genes outperformed the larger subset of genes. This could be due to the effect of the informative genes, as more informative genes contribute to better classification accuracy. For the Leukemia dataset, either the selection of biggest subset of genes or limiting the range of the minimum number of genes to be selected in a particular subset has reduced the prediction accuracy. This is due to the fact that low number of informative genes contributes less to the overall classification accuracy. The highest accuracy achieved for this dataset is by selecting smallest subset of genes which has only two genes selected as the subset. Hence, the gene selection options vary according to the dataset used. Based on the three different options presented for the enhanced random forest gene selection, the first option which is selection of smallest subset of genes based on lowest OOB error rates is suitable for Breast 2 and Leukemia dataset as it provided the highest accuracy compared to other options. The second option using selection of biggest subset of genes based on lowest OOB error rates is suitable for Brain, Breast 3, Colon, Lymphoma, Prostate and SRBCT as it manages to achieve highest accuracy for these datasets using this option. Whereas, the third option which performs selection of smallest subset of genes based on lowest OOB error rates with minimum selected genes set to ten is suitable for Adenocarcinoma and NCI60 dataset as the accuracy achieved for these datasets is highest compared to other options. The highest accuracy achieved for Adenocarcinoma dataset is 0.8371, for Brain dataset is 0.8197, Breast 2 dataset is 0.6718, Breast 3 dataset is 0.6682, Colon dataset is 0.8757, Leukemia dataset is 0.9418, Lymphoma dataset is 0.9620, NCI60 dataset is 0.7271, prostate dataset is 0.9446 and SRBCT dataset is 0.9761. The huge improvement achieved in terms of the error rates differences between the standard random forest gene selection method and enhanced random forest gene selection method is from NCI60 dataset, where the differences of the error rates is 0.0801. Further comparison with other available methods such as Diagonal Linear Discriminant Analysis (DLDA), K nearest neighbor (KNN) and Support Vector Machines (SVM) with Linear Kernel has been done as well. The comparison results have been included as supplementary material.
Figure 1.
Comparison between enhanced variables selection with three different options against standard gene selection method. A higher value indicates lower error rates
Conclusion:
The proposed enhanced random forest gene selection has been tested with ten datasets and the outcome is as presented in the results and discussion section. There is an improvement in terms of prediction accuracy for all datasets compared to the standard random forest gene selection. The option for selecting the smallest subset or bigger subset as well as setting the minimum required number of genes is the key factor in achieving higher accuracy in classification. Hence, this enhanced random forest gene selection method provides the flexibility in determining the range of the genes in the subset as to how small or big is the required subset of genes.
Supplementary material
Acknowledgments
We would like to thank Malaysian Ministry of Higher Education for supporting this research by Fundamental Research Grant Scheme (Vot number: 78679).
Footnotes
Citation:Moorthy & Mohamad, Bioinformation 7(3): 142-146 (2011)
References
- 1.JW Lee, et al. Computational Statistics & Data Analysis. 2005;48:869. [Google Scholar]
- 2.YL Chin, S Deris. Jurnal Teknologi. 2005;43:111. [Google Scholar]
- 3.R Díaz-Uriarte, S Alvarez de Andrés. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L Breiman. Machine Learning. 1996;26:123. [Google Scholar]
- 5.T Li, et al. Bioinformatics. 2004;20:2429. [Google Scholar]
- 6.S Ramaswamy, et al. Nat Genet. 2003;33:49. [Google Scholar]
- 7.SL Pomeroy, et al. Nature. 2002;415:436. [Google Scholar]
- 8.LJ van't Veer, et al. Nature. 2002;415:530. [Google Scholar]
- 9.U Alon, et al. Proc Natl Acad Sci U S A. 1999;96:6745. [Google Scholar]
- 10.TR Golub, et al. Science. 1999;286:531. [Google Scholar]
- 11.AA Alizadeh, et al. Nature. 2000;403:503. [Google Scholar]
- 12.DT Ross, et al. Nat Genet. 2000;24:227. [Google Scholar]
- 13.D Singh, et al. Cancer Cell. 2002;1:203. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 14.J Khan, et al. Nat Med. 2001;7:673. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.