Abstract
Ketoacyl synthases (KSs) catalyze condensing reactions combining acyl-CoA or acyl-acyl carrier protein (acyl-ACP) with malonyl-CoA to form 3-ketoacyl-CoA or with malonyl-ACP to form 3-ketoacyl-ACP. In each case, the resulting acyl chain is two carbon atoms longer than before, and CO2 and either CoA or ACP are formed. KSs also join other activated molecules in the polyketide synthesis cycle. Our classification of KSs by their primary and tertiary structures instead of by their substrates and the reactions that they catalyze enhances insights into this enzyme group. KSs fall into five families separated by their characteristic primary structures, each having members with the same catalytic residues, mechanisms, and tertiary structures. KS1 members, overwhelmingly named 3-ketoacyl-ACP synthase III or its variants, are produced predominantly by bacteria. Members of KS2 are mainly produced by plants, and they are usually long-chain fatty acid elongases/condensing enzymes and 3-ketoacyl-CoA synthases. KS3, a very large family, is composed of bacterial and eukaryotic 3-ketoacyl-ACP synthases I and II, often found in multidomain fatty acid and polyketide synthases. Most of the chalcone synthases, stilbene synthases, and naringenin-chalcone synthases in KS4 are from eukaryota. KS5 members are all from eukaryota, most are produced by animals, and they are mainly fatty acid elongases. All families except KS3 are split into subfamilies whose members have statistically significant differences in their primary structures. KS1 through KS4 appear to be part of the same clan. KS sequences, tertiary structures, and family classifications are available on the continuously updated ThYme (Thioester-active enzYme) database.
Keywords: chalcone synthase, fatty acid elongase, ketoacyl synthase, oxoacyl synthase, phylogeny, primary structure, stilbene synthase, tertiary structure, ThYme
Introduction
Ketoacyl synthases (KSs) (more officially 3-oxoacyl synthases and also known as β-ketoacyl synthases) are the condensing enzymes that catalyze the reaction of acyl-coenzyme A (acyl-CoA) or acyl-acyl carrier protein (acyl-ACP) with malonyl-CoA, malonyl-ACP, or occasionally other substrates. This reaction is a key step in the fatty acid synthesis cycle, as in general it adds two carbon atoms to growing acyl chains (Fig. 1). KSs exist as individual enzymes, which are essential components of type II fatty acid and polyketide synthesis; in addition, KS domains are found in large multidomain enzymes such as type I fatty acid synthases (FASs) and polyketide synthases (PKSs).1
Figure 1.
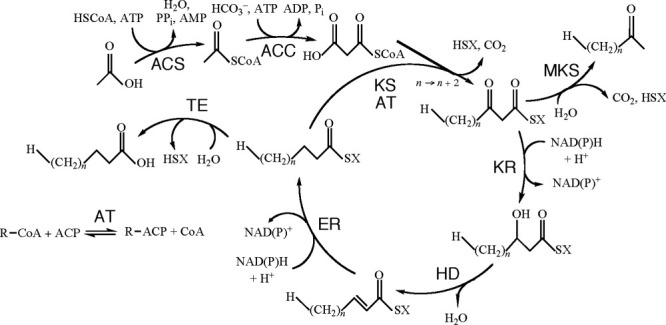
The fatty acid synthesis cycle and the enzyme groups that are part of it. ACC, acetyl-CoA carboxylase; ACS, acyl-CoA synthase; AT, acyl transferase; ER, enoyl reductase; HD, hydroxyacyl dehydratase; KR, ketoacyl reductase; KS, ketoacyl synthase; MKS, methylketone synthase; TE, thioesterase; SX, Coenzyme A or acyl carrier protein. Reprinted with permission from Cantu et al., Nucleic Acids Res, 2011, 39, D342–D346, © Oxford University Press.
We have gathered KS amino acid sequences (primary structures) and three-dimensional (tertiary) structures, along with those of other members of the fatty acid synthesis cycle, into the continually updated ThYme (Thioester-active enzYme) database.2,3 At present, ThYme has 21,028 KS primary and 150 KS tertiary structures. In doing this, we divided each of these enzyme groups into different families based on their primary structural differences. In general, single families contain enzyme members that are related to each other by primary and tertiary structure and mechanism, suggesting that they have a common ancestor. Sometimes members of different families are sufficiently related by primary and tertiary structures and by mechanism that they can be classified as part of a clan, implying that they are descended from a more distant common ancestor. Furthermore, we can divide members of a single family into subfamilies by more subtle primary structural differences.
This article is an account of our division of KSs into families, the gathering of some of the families into clans, and the separation of families into subfamilies. We have done this so that, with the help of known KS crystal structures, mechanisms, and substrate specificities, we could rationally predict the properties of KSs according to the phylogenetic trees that we constructed, and so that we could logically choose KSs to produce and study. Furthermore, we have related properties of KSs with the families in which they are located.
A number of small-scale phylogenetic trees of KSs have already been built. An early tree based on seven tertiary structures showed that ketoacyl-ACP synthase from Synechocystis and Escherichia coli ketoacyl-ACP synthases I and II are very similar, as are Saccharomyces cerevisiae degradative thiolase and Zoogloea ramigera biosynthetic thiolase. More distant from each other and from the other two groups are alfalfa chalcone synthase and E. coli ketoacyl-ACP synthase III.4 A phylogenetic tree of 18 known Arabidopsis ketoacyl-CoA synthases and putative genes has been produced to identify these moieties in putative enzymes.5 A phylogenetic study of 40 β-ketoacyl-ACP synthase III enzymes showed that those produced by bacteria (proteobacteria, firmicutes, and bacteroidetes) and an apicomplexans protist species are widely separated from those produced by monocots, dicots, diatoms, cyanobacteria, and red and green algae.6 Phylogeny of mainly mammalian elongases, but with a few from fungi and other eukaryotes, has been published.7 A detailed tree of protozoal and animal fatty acid elongases as part of a much less detailed tree of elongases from many different phyla has also appeared.8 Lee et al.9 assembled a tree of elongases from protozoal parasites and a few yeasts. Finally, polyunsaturated fatty acid elongases, mainly from marine protists, algae, and diatoms, but including those from a few vertebrates, are found in a phylogenetic tree constructed by Iskandarov et al.10
Family Identification
Basic Local Alignment Search Tool (BLAST)11 and multiple sequence alignment (MSA) were used to classify KSs into different families based on primary structure similarities, while crystal structure superpositions and root-mean-square deviation (RMSD) calculations were used on tertiary structures. More complete descriptions of these methods are found in the Supporting Information of an earlier article.2
The query sequences used for BLAST were KSs with evidence at protein level in the UniProt database,12 ensuring that families are based on sequences with experimental data. Twenty of 187 entries in Enzyme Commission (EC) 2.3.113 are KSs, but four of them have no sequences with evidence at protein level, leaving query sequences to be retrieved from the UniProt database for the remaining 16 EC numbers. Only the KS catalytic domain of each enzyme, obtained from the Pfam database,14 was used. If no Pfam entry appeared, then a hidden Markov model built using HMMER 3.015 was used to find the KS catalytic domain.
BLAST (version 2.2.19) was downloaded and used to populate families with sequences related to the queries in the nonredundant (nr) protein sequence database,16 using an E-value of 0.001. A script automated successive BLAST runs.
MSAs with MUSCLE 3.617 and ClustalX 2.0.1218 using default parameters were conducted on a sample of sequences from each potential family or between different potential families, to determine whether the former should be split or whether the latter should be merged.
The KS catalytic domains of all KS crystal structures in each family, obtained from the RCSB Protein Data Bank,19 were superimposed in MultiProt.20 Then, using MATLAB,21 the RMSDs of the distances between α-carbon atoms of different tertiary structures were calculated.2 Tertiary structures of enzymes in the same family differ in size and number of α-carbon atoms. Therefore Pave values, indicating the average percentage of α-carbon atoms compared, were also recorded. Furthermore, superimposed tertiary structures were visually checked using PyMOL.22
Subfamily Identification
All subfamilies except those in KS3, a very large family (see below), were identified as follows. MSAs of the KS catalytic domains of all sequences in each family were constructed using MUSCLE 3.6. Phylogenetic trees were made using MEGA 4.1 or 5.0.23 They are based on the minimum evolution method24 using complete deletion of sequences, a range of 250–1000 bootstrap iterations, and Jones–Taylor–Thornton (JTT) model values.25 The number of bootstrap iterations was established on a family-by-family basis, the iteration number being reduced in some cases primarily to save computational time while still maintaining an adequate level of rigor in constructing trees of the larger KS subfamilies.
After the phylogenetic tree was complete, subfamilies were chosen based on the visual divergence of one cluster from another, as justified by bootstrap values.24 These manually chosen subfamilies were then subjected to statistical tests to determine each subfamily's z-value26 with respect to another's. The z-value is defined as:
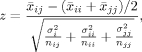 |
where subscripts i and j denote subfamilies i and j,  denotes the JTT distance, σ denotes the variance, and n denotes the number of data points for each
denotes the JTT distance, σ denotes the variance, and n denotes the number of data points for each  point. This z-value determines the likelihood that a certain subfamily is part of another (the higher the z-value, the less likely that two subfamilies overlap). The minimal z-value necessary for every subfamily pairing was 3.33, equivalent to a 0.001 probability of two subfamilies being grouped together in a two-tailed test.
point. This z-value determines the likelihood that a certain subfamily is part of another (the higher the z-value, the less likely that two subfamilies overlap). The minimal z-value necessary for every subfamily pairing was 3.33, equivalent to a 0.001 probability of two subfamilies being grouped together in a two-tailed test.
To refine each subfamily, an MSA of its sequences was made after adding three to five out-group sequences from the subfamily with the highest z-value. Individual subfamily trees were constructed using MEGA 4.1 or 5.0, with tree construction based on the minimum evolution method using pair-wise deletion of sequences, 1000 bootstrap iterations, and JTT distance matrix values. These parameters were used for all KS subfamilies regardless of the bootstrap value used to construct the initial family tree. The subfamily trees were visually inspected to ensure that the out-group sequences appeared as roots. If this were not so, the subfamily was modified by either removing outlier sequences clustered near the out-group sequences or splitting it into two distinct subfamilies. JTT distance values were computed for the refined subfamilies with respect to all other subfamilies. Once this was complete, z-values were then calculated for these refined subfamilies, and they were compared with the z-values before refinement. The procedure was repeated until the required criteria were met.
KS3 has many more sequences (9585 when the tree was produced and 14,098 at present) than the other four KS families (˜3000 or fewer each). MUSCLE 3.6 created an MSA through seven iterations, short of complete convergence, evidently caused by the high number of sequences. This alignment was passed to FastTree27 rather than to MEGA to create a cladogram.
Ketoacyl Synthase Families and Subfamilies
Based on these techniques, KSs are divided into five families (Table I).
Table I.
Ketoacyl Synthase Families and Common Names of Their Members
| Family | Producing organismsa | Dominant EC numbers | Number of subfamilies | Number of sequencesb | Dominant enzyme names |
|---|---|---|---|---|---|
| KS1 | A, B, E | 2.3.1.41, 2.3.1.180 | 12 | 2308 | 3-Ketoacyl-ACP synthase III |
| KS2 | E | 2.3.1.−, 2.3.1.119 | 10 | 359 | 3-Ketoacyl-CoA synthase, fatty acid elongase, very long-chain fatty acid condensing enzyme |
| KS3 | A, B, E | 2.3.1.41, 2.3.1.179 | 14c | 9585 | 3-Ketoacyl-ACP synthase I and II, KS domain of FAS or PKS |
| KS4 | B, E | 2.3.1.74 | 10 | 1191 | Chalcone synthase, stilbene synthase, naringenin-chalcone synthase |
| KS5 | E | — | 11 | 704 | Elongation of very long-chain fatty acid protein, fatty acid elongase |
A, archaea; B, bacteria; E, eukaryota. Most prevalent producers bolded.
At time of producing phylogenetic trees. Number includes outliers.
Manually separated groups.
KS1
Nearly all KS1 members are produced by bacteria, with a few formed by eukaryota and only one from an archaeon.3 The dominant enzyme in this family is 3-ketoacyl-ACP synthase III (KAS III), also called 3-oxoacyl-ACP synthase III and β-ketoacyl-ACP synthase III, which is denoted by EC 2.3.1.180 and whose characteristic reaction is malonyl-ACP + acetyl-CoA → acetoacetyl-ACP + CO2 + CoA (Table II). KAS III enzymes are discrete proteins (not covalently linked to other FAS and PKS enzymes) that catalyze the initial condensation reaction in the type II (dissociated) fatty acid elongation cycle and are known as “loading KSs.” However, 388 sequences named this way are instead labeled EC 2.3.1.41, standing for 3-ketoacyl-ACP synthase I, whose characteristic reaction is malonyl-ACP + acyl-ACP → 3-ketoacyl-ACP + CO2 + ACP, with the product 3-ketoacyl-ACP molecule two carbon atoms longer than the reactant acyl-ACP molecule.
Table II.
Ketoacyl Synthases Commonly Found in ThYme
| EC number | Enzyme name | Catalyzed reaction |
|---|---|---|
| 2.3.1.41 | β-Ketoacyl-ACP synthase I | Malonyl-ACP + acyl-ACP → 3-ketoacyl-ACP + CO2 + ACP |
| 2.3.1.74 | Naringenin-chalcone synthase | 3 Malonyl-CoA + 4-coumaroyl-CoA → naringenin chalcone + 3 CO2 + 4 CoA |
| 2.3.1.119 | Icosanoyl-CoA synthase | Malonyl-CoA + stearoyl-CoA + 2 NAD(P)H + 2 H+ → icosanoyl-CoA+ 2 NAD(P)+ + CO2 + CoA + H2O |
| 2.3.1.179 | β-Ketoacyl-ACP synthase II | Malonyl-ACP + (Z)-hexadec-11-enoyl-ACP → (Z)-3-oxooctadeca-13-enoyl-ACP + CO2 + ACP |
| 2.3.1.180 | β-Ketoacyl-ACP synthase III | Malonyl-ACP + acetyl-CoA → acetoacetyl-ACP + CO2 + CoA |
KS1 is divided into 12 statistically significant subfamilies (Supporting Information Table S1A, with all tables and figures denoted by S found in the Supporting Information) consisting of KSs (overwhelmingly 3-ketoacyl-ACP synthases III) produced only by bacteria, except those in Subfamily 1C, which are produced by cyanobacteria and plants (Table III). Of 2308 aligned KS1 sequences, 128 are outliers. Phylogenetic trees of the 12 subfamilies are found as Supporting Information Figures S1A–S1L.
Table III.
Number of Sequences and Phyla of Producing Species within KS1 Subfamilies
| Subfamily | Number | Dominant phyla |
|---|---|---|
| 1A | 375 | Proteobacteria |
| 1B | 139 | Proteobacteria |
| 1C | 108 | Cyanobacteria, Streptophyta |
| 1D | 112 | Bacteroidetes |
| 1E | 289 | Actinobacteria, Proteobacteria |
| 1F | 286 | Firmicutes, Fusobacteria, Proteobacteria |
| 1G | 41 | Bacteroidetes |
| 1H | 63 | Firmicutes |
| 1I | 143 | Firmicutes, Proteobacteria |
| 1J | 196 | Firmicutes, Proteobacteria |
| 1K | 208 | Bacteroides, Firmicutes, Proteobacteria |
| 1L | 217 | Actinobacteria, Firmicutes, Proteobacteria |
KS1 subfamilies except subfamily 1C have members produced by one to three bacterial phyla (Table III). When members of a single subfamily are produced by bacteria in more than one phylum, some phyla (actinobacteria and firmicutes) contain Gram-positive bacteria and others (bacteroidetes, fusobacteria, and proteobacteria) contain Gram-negative bacteria.
KS2
All KS2 enzymes are from eukaryota, with nearly all from plants. 3-Ketoacyl-CoA synthases, fatty acid elongases, and very long-chain fatty acid condensing enzymes are the most common enzymes in this family. Some are defined as EC 2.3.1.119 (Table II), but the general characterization as EC 2.3.1.– is much more common. Most enzymes in this family catalyze reactions to produce very long-chain fatty acids, whose unbranched chains are longer than 18 carbon atoms.
KS2 can be divided into 10 subfamilies (Table IV, Supporting Information Tables S1B and S2). All but subfamily 2J are composed of enzymes from plants, specifically streptophyta (Table IV); subfamily 2J, on the other hand, has representatives produced by amoeboza and dinoflagellata. All but subfamily 2D have members named as above (although most sequences are undefined); Subfamily 2D has a majority of fiddlehead enzymes.
Table IV.
Number of Sequences and Phyla of Producing Species within KS2 Subfamilies
| Subfamily | Number | Dominant phyla |
|---|---|---|
| 2A | 43 | Streptophyta |
| 2B | 30 | Streptophyta |
| 2C | 45 | Streptophyta |
| 2D | 30 | Streptophyta |
| 2E | 27 | Streptophyta |
| 2F | 22 | Streptophyta |
| 2G | 23 | Streptophyta |
| 2H | 34 | Streptophyta |
| 2I | 48 | Streptophyta |
| 2J | 23 | Amoebozoa, Dinoflagellata |
KS3
KS3 is the largest KS family, containing at present 14,098 sequences. KSs here include the KS domains of large multidomain enzymes such as iterative type I FASs and modular type I PKSs. Many different enzymes are included in this family, but the largest number of members are 3-ketoacyl-ACP synthases I and II (KAS I and KAS II) and undifferentiated PKSs, with EC 2.3.1.41 and EC 2.3.1.179 as the most common EC numbers (Table II). Bacteria produce most KS3 members; eukaryota are substantial producers, and a few KS3 enzymes are of archaeal origin.
Because of the large number of sequences and the program being used, division of the KS3 family into smaller groups had to be carried out by hand from the FastTree cladogram. After controlled sampling, 14 groups were defined, with groups being composed of sequences produced by single or several phyla and usually being composed of a preponderance of enzymes with similar names (Table V and Supporting Information Table S3).
Table V.
Number of Sequences and Phyla of Producing Species within KS3 Groups
| Group | Number | Dominant phyla |
|---|---|---|
| 3A | 1306 | Actinobacteria |
| 3B | 907 | Cyanobacteria, Proteobacteria |
| 3C | 172 | Arthropoda, Chordata |
| 3D | 52 | Amoebozoa |
| 3E | 503 | Ascomycota |
| 3F | 1249 | Actinobacteria, Firmicutes, Proteobacteria |
| 3G | 1376 | Proteobacteria |
| 3H | 93 | Amoebozoa, Arthropoda, Chordata, Echinodermata, Euglenozoa, Placozoa |
| 3I | 73 | Ascomycota |
| 3J | 100 | Streptophyta |
| 3K | 408 | Proteobacteria |
| 3L | 2138 | Actinobacteria, Firmicutes, Proteobacteria, and other bacterial phyla |
| 3M | 453 | Ascomycota |
| 3N | 722 | Actinobacteria, Proteobacteria |
Group 3A members are produced overwhelmingly by actinobacteria, with a large majority named as PKSs (Table V). Group 3B members, on the other hand, have a mixture of names and come from mainly cyanobacteria and proteobacteria. Members of the three smaller groups 3C, 3D, and 3E are composed of a mixture of enzymes and are from animals, protozoa, and fungi, respectively. Group 3F sequences are from several bacterial phyla and have been designated with a number of names. In contrast, enzymes in 3G are overwhelmingly named 3-ketoacyl-ACP synthase I and II and are produced by proteobacteria. Group 3H has enzymes with several names from a number of protozoal and animal phyla. The three small groups 3I, 3J, and 3K contain 3-ketoacyl-ACP synthases from fungi, plants, and proteobacteria, respectively, while Group 3L members are named 3-ketoacyl-ACP synthase II and PKS and come from several bacterial phyla. Finally, Groups 3M and 3N have mainly PKSs from fungi and bacteria, respectively.
KS4
A large fraction of KS4 enzymes are from eukaryota, while the remaining ones are from bacteria. They are classified as chalcone synthases, stilbene synthases, type III PKSs, and naringenin-chalcone synthases, and overwhelmingly those that have EC numbers are listed as EC 2.3.1.74 (Table II).
There are 10 subfamilies in KS4 (Table VI, Supporting Information Tables S1C and S4). Subfamilies 4A–4C are made up of plant (streptophytal) enzymes, with members of 4D being produced by actinobacteria and phaeophyceae (brown algae), 4E coming from ascomycotal fungi, and 4F–4J being enzymes from various bacterial phyla (Table VI). Subfamily 4A has many more sequences than the other subfamilies. All subfamilies except 4B have a wide variety of synthases; 4B, on the other hand, is composed almost exclusively of chalcone synthases.
Table VI.
Number of Sequences and Phyla of Producing Species within KS4 Subfamilies
| Subfamily | Number | Phylum |
|---|---|---|
| 4A | 914 | Streptophyta |
| 4B | 85 | Streptophyta |
| 4C | 39 | Many bacterial phyla |
| 4D | 15 | Actinobacteria, Phaeophyceae |
| 4E | 16 | Ascomycota |
| 4F | 10 | Actinobacteria, Proteobacteria |
| 4G | 30 | Actinobacteria, Proteobacteria |
| 4H | 11 | Bacteroidetes |
| 4I | 27 | Acidobacteria, Actinobacteria, Proteobacteria |
| 4J | 35 | Actinobacteria, Firmicutes, Proteobacteria |
KS5
KS5 members are all from eukaryota, and most are produced by animals. Those that are characterized are almost exclusively fatty acid elongases and elongation of very long-chain (ELOVL) fatty acid proteins. At present, none has an EC number corresponding to an elongase.
KS5 has 11 subfamilies (Table VII, Supporting Information Tables S1D and S5). These subfamilies often have members from several phyla over a wide spectrum (Table VII). Only Subfamily 5B has most of its enzymes with names more specific than the two listed above; that subfamily is populated almost exclusively by fatty acid-CoA elongases. A number of vertebrates produce Subfamily 5C ELOVL1 or ELOVL7 enzymes. Insect and vertebrate ELOVL4 enzymes are found in Subfamily 5F. Subfamily 5G has many vertebrate polyunsaturated fatty acid elongases, some ELOVL5 enzymes, and a few ELOVL2 enzymes. Polysaturated fatty acid elongases from a number of phyla are also found in Subfamily 5H. Some vertebrate ELOVL6 enzymes occur in Subfamily 5K. In mammals, ELOVL1, 3, and 6 catalyze the elongation of saturated and monounsaturated long-chain fatty acids, while ELOVL2, 4, and 5 elongate polyunsaturated long-chain fatty acids.28
Table VII.
Number of Sequences and Phyla of Producing Species within KS5 Subfamilies
| Subfamily | Number | Phylum |
|---|---|---|
| 5A | 110 | Arthropoda |
| 5B | 40 | Arthropoda |
| 5C | 41 | Chordata, Echinodermata, Platyhelminthes |
| 5D | 39 | Arthropoda |
| 5E | 49 | Arthropoda |
| 5F | 46 | Arthropoda, Chordata |
| 5G | 72 | Chordata, Cnidaria |
| 5H | 22 | Several phyla of diatoms, brown algae, green algae, protozoa, and higher plants |
| 5I | 9 | Ascomycota |
| 5J | 106 | Ascomycota, Basidiomycota |
| 5K | 147 | Many phyla of protists, diatoms, dinoflagellates, protozoa, brown algae, and lower and higher animals |
Correspondence with Earlier Ketoacyl Synthase Phylogenetic Trees
Of the seven enzymes arranged in a phylogenetic tree by Moche et al.,4 E. coli ketoacyl-ACP synthases I and II and Synechocystis sp. ketoacyl-ACP synthase II are found in KS3, alfalfa chalcone synthase is in KS4, and E. coli ketoacyl-ACP synthase III is located in KS1. S. cerevisiae degradative thiolase and Z. ramigera biosynthetic thiolase are not KSs, although they also have thiolase-like folds and slight sequence similarity with the KSs in the tree. The relative distances among the different KSs in this work are similar to those found by Moche et al.
The 20 A. thaliana KSs arranged by Blacklock and Jaworski5 all appear to be part of KS2.
All 40 of the 3-ketoacyl-ACP synthase III proteins classified by González-Mellado et al.6 are found in KS1. The enzymes produced by eudicots, monocots, diatoms, and cyanobacteria are all in Subfamily 1C, mostly in the same order as in Supporting Information Figure S1C. The other bacterial enzymes and the one protist enzyme are in various other KS1 subfamilies, as are the four algal proteins.
The mammalian elongases arranged by Leonard et al.7 are found in KS5, Subfamilies 5F, 5G, and 5K, with fungal elongases in Subfamilies 5H and 5J. All protozoal and animal fatty acid elongases in the tree published by Fritzler et al.8 are located in KS5, Subfamily 5K. Also found in KS5, Subfamily 5K, are most of the protozoal parasite elongases arranged by Lee et al.,9 although a few from yeast are in Subfamily 5J. Finally, many protist, algal, and diatom polyunsaturated fatty acid elongases are found in Subfamily 5H, with related vertebrate enzymes in Subfamily 5G.10
KS Specificities
Of the four KS families whose members have assigned EC numbers, KS1 and KS3 members use malonyl-ACP as a chain-elongating agent, whereas KS2 and KS4 enzymes use malonyl-CoA (Tables I and II). KS1, KS2, and KS4 members add these to acyl-CoA moieties, while KS3 members add them to acyl-ACP molecules. The fatty acid elongases in KS5, so far without EC numbers, condense malonyl-CoA with acyl-CoA.28
Although KSs of various types have 20 EC entries, only five comprise the great majority of enzymes gathered by using BLAST with query sequences taken from enzymes with evidence at protein level (Table I). These numbers, assigned by groups working on KSs, are EC 2.3.1.41 (3-ketoacyl-ACP synthase I), EC 2.3.1.74 (naringenin-chalcone synthase), EC 2.3.1.119 (icosanoyl-CoA synthase), EC 2.3.1.179 (3-ketoacyl-ACP synthase II), and EC 2.3.1.180 (3-ketoacyl-ACP synthase III).13 The reactions that they characteristically catalyze are shown in Table II. These factors suggest that KS1 and KS3 contain enzymes that catalyze elongating reactions specific to short (fewer than six carbon atoms) to long (12–20 carbon atoms) acyl chain lengths, whereas enzymes in KS2 and KS5 elongate longer (usually >18 carbon atoms) acyl chains, and KS4 enzymes specifically produce chalcones and related molecules. Moreover, KS2 enzymes are produced almost exclusively by plants and KS5 enzymes come mainly from animals.
Ketoacyl Synthase Crystal Structures
All known tertiary structures of members of KS1, KS3, and KS4 have thiolase-like folds (Fig. 2), with five layers of α-β-α-β-α structure.29 KS2 and KS5 presently have no crystal structures.3 KS1 has 38 crystal structures, with an RMSDave values obtained by superposition of these structures of 1.22 Å and a Pave value of 82.7%. The corresponding values for KS3 are 71 structures, 1.42 Å, and 67.5%, whereas those for KS4 are 41 structures, 1.18 Å, and 93.2%.
Figure 2.
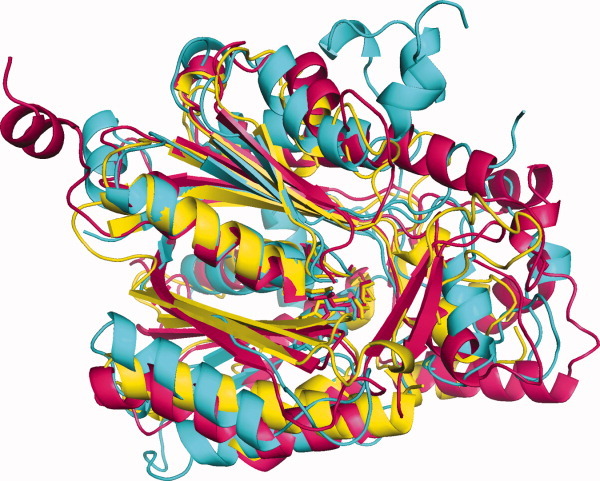
Superimposed KS crystal structures. KS1 (yellow): 1EBL from Escherichia coli β-ketoacyl-ACP synthase III; KS3 (cyan): 2QO3 from Saccharopolyspora erythraea DEBS 2; KS4 (pink): 1Z1E from Arachis hypogaea stilbene synthase.
Crystal structures from KS1, KS3, and KS4, one from each family, were superimposed (Fig. 2). The RMSD of the superimposed structures is 1.96 Å, with a Pave of 68.5%.
Ketoacyl Synthase Catalytic Residues and Mechanisms
Based on crystal structures and consistent with previous results with thioesterases,2 catalytic residues are well conserved within KS1, KS3, and KS4 (Table VIII). This leads us to assume that all members of a family have the same ping-pong kinetic mechanism,30 using cysteine, histidine, and either histidine or asparagine as a catalytic triad.
Table VIII.
Catalytic Residues of Ketoacyl Synthase Families
| Family | Producing organism | Gene | Catalytic residuesa | PDB file |
|---|---|---|---|---|
| KS1 | Escherichia coli | fabH | Cys112, His244, Asn274 | 1EBL, 1HND, 1HNH, 1HNJ, 1HNK |
| KS1 | Pseudomonas aeruginosa | pqsD | Cys112, His257, Asn287 | 3H76, 3H77, 3H78 |
| KS1 | Staphylococcus aureus | fabH | Cys112, His238, Asn268 | 1ZOW |
| KS2 | Arabidopsis sp. | FAE1 | Cys223, His391, Asn424 | — |
| KS3 | E. coli | fabB | Cys163, His298, His333 | 1FJ4, 1FJ8 |
| KS3 | Homo sapiens | OXSM | Cys209, His348, His385 | 2IWY, 2IWZ |
| KS3 | Mycobacterium tuberculosis | kasB | Cys170, His311, His346 | 2GP6 |
| KS3 | M. tuberculosis | kasA | Cys171, His311, His345 | 2WGD, 2WGE, 2WGF, 2WGG |
| KS3 | Saccharomyces cerevisiae | FAS2 | Cys1305, His1542, His1583 | 2PFF, 2UV8 |
| KS3 | Saccharopolyspora erythraea | eryA | Cys199 | 2HG4 |
| KS3 | S. erythraea | eryA | Cys202, His337, His377 | 2QO3 |
| KS3 | Streptococcus pneumoniae | fabF | Cys164, His303, His337 | 1OX0, 1OXH, 2ALM |
| KS3 | Thermus thermophilus | fabF | Cys161, His301, His338 | 1J3N |
| KS4 | Aloe arborescens | — | Cys174, His316, Asn349 | 2D3M, 2D51, 2D52 |
| KS4 | Arachis hypogaea | — | Cys164, His303, Asn338 | 1Z1E, 1Z1F |
| KS4 | Medicago sativa | CHS | Cys164, His303, Asn336 | 1JWX |
| KS4 | M. tuberculosis | — | Cys175, His313, Asn346 | 1TED, 1TEE |
| KS4 | Neurospora crassa | — | Cys152, His305, Asn338 | 3E1H, 3EUO, 3EUQ, 3EUT |
| KS4 | Pinus sylvestris | — | Cys164, His303, Asn336 | 1U0U |
| KS4 | Rheum palmatum | Bas | Cys157, His296, Asn329 | 3A5Q, 3A5R, 3A5S |
| KS4 | Streptomyces coelicolor | — | Cys138, His270, Asn305 | 1U0M |
With one exception, catalytic residues were gathered from literature associated with the listed PDB structures. Those for KS2 came from a mutagenesis study.
Cysteine, histidine, and asparagine form the catalytic traid in KS1. Qiu et al.31 proposed that Cys112 in E. coli β-ketoacyl-ACP synthase III (PDB entry 1HN9) donates a proton to His244 and attacks acetyl-CoA. Then, malonyl-ACP is attached to His244 and Asn274 to be decarboxylated, forming a carbanion. Finally, the carbanion attacks the acetyl moiety to form acetoacetyl-ACP.
In KS2, mutagenetic analysis of Arabidopsis FAE1 β-ketoacyl-CoA synthase strongly suggested that it shares the same ping-pong mechanism and putative Cys-His-Asn/His catalytic residues as members of KS1, KS3, and KS4, but in this case joining malonyl-CoA with a long-chain acyl-CoA.32 Although no crystal structure is yet available to provide confirmation, it appears that the catalytic residues are Cys223, His391, and Asn424, congruent with the identity and spacing of the catalytic residues in the other three families (Table VIII). No NADPH was necessary for wild-type enzyme activity, indicating that EC 2.3.1.119 is an incorrect designation for this enzyme family (Tables I and II).
The KS3 active site has a Cys-His-His triad (Table VIII). In Streptococcus pneumoniae KASII (2ALM), acyl-ACP transfers its acyl moiety to Cys164. It is proposed that a water molecule activated by His303 then attacks malonyl-ACP to form a carbanion. His337 also stabilizes the malonyl moiety. Last, the carbanion attacks the acyl moiety and forms β-ketoacyl-ACP.33
KS4 members have a Cys-His-Asn catalytic triad, the same as KS1 members (Table VIII). The chalcone synthase/stilbene synthase superfamily catalyzes the same acyl transfer, decarboxylation, and condensation steps as KS1, plus further cyclization and aromatization reactions before it forms the final chalcone product.34
Little is yet known about the catalytic mechanism of KS5 enzymes. It appears that no catalytic amino acid residues have yet been identified. MSAs have identified conserved histidine and asparagine residues, the former however in a membrane-spanning region,7 but no conserved cysteine residues (Supporting Information Fig. S2).
Ketoacyl Synthase Clans
Although amino acid sequences of members of different families may completely or almost completely differ from each other, if their crystal structures, catalytic residues, and mechanisms are conserved, they could be part of the same clan, maybe having a distant common ancestor.
KS1, KS2, and KS4 have some members whose sequences are similar. In addition, the tertiary structures of members of KS1, KS3, and KS4 may be superimposed (Fig. 2), with their presumed catalytic residues in the same positions (Fig. 3). Furthermore, their secondary structure elements are found in the same order, although with some gaps (Table IX).
Figure 3.
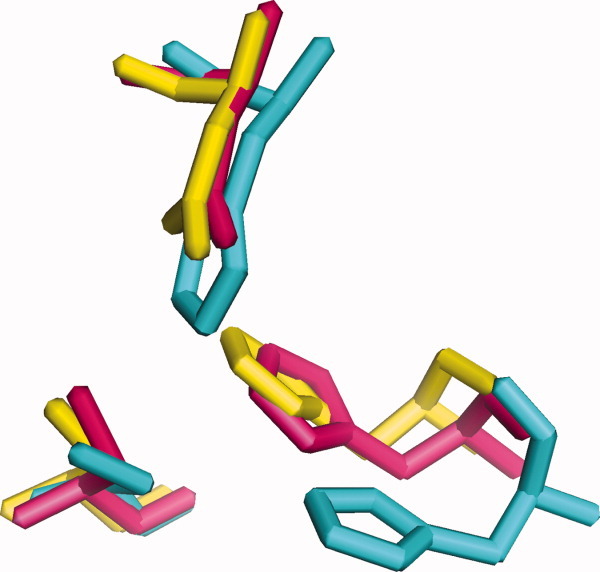
Superimposed KS active sites. KS1 (yellow): 1EBL from Escherichia coli β-ketoacyl-ACP synthase III; KS3 (cyan): 2QO3 from Saccharopolyspora erythraea DEBS 2; KS4 (pink): 1Z1E from Arachis hypogaea stilbene synthase. Bottom left: cysteine; bottom right: histidine; top: asparagine or histidine.
Table IX.
Secondary Structure Elements of Ketoacyl Synthase Families
| Family | Secondary structural elementa |
|---|---|
| KS1 | − β β α α β α α β α β − α β α β − β − − α β α α − β β |
| KS3 | − β − α − − α α β α α α α β α β α β − − α β α α α β β |
| KS4 | α β β α α β α α β α β − α β α β − β β β α β α α α β β |
α, α-helix; β, β-strand.
KS1, KS2, KS3, and KS4 members have similar catalytic triads, indicating that they catalyze essentially the same basic reaction by the same or a similar mechanism. Thus, these four families fall into one clan. There is no indication at present that KS5 enzymes are part of this clan; more specifically, the fatty acid elongases in KS2, nearly all from higher plants, and those in KS5, mainly from animals and almost none from plants, appear not to be related.
Conclusions
The over 20,000 primary structures of the KSs have been sorted into five families, separated by different amino acid sequences and by the characteristic reactions that they catalyze. Four of the families (KS1–KS4) appear to be part of one clan because of their slight similarities of primary structure and strong similarities of secondary structure element orders, tertiary structures, placement and identity of catalytic residues, and implied mechanisms. Four of the families (KS1, KS2, KS4, and KS5) have been further split into 10–12 subfamilies each by their statistically significant differences in primary structure. Sequences of the fifth family (KS3) have been separated manually into 14 groups based on the organisms that produce them and sometimes by the reactions that they catalyze. This information should be useful to researchers in choosing specific KSs to study further.
Acknowledgments
The National Science Foundation Engineering Research Center for Biorenewable Chemicals is headquartered at Iowa State University and includes Rice University, the University of California, Irvine, the University of New Mexico, the University of Virginia, and the University of Wisconsin-Madison. R.P.M. from the University of Michigan was part of a Research Experiences for Undergraduates program associated with this Engineering Research Center. The authors thank Professor Derrick Rollins (Iowa State University) for providing the z-value equation. They also thank Professor Basil Nikolau and the members of his research group for helpful advice.
Supplementary material
References
- 1.Smith S, Tsai S. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cantu DC, Chen Y, Reilly PJ. Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantu DC, Chen Y, Lemons ML, Reilly PJ. ThYme: a database for thioester-active enzymes. Nucleic Acids Res. 2011;39:D342–D346. doi: 10.1093/nar/gkq1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moche M, Dehesh K, Edwards P, Lindqvist Y. The crystal structure of β-ketoacyl-acyl carrier protein synthase II from Synechocystis sp. at 1.54 Å resolution and its relationship to other condensing enzymes. J Mol Biol. 2001;305:491–503. doi: 10.1006/jmbi.2000.4272. [DOI] [PubMed] [Google Scholar]
- 5.Blacklock BJ, Jaworski JG. Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem Biophys Res Commun. 2006;346:583–590. doi: 10.1016/j.bbrc.2006.05.162. [DOI] [PubMed] [Google Scholar]
- 6.González-Mellado D, Wettstein-Knowles P, Garcés R, Martínez-Force E. The role of β-ketoacyl-acyl carrier protein synthase III in the condensation steps of fatty acid biosynthesis in sunflower. Planta. 2010;231:1277–1289. doi: 10.1007/s00425-010-1131-z. [DOI] [PubMed] [Google Scholar]
- 7.Leonard AE, Pereira SL, Sprecher H, Huang YS. Elongation of long-chain fatty acids. Prog Lipid Res. 2004;43:36–54. doi: 10.1016/s0163-7827(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 8.Fritzler JM, Millership JJ, Zhu G. Cryptosporidium parvum fatty acid elongase. Eukaryot Cell. 2007;6:2018–2028. doi: 10.1128/EC.00210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SH, Stephens JL, Englund PT. A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol. 2007;5:287–297. doi: 10.1038/nrmicro1617. [DOI] [PubMed] [Google Scholar]
- 10.Iskandarov U, Khozin-Goldberg I, Ofir R, Cohen Z. Cloning and characterization of the Δ6 polyunsaturated fatty acid elongase from the green microalga Parietochloris incisa. Lipids. 2009;44:545–554. doi: 10.1007/s11745-009-3301-y. [DOI] [PubMed] [Google Scholar]
- 11.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–2402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC–IUBMB) Enzyme nomenclature. San Diego, CA: Academic Press; 1992. [Google Scholar]
- 14.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 16.National Center for Biotechnology Information (NCBI) 2011. RefSeq. Available at: http://www.ncbi.nlm.nih.gov/RefSeq/
- 17.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 19.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlić A, Quesada M, Quinn GB, Westbrook JD, Young J, Yukich B, Zardecki C, Berman HM, Bourne PE. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 21.The MathWorks, Inc. 2011. Available at: http://www.mathworks.com/matlabcentral/
- 22.Schrödinger, LLC. 2011. The PyMOL molecular graphics system, version 1.3. Available at: http://www.pymol.org/
- 23.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 24.Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 25.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 26.Mertz B, Kuczenski RS, Larsen RT, Hill AD, Reilly PJ. Phylogenetic analysis of family 6 glycoside hydrolases. Biopolymers. 2005;79:197–206. doi: 10.1002/bip.20347. [DOI] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Huang W, Jia J, Edward P, Dehesh K, Schneider G, Lindqvist Y. Crystal structure of β-ketoacyl-acyl carrier protein synthase II from E. coli reveals the molecular architecture of condensing enzymes. EMBO J. 1998;17:1183–1191. doi: 10.1093/emboj/17.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plowman KM. Enzyme kinetics. New York: McGraw-Hill; 1972. pp. 41–42. [Google Scholar]
- 31.Qiu X, Janson CA, Konstantinidis AK, Nwagwu S, Silverman C, Smith WW, Khandekar S, Lonsdale J, Abdel-Meguid SS. Crystal structure of β-ketoacyl-acyl carrier protein synthase. III. A key condensing enzyme in bacterial fatty acid biosynthesis. J Biol Chem. 1999;274:36465–36471. doi: 10.1074/jbc.274.51.36465. [DOI] [PubMed] [Google Scholar]
- 32.Ghanevati M, Jaworski JG. Engineering and mechanistic studies of the Arabidopsis FAE1 β-ketoacyl-CoA synthase, FAE1 KCS. Eur J Biochem. 2002;269:3531–3539. doi: 10.1046/j.1432-1033.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Hulbert J, White SW, Rock CO. Roles of the active site water, histidine 303, and phenylalanine 396 in the catalytic mechanism of the elongation condensing enzyme of Streptococcus pneumoniae. J Biol Chem. 2006;281:17390–17399. doi: 10.1074/jbc.M513199200. [DOI] [PubMed] [Google Scholar]
- 34.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


