Abstract
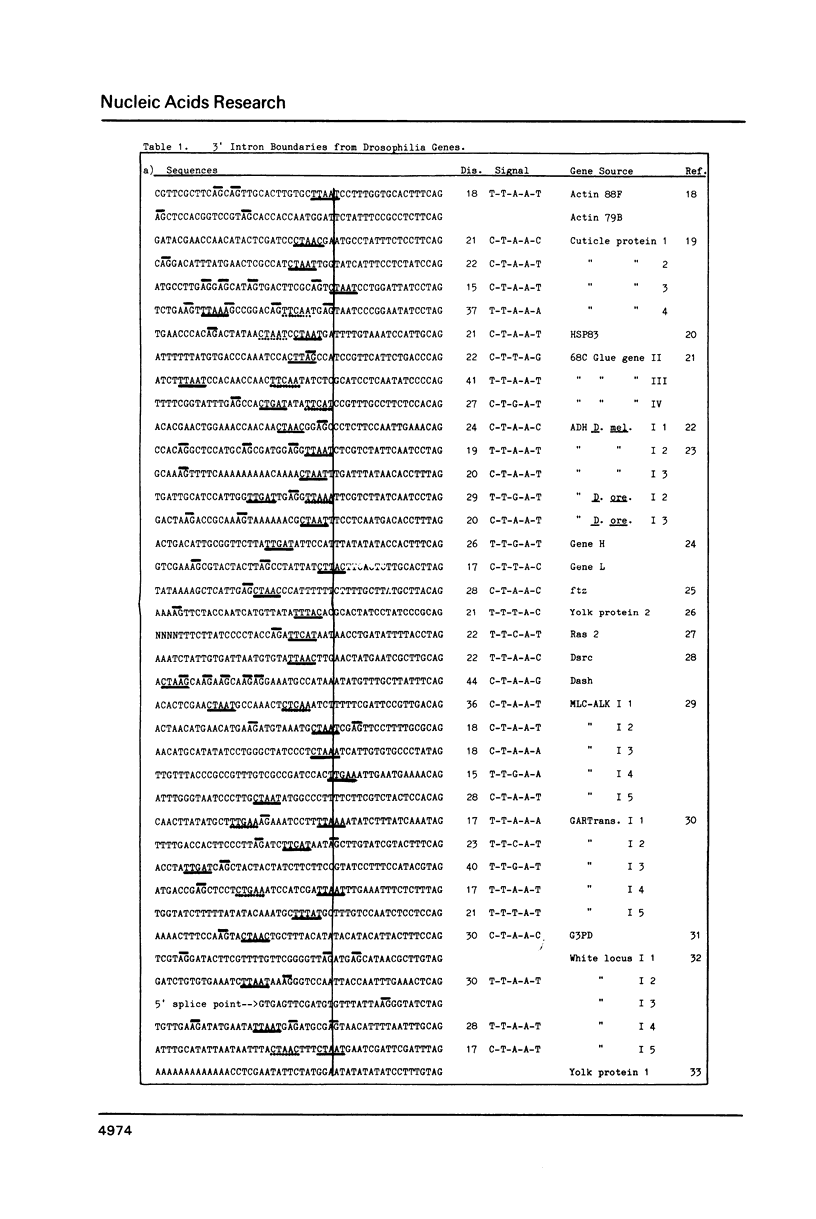
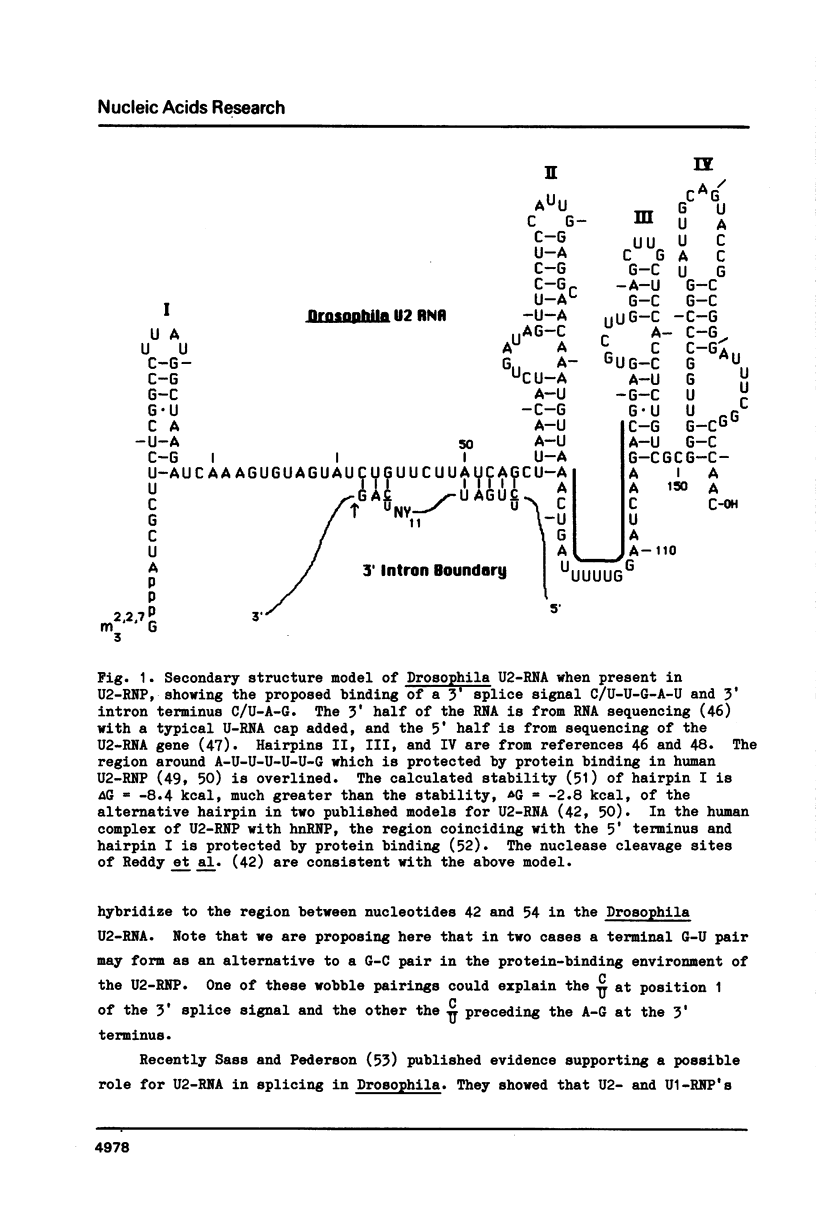
The introns of Drosophila pre-mRNAs have been analysed for conserved internal sequence elements near the 3' intron boundary similar to the T-A-C-T-A-A-C in yeast introns and the C/T-T-A/G-A-C/T in introns of other organisms. Such conserved internal elements are the 3' splice signals recognized in intron splicing. In the lariat splicing mechanism, the G at the 5' end of an intron joins covalently to the last A of a 3' splice signal to form a branch point in a splicing intermediate. Analysis of 39 published sequences of Drosophila introns reveals that potential 3' splice signals with the consensus C/T-T-A/G-A-C/T are present in 18 cases. In 17 of the remaining cases signals are present which vary from this consensus just in the middle or last position. In Drosophila introns the 3' splice signal is usually located in a discrete region between 18 and 35 nucleotides upstream from the 3' splice point. We note that the Drosophila small nuclear U2-RNA has sequences complementary to C-T-G-A-T, one variant of the signal, and to C-A-G, one variant of the 3' terminus of an intron. We also note that the absence of any A-G between -3 and -19 from the 3' splice point may be an essential feature of a strong 3' boundary.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso A., Jorcano J. L., Beck E., Spiess E. Isolation and characterization of Drosophila melanogaster U2 small nuclear RNA genes. J Mol Biol. 1983 Sep 25;169(3):691–705. doi: 10.1016/s0022-2836(83)80165-x. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Spoerel N., Haymerle H., Ashburner M. The messenger RNA for alcohol dehydrogenase in Drosophila melanogaster differs in its 5' end in different developmental stages. Cell. 1983 May;33(1):125–133. doi: 10.1016/0092-8674(83)90341-0. [DOI] [PubMed] [Google Scholar]
- Bodmer M., Ashburner M. Conservation and change in the DNA sequences coding for alcohol dehydrogenase in sibling species of Drosophila. 1984 May 31-Jun 6Nature. 309(5967):425–430. doi: 10.1038/309425a0. [DOI] [PubMed] [Google Scholar]
- Boel E., Hansen M. T., Hjort I., Høegh I., Fiil N. P. Two different types of intervening sequences in the glucoamylase gene from Aspergillus niger. EMBO J. 1984 Jul;3(7):1581–1585. doi: 10.1002/j.1460-2075.1984.tb02014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Ebel J. P., Lazar E., Haendler B., Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1(10):1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Calvet J. P., Meyer L. M., Pederson T. Small nuclear RNA U2 is base-paired to heterogeneous nuclear RNA. Science. 1982 Jul 30;217(4558):456–458. doi: 10.1126/science.6178162. [DOI] [PubMed] [Google Scholar]
- Calvet J. P., Pederson T. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell. 1981 Nov;26(3 Pt 1):363–370. doi: 10.1016/0092-8674(81)90205-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Tanner N. K., Tinoco I., Jr, Weir B. R., Zuker M., Perlman P. S. Secondary structure of the Tetrahymena ribosomal RNA intervening sequence: structural homology with fungal mitochondrial intervening sequences. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3903–3907. doi: 10.1073/pnas.80.13.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Dons J. J., Mulder G. H., Rouwendal G. J., Springer J., Bremer W., Wessels J. G. Sequence analysis of a split gene involved in fruiting from the fungus Schizophyllum commune. EMBO J. 1984 Sep;3(9):2101–2106. doi: 10.1002/j.1460-2075.1984.tb02097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenthal S., Parker V. P., Davidson N. Developmental variations in the splicing pattern of transcripts from the Drosophila gene encoding myosin alkali light chain result in different carboxyl-terminal amino acid sequences. Proc Natl Acad Sci U S A. 1985 Jan;82(2):449–453. doi: 10.1073/pnas.82.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel M. D., Pruitt R. E., Meyerowitz E. M. DNA sequences, gene regulation and modular protein evolution in the Drosophila 68C glue gene cluster. J Mol Biol. 1983 Aug 25;168(4):765–789. doi: 10.1016/s0022-2836(83)80074-6. [DOI] [PubMed] [Google Scholar]
- Hackett R. W., Lis J. T. Localization of the hsp83 transcript within a 3292 nucleotide sequence from the 63B heat shock locus of D. melanogaster. Nucleic Acids Res. 1983 Oct 25;11(20):7011–7030. doi: 10.1093/nar/11.20.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Sloan J. S., Kelly J. D. A Drosophila metabolic gene transcript is alternatively processed. Cell. 1983 Sep;34(2):405–414. doi: 10.1016/0092-8674(83)90374-4. [DOI] [PubMed] [Google Scholar]
- Hoffmann F. M., Fresco L. D., Hoffman-Falk H., Shilo B. Z. Nucleotide sequences of the Drosophila src and abl homologs: conservation and variability in the src family oncogenes. Cell. 1983 Dec;35(2 Pt 1):393–401. doi: 10.1016/0092-8674(83)90172-1. [DOI] [PubMed] [Google Scholar]
- Hovemann B., Galler R., Walldorf U., Küpper H., Bautz E. K. Vitellogenin in Drosophila melanogaster: sequence of the yolk protein I gene and its flanking regions. Nucleic Acids Res. 1981 Sep 25;9(18):4721–4734. doi: 10.1093/nar/9.18.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M. C., Wensink P. C. Sequence and structure conservation in yolk proteins and their genes. J Mol Biol. 1983 Mar 15;164(4):481–492. doi: 10.1016/0022-2836(83)90046-3. [DOI] [PubMed] [Google Scholar]
- Karn J., Brenner S., Barnett L. Protein structural domains in the Caenorhabditis elegans unc-54 myosin heavy chain gene are not separated by introns. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4253–4257. doi: 10.1073/pnas.80.14.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird J. H., Fincham J. R. The complete nucleotide sequence of the Neurospora crassa am (NADP-specific glutamate dehydrogenase) gene. Gene. 1983 Dec;26(2-3):253–260. doi: 10.1016/0378-1119(83)90195-6. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Grabowski P. J., Padgett R. A., Sharp P. A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985 Feb 14;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Kramer J. M., Cox G. N., Hirsh D. Comparisons of the complete sequences of two collagen genes from Caenorhabditis elegans. Cell. 1982 Sep;30(2):599–606. doi: 10.1016/0092-8674(82)90256-2. [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Liautard J. P., Sri-Widada J., Brunel C., Jeanteur P. Structural organization of ribonucleoproteins containing small nuclear RNAs from HeLa cells. Proteins interact closely with a similar structural domain of U1, U2, U4 and U5 small nuclear RNAs. J Mol Biol. 1982 Dec 15;162(3):623–643. doi: 10.1016/0022-2836(82)90392-8. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., De Robertis E. M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985 Jan;40(1):111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., Zeller R. Xenopus laevis U2 snRNA genes: tandemly repeated transcription units sharing 5' and 3' flanking homology with other RNA polymerase II transcribed genes. EMBO J. 1983;2(11):1883–1891. doi: 10.1002/j.1460-2075.1983.tb01675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H., Sundaralingam M. Stacking of Crick Wobble pair and Watson-Crick pair: stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon-anticodon Wobble interaction. Nucleic Acids Res. 1978 Nov;5(11):4451–4461. doi: 10.1093/nar/5.11.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myslinski E., Branlant C., Wieben E. D., Pederson T. The small nuclear RNAs of Drosophila. J Mol Biol. 1984 Dec 25;180(4):927–945. doi: 10.1016/0022-2836(84)90264-x. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F. S., Schejter E., Hoffmann F. M., Shilo B. Z. The Drosophila ras oncogenes: structure and nucleotide sequence. Cell. 1984 Jul;37(3):1027–1033. doi: 10.1016/0092-8674(84)90437-9. [DOI] [PubMed] [Google Scholar]
- Nojima H., Kornberg R. D. Genes and pseudogenes for mouse U1 and U2 small nuclear RNAs. J Biol Chem. 1983 Jul 10;258(13):8151–8155. [PubMed] [Google Scholar]
- O'Hare K., Murphy C., Levis R., Rubin G. M. DNA sequence of the white locus of Drosophila melanogaster. J Mol Biol. 1984 Dec 15;180(3):437–455. doi: 10.1016/0022-2836(84)90021-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Mount S. M., Steitz J. A., Sharp P. A. Splicing of messenger RNA precursors is inhibited by antisera to small nuclear ribonucleoprotein. Cell. 1983 Nov;35(1):101–107. doi: 10.1016/0092-8674(83)90212-x. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Reddy R., Busch H. Small nuclear RNAs and RNA processing. Prog Nucleic Acid Res Mol Biol. 1983;30:127–162. doi: 10.1016/s0079-6603(08)60685-6. [DOI] [PubMed] [Google Scholar]
- Reddy R., Henning D., Epstein P., Busch H. Primary and secondary structure of U2 snRNA. Nucleic Acids Res. 1981 Nov 11;9(21):5645–5658. doi: 10.1093/nar/9.21.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. R., Pikielny C. W., Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984 Dec;39(3 Pt 2):603–610. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sanchez F., Tobin S. L., Rdest U., Zulauf E., McCarthy B. J. Two Drosophila actin genes in detail. Gene structure, protein structure and transcription during development. J Mol Biol. 1983 Feb 5;163(4):533–551. doi: 10.1016/0022-2836(83)90111-0. [DOI] [PubMed] [Google Scholar]
- Sass H., Pederson T. Transcription-dependent localization of U1 and U2 small nuclear ribonucleoproteins at major sites of gene activity in polytene chromosomes. J Mol Biol. 1984 Dec 25;180(4):911–926. doi: 10.1016/0022-2836(84)90263-8. [DOI] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. BKV splice sequences based on analysis of preferred donor and acceptor sites. Nucleic Acids Res. 1979 Jul 25;6(10):3387–3398. doi: 10.1093/nar/6.10.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Davidson N. Two gene families clustered in a small region of the Drosophila genome. J Mol Biol. 1983 May 15;166(2):101–118. doi: 10.1016/s0022-2836(83)80001-1. [DOI] [PubMed] [Google Scholar]
- Snyder M., Hunkapiller M., Yuen D., Silvert D., Fristrom J., Davidson N. Cuticle protein genes of Drosophila: structure, organization and evolution of four clustered genes. Cell. 1982 Jul;29(3):1027–1040. doi: 10.1016/0092-8674(82)90466-4. [DOI] [PubMed] [Google Scholar]
- Sri-Widada J., Liautard J. P., Brunel C., Jeanteur P. Interaction of snRNAs with rapidly sedimenting nuclear sub-structures (hnRNPs) from HeLa cells. Nucleic Acids Res. 1983 Oct 11;11(19):6631–6646. doi: 10.1093/nar/11.19.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell S. W., Weiner A. M. Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol. 1984 Mar;4(3):492–499. doi: 10.1128/mcb.4.3.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G., Zabielski J., Hammarström K., Monstein H. J., Bark C., Pettersson U. Clustered genes for human U2 RNA. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3811–3815. doi: 10.1073/pnas.81.12.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Woudt L. P., Pastink A., Kempers-Veenstra A. E., Jansen A. E., Mager W. H., Planta R. J. The genes coding for histone H3 and H4 in Neurospora crassa are unique and contain intervening sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5347–5360. doi: 10.1093/nar/11.16.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]



