Abstract
Purpose
To investigate the frequency of xenotropic murine leukemia virus (MLV) presence in human cell lines established from mouse xenografts and to search for the evidence of horizontal viral spread to other cell lines.
Results
Six of 23 (26%) mouse DNA free xenograft cultures were strongly positive for MLV and their sequences had greater than 99% homology to known MLV strains. Four of five available supernatant fluids from these viral positive cultures were strongly positive for RT activity. Three of these supernatant fluids were studied to confirm the infectivity of the released virions for other human culture cells. Of the 78 non-xenograft derived cell lines maintained in the xenograft culture-containing facilities, 13 (17%) were positive for MLV, including XMRV, a virus strain first identified in human tissues. By contrast, all 50 cultures maintained in a xenograft culture-free facility were negative for viral sequences.
Methodology
We examined xenograft tumor cell lines from seven independent laboratories and 128 non-xenografted tumor cell lines. Cell line DNA was examined for mouse DNA contamination, and by 3 Taqman qPCR assays targeting the gag, env or pol regions of MLV. Sequencing was used for viral strain identification. Supernatant fluids were tested for reverse transcriptase (RT) activity.
Conclusions
Human cultures derived after mouse xenografting frequently contain and release highly infectious xenotropic MLV viruses. Laboratories working with xenograft-derived human cultures should be aware of the risk of contamination with potentially biohazardous human-tropic mouse viruses and their horizontal spread to other cultures.
Key words: xenograft cultures, xenotropic murine leukemia virus, retrovirus, XMRV virus, cell cultures, lung cancer, prostate cell line
Introduction
Retroviruses are enveloped viruses possessing an RNA genome and replicate via a DNA intermediate. Retroviruses rely on the enzyme reverse transcriptase to perform reverse transcription of its genome from RNA into DNA, which can then be integrated into the host cell's genome with an integrase enzyme. Retroviral DNA can remain in a latent form in the genome (provirus) or its RNA can be expressed intermittently as infectious virions.1,2 Examination of the mouse genome provides evidence that a substantial proportion of the genome is comprised of transposable elements (37.5%).3 Most (> 90%) of these transposable elements are retroelements that require reverse transcriptase to reverse transcribe RNA into DNA prior to integration into the genome. Endogenous retroviral sequences (ERV) belong to long-terminal repeats (LTRs) containing retroelements which comprise about 10% of the mouse genome.3 ERVs represent remnants of ancestral germline infection by exogenous retrovirues and after integration into the genome are transmitted vertically as proviruses. Murine leukemia viruses (MLV) as ERV provirus forms are present at about 60 copies per mouse genome3 from which up to 15 copies are related to infectious xenotropic murine leukemia viruses (XMLV).4 The XMLV are type C retroviruses from mouse which can infect human or other foreign species.5–7 The XMLV virus related to mouse endogenous provirus in mice can be readily isolated or induced after chemical or immunological stimulation. For example, Bxv1 (also termed Xmv43) provirus is found in several common inbred mice can be reliably induced to produce virus and was found to contribute to production of pathogenic recombinant leukemogenic polytropic MLVs.4,8,9 Thus, active mouse ERV provirus present in common inbred mouse tissues can be the origin of XMLV or recombinant poly-tropic MLVs which are infectious to human tissues implanted in laboratory mice.
Earlier studies have documented that XMLV type-C retrovirus particles were indentified in human xenograft cultures derived after xenografting in immune-compromised mice.10–13 For example, XMLV was found in a small cell lung cancer (SCLC) line out of 14 xenograft derived cell lines from varied tumor types.13 NCI-N417 SCLC cell line was established from a mouse xenograft by the Gazdar lab at the National Cancer Institute (NCI) in the early 1980s and this cell line was subsequently found to contain XMLV a few years later.14,15 Accordingly further distribution of NCI-N417 was stopped and it was no longer maintained in the lab. In the early 1990s, several human hematologic tumor cell lines were found to acquire XMLV after passage through nude mice.16 Recently, human prostate cancer xenograft cell line 22Rv1 which was established after xenografting in athymic nude mice, was found to contain the XMLV-related (XMRV) virus.17,18 XMRV, originally identified in human tissues, is a retrovirus closely related or derived from XMLV19 and believed to derive from the recombination of PreXMRV-1 and PreXMRV-2 proviruses present in mice.20 These findings clearly demonstrated that xenograft cell lines, currently widely used as experimental models, can potentially acquire XMLV including XMRV after mouse xenografting. However, the majority of these studies studied a limited number of samples and were conducted with relatively crude detection methods and mouse cell contamination of the human cultures was not tested. Thus it is difficult to determine the frequency of XMLV infection in xenograft cultures. Further concerns are that the infected-xenograft cell lines may be capable of producing high titers of XMLV which may potentially spread horizontally to other non-xenograft cultures.12 The spread of XMLV from NCI-N417 to other non-xenograft cultures was reported at least in one laboratory.21 Recently contamination of MLV gammaretroviruses was found in the cell lines used for human immunodeficiency virus research22 and the MLV sequence was found in 2.2% of 411 human cell line DNA screened.23 However, there is a lack of studies to address how frequent the potential contamination of XMLV is in the laboratories involved in the establishment and/or handling of xenograft cultures.
Our study primarily addresses the following questions: (1) How frequently are xenograft cancer cell lines infected with XMLV? and (2) How frequently does XMLV virus spread horizontally to non-xenograft derived cell lines? The questions we asked are important not only to scientific community establishing or involving in xenograft cultures but also to document the frequency of contamination of XMLV and XMLV-related retroviruses that have a potential biohazard to infect human cells and may transmit to humans.24 In this report, we use the term of “xenograft cultures (or cell line)” to refer to human cultures or cell lines which are derived after mouse xenografting, and “non-xenograft cultures” to refer to human cultures or cell lines which were established without mouse xenografting. In order to achieve our aims we used primers that detect most MLV strains including all known mouse xenotropic strains and we tested for the presence of residual mouse cell DNA which may give spurious positive data.
Results
Design and validation of Taqman qPCR assays for detection MLV sequences.
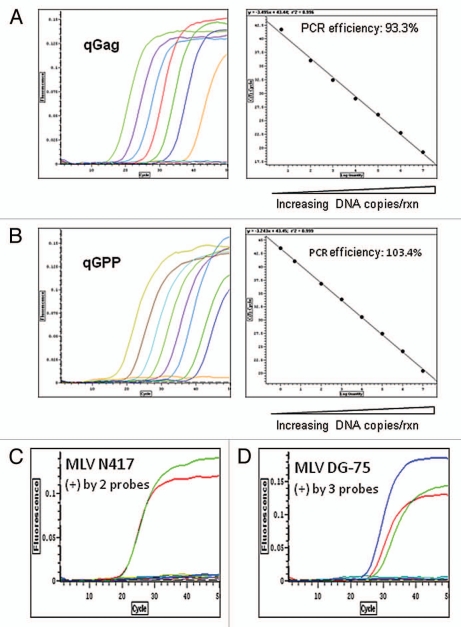
To detect MLV provirus in the human DNA samples, we validated and/or designed novel multiple Taqman qPCR assays specific to MLV sequence. At the start of this study, we adopted two sets of primers/probe Taqman qPCR sequences specific to XMRV strain or most XMLV strains targeting MLV gag or env gene regions respectively as described previously (Fig. S1 and Table S3).25,26 The assays were designated as MLV qGag and MLV qEnv. We also designed a third novel MLV qPCR, namely qGPP, targeting gap-pro-pol region of MLV which has broader specificity to most known MLV viruses as suggested previously in reference 27 (Fig. S1). A non-specific amplification PCR product in qGPP was observed but we validated that it did not appear to affect the sensitivity and specificity of Taqman qGPP probe detection. Under our defined MLV qPCR conditions using XMRV sequence-containing VP62 plasmids, the MLV sequences were quantitatively detected at a dynamic range of 1 to 1 × 107 copies and the sensitivity was 1–5 copies of MLV sequences per 100 ng DNA (Fig. 1A and B). The MLV N417 provirus was detected by both qEnv and qGPP probe at equal sensitivity and not detected by qGag, confirming that qGag is specific to XMRV sequences detection (Figs. 1C and S1 and Table S1). In all three MLV qPCR assays, MLV detection was not present in negative controls including pooled human leukocyte DNA, early passage of NCI-H60 culture or in water (Fig. 1C). The MLV DG-75 provirus in CAK1 pancreatic carcinoma xenograft line (see below) was strongly positive by all three viral probes, which agreed with the specificity of their viral sequences (Figs. 1D, S1 and S5).
Figure 1.
Detection of MLV-related sequences by multiple MLV specific Taqman qPCR assays. (A and B) Validation of MLV qGag and qGPP assays. XMRV VP62 containing plasmids as standard references were serially diluted in an range of 1 × 107 copies to one copy/reaction (copies/rxn) with salmon sperm carrier genomic DNA (100 ng/rxn) and detected in a quantitative range of over 6 logs by MLV qGag or qGPP PCR. The lowest detection limit is one to five copies of MLV 100 ng DNA. The validation of MLV qEnv for xenotropic MLV detection was described previously (see Materials and Methods). Note: validation of the third MLV assay, MLV qEnv, has already been published. (C) Detection of MLV N417 provirus by two viral probes in the DNA of late passage of MLV N417 virus-containing NCI-H60 culture. The DNA of late passage of NCI-H60 (50 ng) culture was strongly positive by qGPP (green) and qEnv (red) probes but not qGag (blue). These findings are consistent with the specificity of MLV N417 viral sequence present in the reference NCI-N417 cell line (Fig. S1 and Table 1). By contrast, the viral free early passage of NCI-H60 culture, pooled human WBC DNA and water controls were all negative by all three MLV qPCR assays. (D) Detection of MLV DG-75 provirus by all three viral probes in CAK1 pancreatic carcinoma xenograft line. The DNA of CAK1 cells (100 ng) was strongly positive by all three MLV qPCR assays as indicated as qGag (blue), qGPP (green) and qEnv (red) (Table 1). These findings are consistent with the specificity of the sequences which were identified in CAK1 and homologous (> 99.0%) with MLV DG-75 virus (Fig. S1 and 5). Water controls in triplets were negative in each assay.
Rxn = reaction.
We applied multiple MLV specific qPCR (Fig. S1) in order to increase the sensitivity and specificity as MLV sequences include a wide spectrum of viral strains. In principle, we expect most or all XMLV viruses to be positive with qEnv and qGPP probes, and XMRV related viruses to be positive with all three strains.
Presence of mouse DNA in xenograft cell lines.
Because the xenograft tumors consist of human cells supported by stromal, vascular and inflammatory cells of mouse origin, initial cultures consist of cells derived from both species. The persistence of mouse cells, which contain multiple copies of MLV-related pro-viruses, may result in false-positive results. Thus we tested all xenograft derived cultures for the presence of human and mouse DNA.
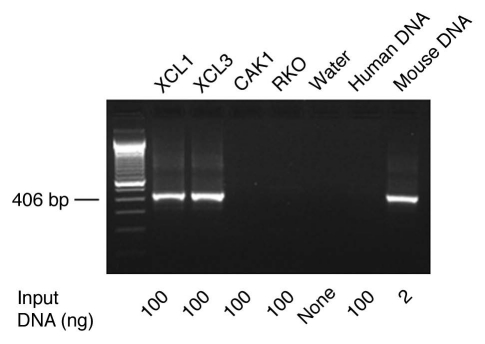
To detect mouse DNA in human cultures, we applied a highly sensitive PCR assay using primers specific to mouse GAPDH gene and GAPDH pseudogenes (over 250 copies/genome28), and the primer sequences were described previously in reference 29 (Table S3). Under our PCR conditions, the mouse GAPDH PCR is highly sensitive and able to detect 0.1–0.01 pg of mouse DNA (Fig. S2), which is comparable to the sensitivity level of mouse DNA detection by the highly sensitive IAP (intracisternal A particle) assay recently described.30 The mouse GAPDH PCR is also highly specific for mouse DNA detection, based on the finding that human control DNA, salmon sperm carrier control DNA and water controls were all negative (Figs. 2 and S2).
Figure 2.
Identification of mouse DNA in human pancreatic carcinoma cultures established from mouse xenografts by PCR. A sensitive PCR assay specific for mouse GAP DH and its pseudogenes was used (Fig. S2 and Materials and Methods). The genomic DNA of human pancreatic carcinoma XCL1 and XCL3 xenograft cultures were found positive for mouse DNA. By contrast, the DNA of human pancreatic carcinoma CAK1 and human colorectal carcinoma RKO culture as well as the control pooled human WBC DNA were free mouse DNA (see Materials/Methods and Table S1–3).
All 26 of the xenograft derived human cell cultures were positive for the human GAPDH probe (Tables 1 and S1) with comparable amount of DNA tested to make sure that any negative detection of MLV sequences are not due to inadequate DNA or presence of DNA polymerase inhibitors in the samples tested. However three of the lines, LX44, XCL1 and XCL3, were found to contain residual mouse DNA (Fig. 2). Sequencing demonstrated evidence of multiple MLV-related sequences, consistent with mouse DNA which contains multiple endogenous MLV proviral sequences. Thus these three cell lines were excluded from further analysis.
Table 1.
Identification of xenotropic murine leukemia viruses (XMLV) and MLV-related viruses in xenograft cell lines
| Cancer type | Cell line | Input DNA (ng) | Ct of human qGAPDH | Ct of MLV qGag1 | Ct of MLV qEnv1 | Ct of MLV qGPP1 | Sequence homology2 | RT enzyme (nU/µl)3 | Mouse DNA5 | Comments | Source: lab PI |
| Lung NSCLC | 1065met | 50 | 29.2 | 34.0 | 29.8 | 34.2 | XMLV NZB | 2.8 × 105 | − | J. Minna | |
| Lung SCLC | NCI-N417 | 50 | 28.9 | neg. | 24.0 | 24.3 | MLV N417 | 8.2 × 105 | − | G. Bepler and A. Gazdar | |
| Lung SCLC | LX47 | 100 | 29.6 | 30.3 | 28.6 | 36.2 | XMLV NZB | 2.9 × 102 | − | C. Rudin | |
| Lung SCLC | LX48 | 100 | 31.9 | neg. | 30.8 | 31.3 | MLV MCF1233 | Negative | − | C. Rudin | |
| Pancreas | CAK1 | 100 | 30.5 | 27.9 | 30 | 31.8 | XMLV DG-75 | ND | − | A. Maitra | |
| Prostate | LAPC-4 | 100 | 29.7 | 28.2 | 28.6 | 28.1 | MLV N417, MLV SP(B)1–2 | up to 4.1 × 105 | − | C. Sawyers and J.T. Hsieh | |
| Lung SCLC | LX44 | 100 | 35.9 | 24.1 | 26.9 | 23.8 | mutiple MLV strains4 | 1.5 × 102 | +++ | Residual mouse cells | C. Rudin |
| Pancreas | XCL1 | 100 | 29.1 | 24.4 | 26.4 | 24.4 | mutiple MLV strains4 | ND | +/− | Residual mouse cells | A. Maitra |
| Pancreas | XCL3 | 100 | 29.7 | 31.2 | 32.3 | 29.7 | mutiple MLV strains4 | ND | +/− | Residual mouse cells | A. Maitra |
| Control tissues | |||||||||||
| Mouse spleen | 10 | neg. | 28.5 | 29.6 | 26.2 | ND | ND | ++++ | A. Gazdar | ||
| XMRV VP62 Plasmids | 1 × 105 copies | neg. | 28.6 | 39.4 | 30.3 | N/A | N/A | N/A | A. Gazdar | ||
| Mouse neural stem cells | DHH C5(wt) mNSC | 100 | neg. | 23.5 | ND | 22.9 | N/A | Negative | ++++ | A. Gazdar | |
| Pooled human WBC | 100 | 27.1 | neg. | neg. | neg. | N/A | N/A | neg. | A. Gazdar | ||
| Negative c ontrol (water) | neg. | neg. | neg. | neg. | N/A | N/A | neg. | A. Gazdar |
The genomic DNA of xenograft cultures were examined by the 3 qPCR assays, namely qGag, qEnv and qGPP, targeting the three major regions of MLV viral genome (gag, env and gag-pro-pol), respectively (see Materials and Methods).
The MLV strain(s) confirmation was based on sequence homology analysis of the partial sequences of gag and env regions or full viral genome sequences (NCI-N417) to known XMLV or MLV-related virus strains (see Table S3 and Figs. S3–7 for the details).
The reverse transcriptase (RT) activity in culture supernatant fluids was measured by TM-PERT assay (see Materials and Methods). One nU of reverse transcriptase activity was estimated to correspond to one MLV viral particle.
Multiple MLV sequences due to mouse DNA contamination.
−, no mouse DNA detected by PCR using the mouse specific primers; +/− to +++, mouse DNA detected from minor to strong positivity. ND, not done; N/A, not applied.
Detection of MLV sequences in xenograft cell lines.
Six of 23 (26%) of the remaining xenograft cell lines free of mouse DNA contamination were strongly positive when tested with two or all three MLV probes. Their qPCR Ct values mostly fell in the range of 24–30, which correspond to about 1.4 × 104 to 1 × 106 MLV copies in the input DNA (50 or 100 ng), as estimated by the MLV qGPP standard curve (Table 1 and Fig. 1). Using an estimate of 6 pg as the average DNA content of a human genome, the overall positive cultures were estimated to contain from 0.1–96 viral copies/cell (Fig. 1 and Table 1). The six positive cell lines (Table 1) originated from six of the seven independent labs. The positive tumor types included non-small cell lung cancer (NSCLC), SCLC, pancreas and prostate. Mouse spleen DNA and neural stem cell culture (positive controls) contained estimated 130–132 copies/cell, which include the provirus sequences for both ecotropic and xenotropic MLV.
The MLV sequences in the viral probe positive cultures were further confirmed by DNA sequencing of partial MLV gag and/or env genes or full viral genome (NCI-N417 line). These sequencing data and analyses are provided in Figures S3–7. The viral sequences contained in each infected culture had greater than 99% of homology to one or two MLV strains (Table 1).
To further determine whether these MLV sequence-positive cultures released virus particles, we examined the level of reverse transcriptase activity of culture supernatant fluids with a highly sensitive and specific qRT-PCR assay (Fig. S8 and Table 1). Of six available supernatant fluids from viral positive cultures, five were strongly positive for reverse transcriptase activity, indicating release of potentially infectious viral particles (vp) (estimated range of 2.9 × 102 to 8.2 × 105 vp/µl).
In summary, of 23 xenograft culture free of mouse DNA, we identified six xenograft cell cultures containing five distinct viral strains. Two of the positive cell lines and their viral genomes are discussed in greater detail in the next section.
Characterization of viral genomes in xenograft NCI-N417 and LAPC-4 cell lines.
The NCI-N417 cell line was one of the first xenograft derived cell lines found to be shedding MLV particles (Gazdar AF, unpublished data).14,15 After this discovery in the Gazdar lab (then located at the NCI, Bethesda, MD), existing stocks were destroyed. However, previously the cell line had been distributed to several investigators. We obtained a frozen vial from Dr. Gerold Bepler, Tampa, FL and completely sequenced the contained virus. We confirmed that the virus MLV strain N417 (GeneBank Accession number HQ246218) was consistent with the partial sequence of an isolate from the same cell lines as performed in another laboratory (MLV env gene: GeneBank Accession number AF064089). Full viral genome sequencing indicated the presence of the full length viral genome (8,189 bp) as well as an apparently defective form of the same strain containing a frame shift deletion of 55 nt in the gag region (nt 964–1,018, Fig. S7A).
The LAPC-4 cell line was established from human prostate tumor explants after serially passing through SCID mice31 in Dr. Charles Sawyers' lab around 1997 and has been widely used in prostate cancer research as a model of androgen independent growth.32 MLV sequences were found in DNA of early passage of LAPC-4 cells obtained from Dr. Sawyers and confirmed in late passage of LAPC-4 cells from both Dr. Sawyers' lab and Dr. Hsieh's lab (Table 2) and all three subcultures of LAPC-4 were free of mouse DNA. All three cultures of LAPC-4 were strongly positive by all three MLV probes (gag, pol and env) (Table 2).
Table 2.
Detection of mouse leukemia virus (MLV) in both early and later passage of prostate LAP C-4 cell line
| Cell lines | Passage | MLV sequence homology1 | Copies/genome2 | RT enzyme (nU/µl)3 | Mouse DNA | Source: lab PI |
| LAP C-4 | Early passage | N417, SP(B)1–2 | 1.1 (0.25 to 1.98) | 1.4 × 104 | Negative | C. Sawyers |
| LAP C-4 | Late passage | N417, SP(B)1–2 | 0.5 (0.07 to 1.0) | 5.4 × 104 | Negative | C. Sawyers |
| LAP C-4 | Late passage | N417, SP(B)1–2 | 9.4 (5.61 to 13.17) | 4.1 × 105 | Negative | J.T. Hsieh |
The MLV strain(s) confirmation was based on sequence homology analysis of the partial sequences of gag and env regions to known XMLV or MLV-related virus strains (Figs. S7A–E).
The copies of MLV were estimated based on MLV qGag and MLV qGPP standard curves (Fig. S1) and amount of input DNA tested and estimate of 6 pg per human genome.
One nU of reverse transcriptase activity was estimated to correspond to 1 MLV viral particle (vp) (see Materials and Methods).
The PCR products using XS3/XS4 primers (Table S3) which cover qGag probe gag region were detected in both early and late passage of LAPC-4 cell lines. The PCR products using XS14/XS15 primers cover env gene region (Table S3). The sequences of both XS3/XS4 and XS14/XS15 PCR products demonstrated that the sequences were identical to MLV N417 virus (Figs. S7A and B). Since our early data demonstrated that MLV N417 pro-virus is negative by our qGag probe due to nucleotide mismatches (Figs. 1C and S1), we suspected that an additional MLV strain sequence may be present in LAPC-4 cells besides harboring MLV N417 provirus or -like provirus. To explore this possibility, we performed three overlapping PCR reactions around qGag probe region of gag gene (Fig. S1) using primers including qGag PCR primers (XS3/XS4, XS3/GagQ2R2 and GagQ2F2/XS4 in Table S3) followed by sequencing (Fig. S7C). All three overlapping MLV PCR products were detected in LAPC-4 genomic DNA (Fig. S7C). However, the sequences of GagQ2F2/XS4 PCR products had 100% homology to MLV SP(B) 1–2 isolate (GeneBank. Accession number AY349140) which is distinctly different from MLV N417 sequences identified in XS3/XS4 PCR products and specific to MLV qGag detection (Figs. S1, 7E and 7D). Therefore, qGag positivity in LAPC-4 cultures was likely due to the additional presence of MLV SP(B) 1–2 viral sequences (Fig. S1). Although XMRV strain has been associated with prostate cancers, the viral genomes in LAPC-4 cell lines lacked the XMRV-characteristic 24-nt deletion sequence in the gag leader region (data not shown). The MLV sequences detected in all three subcultures of LAPC-4 were near identical. High reverse transcriptase activities were detected in culture supernatant fluids of LAPC-4 (Tables 1 and 2), implying LAPC-4 cultures actively release MLV retroviruses. Taken together, our findings are consistent with LAPC-4 cultures acquiring XMLV virus infection during mouse passage and containing at least two XMLV virus strains related to MLV N417 and MLV SP(B) 1–2.
Detection of MLV sequences in non-xenograft cell lines.
In order to detect horizontal spread of XMLV, we tested non-xenograft cultures (n = 78) maintained in xenegraft culture containing facilities (from five labs) as well as those (n = 50) established and maintained in a xenografte culture free lab (A. Gazdar lab at UTSW) (Tables 3, 4 and S2). The Gazdar/Minna labs established cell lines both during their tenure at the NCI, Bethesda MD (NCI series) and after their move to UT Southwestern Medical Center, Dallas, TX, in 1991 (HCC series). While at the NCI 53 non-xenograft lung cancer lines (37 SCLC and 16 NSCLC) (Table S2) were, until 1985, cultured prior to the identification of the N417 strain of XMLV.15 These cell lines had serial numbers lower than NCI-H1515. After the discovery that xenotropic strains could infect human cultures, all stocks of known contaminant cell lines were destroyed. The other 25 non-xenograft cancer lines tested were from four other labs.
Table 3.
Frequent detection of murine leukemia virus (MLV) contamination of non-xenograft human cultures
| Lab PI | Non-xenograft cell line type | No. of MLV postive cell lines1/No. of total cell lines tested (%) |
| Xenograft culture lab | ||
| A. Gazdar (NCI)2 | NSCLC | 1/16 (5%)* |
| A. Gazdar (NCI)2 | SCLC | 5/37 (13%)* |
| C. Rudin | NSCLC | 1/4 (25%) |
| C. Rudin | SCLC | 2/4 (50%) |
| A. Maitra3 | Pancreas | 0/9 (0%) |
| A. Maitra | Colon | 1/1 (100%) |
| R. Brekken3 | Pancreas | 0/2 (0%) |
| J.T. Hsieh | Prostate | 3/7 (43%) |
| Total: 13/78 (17%) | ||
| Xenograft culture free lab | ||
| A. Gazdar (UTSW)4 | NSCLC | 0/33 (0%)* |
| A. Gazdar (UTSW)4 | SCLC | 0/17 (0%)* |
| Total: 0/50 (0%) |
The cell lines were strongly positive by two or three viral probes by MLV qPCR and confirmed by the partial sequences of gag and env regions or full genome sequence (see Materials/Methods for the details and Table 4). See Table S2 for the list of cell lines tested.
A. Gazdar's former lab at NCI.
The DNA of two cell lines (MIAP aCa-2 and PA NC-1) from both labs was tested.
A. Gazdar's current lab at UTSW.
p = 0.01, Fisher exact, 2-tailed test.
Table 4.
Characterization of murine leukemia viruses (MLV) detected in human non-xenograft cultures in xenograft culture laboratories1
| Cell line type | MLV positive cell lines1 | MLV sequence homology2 | RT enzyme (nU/µl) | Mouse DNA3 | Other sources or passages4 | Source: lab PI |
| NSCLC | NCI-H460 | ND | Negative | − | Negative | C. Rudin |
| NSCLC | NCI-H1155 | MLV N417 | ND | − | ND | A. Gazdar (NCI)6 |
| SCLC | NCI-H60 | MLV N417 | 3.6 × 106 | − | Negative | A. Gazdar (NCI)6 |
| SCLC | NCI-H82 | MLV NZB | 1.3 × 106 | − | Negative | C. Rudin |
| SCLC | NCI-H1092 | MLV N417 | 8.0 × 103 | − | Negative | A. Gazdar (NCI)6 |
| SCLC | NCI-H182 | MLV N417 | ND | − | ND | A. Gazdar (NCI)6 |
| SCLC | NCI-H289 | MLV N417 | ND | − | Negative | A. Gazdar (NCI)6 |
| SCLC | NCI-H1514 | MLV N417 | ND | − | ND | A. Gazdar (NCI)6 |
| Colon | RKO | XMRV | 2.9 × 103 | − | Negative | A. Maitra |
| Prostate | PrEC2 | ND | ND | − | ND | J.T. Hsieh |
| Prostate | LNCaP | Multiple MLV strains5 | ND | ++++ | Negative | J.T. Hsieh |
| Prostate | PC3 | ND | ND | −/+ | Negative | J.T. Hsieh |
| SCLC | NCI-H146 | MLV NZB likely | 7.2 × 105 | −/+ | Negative | C. Rudin |
These cell lines were detected positive for at least two viral probes by 3 Taqman qPCR assays targeting the gag, env or pol regions of MLV, respectively (see Materials and Methods for the details). See Table S2 for the list of cell lines tested.
The MLV strain(s) confirmation was based on the sequencing homology analysis of the partial sequences of gag and env regions or full viral genome sequences for MLV-positive NCI-H60 and NCI-H1092 cultures.
−, no mouse DNA detected by PCR using the mouse specific primers; +/− to +++, mouse DNA detected from minor to strong positivity.
The MLV sequences in the early passage of cultures or the cultures from other sources were tested by the Taqman qPCR assays targeting the gag, env or pol regions of MLV, respectively and confirmed negative (see Materials and Methods).
Not fully identified due to mouse DNA contamination.
A. Gazdar's former lab at NCI. RT, reverse transcriptase; ND, not done due to unavailability of samples or weak MLV positive.
Of 78 non-xenograft cultures maintained in the same facility as xenograft cultures, 13 (17%) were positive for MLV provirus sequences (Table 3). Three of these 13 cultures contained both human and mouse DNA while the other ten cultures that contained only human DNA, based on human GAPDH qPCR and the PCR with the mouse specific primers (Table 4). Thus 10 out of 75 (13%) non-xenograft human cultures free of mouse DNA were found to contain MLV sequences. Four out of five MLV provirus positive human cultures tested had high levels of reverse transcriptase activity in the corresponding supernatant fluids. The provirus positive cultures included those of prostate (n = 3), SCLC (n = 7), NSCLC (n = 2) and colon (n = 1) cell line types. The provirus sequences in eight provirus positive cultures were identified by sequencing. All six lung positive cultures from the former Gazdar NCI-lab were negative in their early passages of cultures and contained an identical N417 strain viral sequences found in NIH-N417 xenograft culture (Table 4). Similarly, NCI-H82 and NCI-146 non-xenograft cultures were free of viral detection in their early passage and contained the same viral sequences identified in the LX47 xenograft culture which was maintained in the same lab (Tables 1 and 4 and Fig. S4). Of interest, the same strains had been identified in the xenograft-derived cultures maintained in the same culture facilities, indicating probable horizontal spread. Another 50 non-xenograft lung cancer cell lines (33 NSCLC, 17 SCLC) cultured in a xenograft culture free lab (A. Gazdar's current lab at UT Southwestern) were negative for XMLV (Table 3). The differences in virus positivity for non-xenograft cultures maintained in xenograft culture free and xenograft culture containing labs were significant (Fisher exact 2-tailed test p = 0.01).
Among 13 XMLV DNA positive lines, human colorectal carcinoma cell line RKO from the Maitra Lab (John Hopkins Medical Center), was identified as containing XMRV virus (Table 4). However other cultures of the same line obtained by the Gazdar lab from the American Type Culture Collection, Manassas, VA, were negative by all three MLV viral probes in agreement of the previous report in reference 23. The viral genome sequence of the first 550 bp had >99.6% homology to all known XMRV isolates (Fig. S6) and is different from 129x 1Svj isolate (GeneBank Accession number AAHY01591888.1) which shares a 24-nt deletion feature. The full viral genome sequence of XMRV RKO (GeneBank Accession number JF274252) had >99.9% homology to all known XMRV isolates (Table S4). Of interest, the sequence of XMRV in the RKO cell line had a higher homology to XMRV 22Rv1 than other isolates (Table S4), suggesting the viral contamination possible resulting from XMRV virus from xenograft prostate cancer 22Rv1 cell line.23 The RKO cell line was negative for mouse DNA and contained high RT activity in culture supernatants (Table 4). Retrospective analysis indicated that there was a short period of time (a few days) during which the 22Rv1 and RKO cell lines were maintained in the same culture facility. Fingerprint genotyping confirmed that the XMRV-positive RKO matches the reference RKO fingerprint and not that of the 22Rv1 cell line. In conclusion, RKO cultures from other sources were negative for XMLV, confirming that the XMRV virus present in the RKO culture from the Maitra lab was not a novel XMRV strain originally residing in RKO, but represented horizontal spread from the XMRV containing 22Rv1 prostate carcinoma cell line maintained in the same culture facility.
Infectivity of XMLV viruses from supernatant fluids of XMLV-positive xenograft cultures.
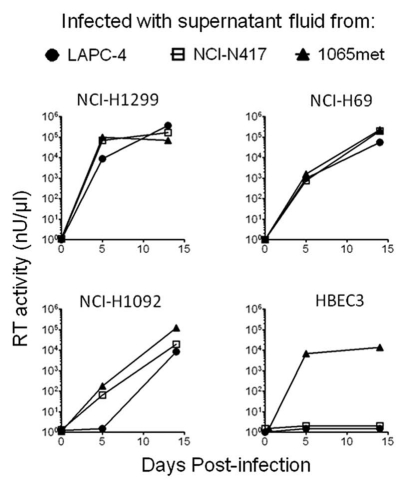
We performed studies to determine whether XMLV-positive supernatant fluids are readily transmissible to other, non-infected human cultures. Supernant fluids from three XMLV-positive xenograft cultures (LAPC-4, NCI-N417 and 1065met) (Table S1) were used to infect virus free early passages of five cell lines (Fig. 3 and Table S5). At days 5 and 14, supernatant fluids were collected and tested for reverse transcriptase activity. On day 14, DNA was extracted from the infected cell lines and tested for the presence of viral DNA sequences using three probes (Table S5). For three cell lines (SCLC lines NCI-H69 and NCI-H1092, and NSCLC line NCI-H1299) were strongly positive for reverse transcriptase activity and genomic DNA was positive for at least two of three viral probes. NSCLC cell line NCI-H460 demonstrated minimal or no reverse transcriptase activity but was positive for viral sequences after infection with virus-positive supernatant fluids from cell lines 1065met and NCI-N417 except LAPC-4, indicating latent infection. Bronchial epithelia cell line HBEC3 was positive for reverse transcriptase activity and viral sequences after infection with 1065met fluid, but not after infection with the other two supernatant fluids. The uninfected controls were negative for presence of XMLV. DNA fingerprinting confirmed the correct provenance of the infected cell lines, and all cultures were free of mouse DNA contamination.
Figure 3.
Infectivity of supernatant fluids of MLV positive xenograft derived cell lines. Five previously XMLV virus-free cultures (2 NSCLC lines: NCI-H1299 and NCI-H460; two SCLC lines: NCI-H69 and NCI-H1092, and an immortalized human bronchial epithelial cell culture HBEC3K-TR53 [HBEC3]), were infected with the supernatant fluids (250 µl) from 3 XMLV-positive xenograft derived cell lines (LAP C-4, NCI-N417 and 1065met) (Table 1), which had the reverse transcriptase (RT) activities of 5.7 × 104 nU/µl, 3.1 × 105 nU/µl and 1.8 × 105 nU/µl, respectively. The supernatants collected at days 5 and 14 post-infection were examined for the RT activity (see Materials/Methods). The supernatants from the infected NSCLC cell line NCI-H460 showed minimal or no RT activities for all three viruses on day 5, 14 and 18 post-infection (data not shown).
Discussion
Reports of XMLV strains being present in human xenograft cultures appeared in the 1970s.10,12,33 While several reports have documented this phenomenon, most reports are sporadic case reports without systematic analysis of true frequency. Two reports examined multiple xenograft cultures12,33 and found frequencies of 7 and 67% respectively. However, because of the relatively crude detection methods available at that time and failure to check for mouse cell contamination, the true frequencies cannot be determined from these reports. Because of our initial failure to culture SCLC tumors directly from tumor samples, we developed long-term cultures from xenografted tumors.34 In the mid 1980s we detected (by electron microscopy) large numbers of retroviral particles released by SCLC cell line NCI-N417. We destroyed stocks of this cell line and persuaded the American Type Culture Collection to stop distribution. However, prior to this finding, the cell line had been distributed extensively to the scientific community and we obtained a culture from Gerold Bepler, Moffitt Cancer Center, Tampa, FL. While we did not report on the viral findings, others have done so14,15 and we refer to this XMLV strain as N417 although it has also been referred to as VB3.2.21 Recent reports of squirrel monkey retroviruses (SMRV) infecting human cultures (by unknown routes or sources) and their possible large scale horizontal spread have also been published,35,36 and several human cell lines for human immunodeficiency virus research were found to release infectious XMLV.22,37 However, there are virtually no reports of contamination of xenograft cultures published during the past 15 y and most scientists appear unaware of the contamination problem (authors' observations). The purpose of our study was to determine the frequency of xenograft culture contamination by xenotropic viruses and their horizontal spread in multiple laboratories. By sequencing the viruses so isolated we could determine whether a single or multiple strains of XMLV were responsible for contamination and horizontal spread.
For our initial study we obtained 26 xenograft cultures from seven independent laboratories. Only two of the laboratory chiefs that participated in this study were previously aware of the XMLV contamination issue. We utilized three primer sets that detected virtually all XMLV stains as well as most MLV strains, covering the three major structural and functional regions of the mouse retroviral genome. Our multiple qPCR approaches had advantage to detect all possible MLV sequences (increased sensitivity) with one or two additional probe confirmation (increased specificity) as compared with a single qPCR assay. Nine of the samples were positive by either two or all three of the probes used. However, three of the nine positive samples contained varying amounts of mouse DNA, presumably as a result of survival of mouse stromal cells from the mouse xenograft. Apparently the mouse stromal cells may persist for lengthy periods in culture, occasionally in excess of one year. These three cultures were removed from further study as the mouse genome contains multiple endogenous MLV provirus sequences which make interpretation difficult. Six of the remaining 23 xenograft cultures (26%) were positive for one (or in one case, two) strains of XMLV or related viruses. These six cultures came from six independent labs indicating the widespread nature of the contamination. Of interest, our viral sequencing homology analysis indicated that multiple strains of XMLV were present in the six positive lines, demonstrating that there are multiple strains of XMLV or MLV-related viruses capable of infecting human cells individually or simultaneously after xenografting. The LAPC-4 cell line, widely used as a model for androgen independent prostate cancer, with over 120 citations in PubMed, contained two strains. Early and late passage cultures from the originator (Dr. Charles Sawyers) as well as a culture distributed to the Hsieh lab contained the same strain mixture.
Supernatant fluids were available from five of the six viral positive xenograft cell lines. Four of these released large numbers of potentially infectious viral particles, indicating possibilities of horizontal spread to other human cultures as well as posing a biohazard of unknown potential to laboratory personnel. We presume that in one of these five cell lines the XMLV virus existed in a latent form. Because of the possibility of spread to non-xenografted human cultures, we examined cultures maintained in the same culture facilities. A total of 78 cultures were obtained from five of the laboratories. Thirteen of these cultures (17%) from four of the laboratories were positive for XMLV. The viruses identified as being responsible for horizontal spread were identical to those identified in the positive xenograft cultures in the respective laboratories. By contrast, 50 cell lines from the Gazdar lab maintained in a facility free of xenograft cultures tested negative for virus. These differences were significant, indicating the potential for horizontal viral spread to other cultures maintained in the same laboratory facility as xenograft cultures. In one case XMRV virus infection to a non-xenograft colorectal carcinoma cell line RKO38 was demonstrated from an XMRV containing prostate xenograft derived cell line 22Rv1 even though the two cell lines had been maintained in the same culture facility for only a few days.
Our results indicated the frequent presence of XMLV viruses in cultures initiated from human xenografted tumors, with integration of the viral genomes and production of abundant virions. We also found evidence of widespread contamination of non-xenografted cultures maintained in the same tissue culture facilities with xenografted cultures. However, to rigorously prove that the virions released by the xenografted cultures were infectious for human cells, we collected supernatant fluids from three XMLV-positive xenografted cultures including LAPC-4 and used them to infect five other human cell lines. All three viruses readily infected two SCLC lines, one NSCLC line and one virus infected a non-tumorigenic immortalized bronchial epithelial cell culture. The fifth cell line, NSCLC line NCI-H460, appeared to undergo an abortive infection, as monitored by reverse transcriptase activity. However genomic analyses demonstrated the presence of relatively low amounts of integrated forms of two of three infected viruses, indicating latent infection. Thus human cells demonstrate heterogeneity for XMLV viral sensitivity and the XMLV viruses demonstrate variable infective potential.
Several reports, have described the finding, usually of partial XMRV or XMLV related sequences at low abundance, in human tumors and tissues.19,39,40 These methods often utilize nested PCR techniques for detection, and multiple other labs have failed to confirm the findings. Considerable recent evidence indicates contamination as the probable cause of these sequences in human cells.41,42 Because of sequence similarities, the presence of XMRV virus in human tumors and cells has been attributed to contamination, especially from the frequently used 22Rv1 prostate cancer cell line41 but it has not been confirmed in any cell lines, to our knowledge.23 Hue et al. screened human tumor 411 cell lines from the COSMIC collection and found XMLV sequences positive in nine cell lines (2.2%), in which five are closely related to DG-75 strains but none of these cell lines are confirmed to be infected with XMRV and the verification of mouse DNA contamination was not addressed.23 Our findings demonstrated that the XMLV virus in colorectal cell line RKO from the Maitra Lab is an XMRV isolate contaminated from the virus containing xenograft derived 22Rv1 prostate cancer culture. The window during which both cell lines were maintained together in the same culture facility was only a few days, indicating that horizontal spread may occur rapidly. This XMRV-positive RKO culture probably represents the first report of horizontal spread of the XMRV virus.
Our results indicate that human tumor cells frequently become infected with MLV virus after xenografting and subsequent culture. We have observed that mouse stromal cells may persist in culture for lengthy periods. Mouse stromal cells, while they contain abundant provirus forms of MLV, including ecotropic, polytropic and xenotropic strains, seldom spontaneously release large amounts of infectious virus (authors' unpublished findings). Virus infection of xenografted cells may require activation of XMLV virus by chemical or immunological induction in mouse and by prolonged mouse and human cell contact. Viral transfer may occur in the mouse host or during subsequent xenograft culture. Our findings of infectivity of XMLV-positive supernatant fluids demonstrated that XMLV can readily infect other human cultures without presence of mouse cells or other aiding factors, indicating that these viruses are highly infectious.
In conclusion, our studies demonstrated that several MLV strains were present in over one fourth of xenograft cell lines. Infected cell lines were identified in most laboratories working with or establishing xenograft cultures, indicating that such contamination was widespread. Infected cultures usually release large numbers of infectious virions, and intra-laboratory spread of MLV virus to other cell lines maintained in the same facilities may occur, confirming the highly infectious nature of MLV virus. Retroviruses have been associated with multiple diseases including solid and hematologic malignancies, AIDS as well as with non-malignant diseases. The high susceptibility of human cells to infection with XMLV, the high levels of reverse transcriptase activity present in culture supernatant fluids and the demonstrated infectivity of the shed virions suggest that such viruses may present potential biohazards to laboratory personnel involved in cell culture facilities or to those handling human xenografts. In addition, the effects of the integrated provirus or the released virions on the biology of infected tumor cells are unknown. Provirus integration into the genome is not random, and occurs preferentially at transcription start sites, CpG islands, DNase-hypersensitive sites and gene-dense regions, suggesting that provirus integration may influence transcription in the host cell.43 Thus laboratories handling or culturing human xenografts should monitor for the presence of MLV, and should consider monitoring personnel for viral antigens or antibodies to them. Laboratories working with xenograft cultures should have full knowledge and understanding of the potential biological and biohazardous risks and should not distribute or publish their findings without full disclosure of the virus status of their xenograft-derived materials.
Materials and Methods
Cell lines.
The 154 cell lines tested were provided from the following seven independent laboratories: Drs. Adi F. Gazdar, John D. Minna/Boning Gao, Jer-Tsong Hsieh and Rolf Brekken at the Univ. of Texas Southwestern Medical Center at Dallas; Drs. Anirban Maitra and Charles M. Rudin at Johns Hopkins University School of Medicine, and Dr. Charles Sawyers at Memorial Sloan-Kettering Cancer Center. The lists of 26 xenograft cell lines and 128 non-xenograft cell lines are provided in Tables S1 and S2, respectively. Genomic DNA or living cells were provided from the labs. The NCI-N417 cell line was originally established from a small cell lung cancer tumor xenografted into an athymic nude mouse in A. Gazdar's former Laboratory at NCI, Bethesda, MD.34 As it was found to contain a retrovirus in the 1980s, it was no longer maintained in the laboratory.14,15 For this study, a vial of the NCI-N417 cell line was obtained in January of 2010 from Dr. Gerold Bepler (who originally obtained it from Dr. Gazdar), the Moffitt Cancer Center, Tampa, FL. Non-xenograft cell lines maintained in the same culture facility as the xenograft cell lines were obtained from five labs. These included cell lines from Dr. Gazdar's lab while he was at the NCI. In addition, Dr. Gazdar provided non-xenograft lines maintained in a facility at UT Southwestern Medical Center that was free of xenograft cultures (Table S2). DHHC5(wt) mNSC mouse neural stem cell line was kindly provided form Dr. Yi Li (Dr. Sandra L. Hoffmann Lab., UT Southwestern Medical Center). Authenticity of MLV positive lung cancer cultures from Gazdar's lab and the RKO from Maitra's lab was confirmed by DNA fingerprint genotyping tests.44 All specimens were obtained following approval from the Johns Hopkins Medicine Institutional Review Boards (protocol 05-04-14-02) or the University of Texas Southwestern Institutional Review Board (protocol 1191-36400).
DNA isolation.
All DNA isolations were performed in a room designated to be free of high-copy of amplicons of MLV DNA. Standard practices for molecular biology were taken during DNA isolation to avoid DNA cross-contaminations. Genomic DNA from cultures was isolated using QIAamp Blood Mini kit (Qiagen) or equivalent DNA isolation kit. To eliminate RNA contamination, genomic DNA was isolated after adding RNase A (Invitrogen) according the procedures described in QIAamp Blood Mini kit (Qiagen). Genomic DNA was obtained from mouse spleen (Black Swiss mice). DNA concentrations were measured using Nanodrop2000 (Thermo Scientific).
PCR.
Pre-PCR preparation was conducted in a designated PCR workstation (AirClean600 PCR Station, Raleigh, NC) with use of UV radiation 15 min before and after each PCR setting. Pre-PCR and Post-PCR samples were handled in separate laboratory rooms. qPCR and standard PCR were performed using the Chromo4 MJ Research Real time PCR system and 9600 thermal cycler (Applied Biosystem Inc.), respectively.
Three sets of Taqman qPCR assays were used as screening tests for XMLV sequences in order to reduce false negativity and increase sensitivity and specificity of XMLV detection. The three qPCR assays, namely qGag, qEnv and qGPP, targeted the three major regions of MLV viral genome (gag, env and gag-pro-pol) and are specific to XMRV strains, most XMLV strains and most known MLV strains, respectively (Table S3). The primer and probe sequences for qGag PCR and qEnv PCR were described previously in reference 25 and 26. The qGPP PCR assay was developed by us for the detection of most known MLV strains including ecotropic and xenotropic MLV strains. qGPP primers and probes are described in Table S3. Since a non-specific PCR amplification product from qGPP PCR was observed, we validated that it did not affect the sensitivity and specificity of qGPP assay for detecting MLV sequences. The MLV qGag and qGPP assays were validated using XMRV sequencing-containing VP62/pcDNA3.1 plasmids as standard references (Fig. 1) (VP62/pcDNA3.1 plasmid, obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: contributor of Dr. Robert Silverman). The viral copy number in a given DNA sample was estimated based on its MLV qPCR Ct value and the validated MLV qPCR standard curves (Fig. 1). Human GAPDH real-time PCR was run as an internal control for DNA input and qualities, and its primers and probes were described previously in reference 45 (Table S3). Either 50 or 100 ng DNA of each sample was examined initially by human qGAPDH and three MLV qPCR assays.
The PCR controls included multiple negative controls (three to six) with water only, human DNA controls with pooled human blood cell DNA from eight individuals, positive controls with mouse spleen DNA (1–10 ng/reaction) or diluted XMRV VP62/pcDNA3.1 plasmids (1 × 105 copies/reaction). The qPCR reaction was performed in 25 µl reaction mixture containing 1x TaqMan PCR buffer, 3.5 mM MgCl2, 0.2 mM dNTP, 0.6 µM of each forward and reverse primers, 0.2 µM of probe, 1.25 U of HotStar Taq DNA polymerase (Qiagen) and 50–200 ng of DNA sample. PCR was performed under the following conditions: 95°C for 15 min, followed by 50 cycles at 95°C for 15 sec and 60°C for 1 min, unless specifics indicated.
To rule out mouse DNA contamination, DNA samples were analyzed by PCR using primers specific to mouse GAPDH gene and GAPDH pseudogenes (over 250 copies/genome28) as described previously in reference 29 (Table S3). The mouse spleen genomic DNA were 1:10 serially diluted in salmon sperm DNA (20 ng/µl) (Invitrogen) and examined by mouse GAPDH PCR at a input DNA range of 10 ng to 0.1 fg of mouse DNA per reaction (Fig. S2). The mouse GAPDH PCR reaction mixture (50 µl) contains 1x TaqMan PCR buffer, 3.5 mM MgCl2, 0.2 mM dNTP, 0.4 µM of each forward and reverse GAPDH primers, 1.65 U of Hot Star Taq DNA polymerase (Qiagen). The PCR was run as 95°C for 12 min, followed by 40 cycles at 94°C for 30 sec and 60°C for 30 sec and 72°C for 30 sec and 72°C for 7 min. These primers do not amplify human DNA (Fig. 2).
DNA sequencing.
To further confirm viral strain sequences, the qPCR MLV positive DNA samples were further amplified by PCR and sequenced for MLV strain verification by using the sequencing primers (Table S3) covering 5′ leader region of gag gene (GagF1/GagR2 primer set), early gag gene (XS3/XS4 primer set) and/or env gene region (XS14/XS15 primer set). The whole MLV genome sequencing was performed on the PCR products overlapping cross the viral genome which were amplified from cDNA of NCI-N417 cell line or genomic DNA of the identified viral positive cell line RKO (Table S2 and S4) using the sequencing primers described previously19 and additional primers described in Table S3. The sequencing reactions were performed as follows: 50 or 100 ng of cDNA or genomic DNA, 25 µL of HotStart-IT FideliTaq Master Mix (USB Corporation), 1.0 µL of each of 10 µM forward and reverse oligonucleotide primers in reaction volumes of 50 µL. PCR were performed as 95°C 2 min, 94°C 30 sec, 58°C 30 sec, 72°C 60 sec, 35 cycles, 68°C 7 min. Total RNA from cell lines were isolated using RNeasy Mini Kit and purified with DNase digestion and RNA Clean-up following the kit instructions (Qiagen). The cDNA were prepared using High Capacity cDNA Reverse Transcription kit and random primers (Applied Biosystems Inc.). The PCR products were purified with ExoI/SAP (USB), gel-purified (Qiagen) or sub-cloned into pCR 2.1-TOPO vector (Invitrogen), prior to sequencing by an ABI 3100 DNA sequencing system (Applied Biosystems Inc.). The alignments of DNA sequencing data were performed using Clustalw software (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalwan.html).46
TM-PERT assay for reverse transcriptase activity.
For reverse transcriptase (RT) activity assays, cell culture supernatant fluids were collected from exponentially growing cultures 2–3 d after a medium change. The cells were removed by centrifuging cell suspension fluids at 300x g for 10 min, and the resultant supernatant fluids were further centrifuged at 2,000x g for 20 min at 10°C to remove any cell debris. The supernatant fluids were stored at −80°C prior to testing. Reverse transcriptase activities were measured using Taqman fluorogenic 5′-nulcease product-enhanced reverse transcriptase assay (TM-PERT) as described previously in reference 47 and 48 with some modifications (Table S3). Briefly, M-MLV reverse transcriptase (Promega, 200 u/µl) was 1:10 serially diluted with RT enzyme dilution buffer (50% glycerol, 2% BSA [Sigma, V fractioned] in 1x RT buffer [Promega]) and used as references to establish quantitative standard curve (Fig. S8). The RT reactions contained 5 µl of sample (1:10 and 1:40 diluted with RT enzyme dilution buffer) or controls, 8 u RNase inhibitor (Promega), 0.32 mM dNTP (Sigma), 360 nM MS2 reverse primer and 8 ng MS2 RNA (Roche) in 25 µl of 1x RT buffer (Promega: 50 mM TRIS-HCl pH 8.3, 5 mM MgCl2 (modified from 3 mM), 75 mM KCl, 10 mM DTT). Following the RT reactions (25°C 10 min, 37°C 120 min and 85°C 5 sec), 5 out of 25 µl RT reactions were transferred to optical 96 well plates containing final volume of 25 µl of PCR mix (RNase 2 U [Progema], 1x TaqMan PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTP, 0.6 µM of each MS2 forward and MS2 reverse primers, 0.2 µM of MS2 probe, 1.25 U of HotStar Taq DNA polymerase [Qiagen]). qPCR reactions were performed at 37°C 30 min, 95°C 15 min, then 95°C 15 sec 60°C 60 sec for 50 cycles using the Chromo4 MJ Research Real time PCR system. Under our defined conditions (Fig. S8), the positive threshold value was set as ≥ 94.6 nU/µL, which was determined as a sum of mean background value (29.9 nU/µL) of culture supernatant fluids from nine different MLV negative cell lines plus three times of Standard Deviation (21.6 nU/µL) as described previously in reference 47 and 48. To correlate functions and viral copy number, 1 nU of reverse transcriptase activity was estimated to correspond to 1 MLV viral particle as described previously in reference 47 and 48.
Infectivity of xenograft derived MLV strains.
To examine whether the supernatants from the XMLV-positive cell lines are infectious for virus free cultures, we collected and tested the supernatants from three cell lines selected simply upon their availability (LAPC-4, p142; NCI-N417 and 1065met) (Table S1). Supernatant fluids were collected as previously described and filtered through with 0.22 µm sterile culture filter units (Millipore) and used to infect four XMLV free lung cancer cultures (NSCLC lines NCI-H1299 and NCI-H460; SCLC lines NCI-H69 and NCI-H1092) and an immortalized human bronchial epithelial cell culture HBEC3KTR53 (HBEC3). All cell lines tested were grown in humidified atmosphere with 5% CO2 at 37°C. NCI-H1299, NCI-H460 and NCI-H69 were grown in RPMI 1640 media (Cellgro) supplemented with 10% fetal bovine serum (FBS) (Hyclone), NCI-H1092 in HITES media supplemented with 10% FBS,44 and HBEC3 in KSFM media supplemented with bovine pituitary extract and recombinant human epidermal growth factor (Invitrogen).49 For infection, 250 µL of supernatant fluid was added to the cultures in growth phase and the medium replaced after 48 h. Supernatant fluids were collected on days 5 and 14, and cell pellets on day 14. Supernatant fluids were tested for reverse transcriptase activity and cell pellets for DNA fingerprinting, presence of mouse DNA and presence of XMLV DNA (three probes as described above previously).
Acknowledgments
We thank Misty Watson, Jason Toombs, Laura Sullivan, Juliet Rivera, Dr. Chun-Xian Huang, Dr. Puja Gupta, Dr. Yi Li, Dr. Philip E. Thorpe and Dr. Yi Yin at UT Southwestern Medical Center for their kind help in procurement of mouse tissues, xenografts and xenograft cell lines, Sara Murphy at Johns Hopkins University School of Medicine for kindly preparing the DNA of cell lines from Dr. Charles M. Rudin Laboratory, Dr. Charles Sawyers and John Wongvipat at Memorial Sloan-Kettering Cancer Center for generously providing LAPC-4 cell line and its related discussions. We acknowledge the assistance from Dr. James Eshleman at Johns Hopkins University School of Medicine in establishing the pancreatic carcinoma cell line.
Funding was provided by the Texas Specialized Program of Research Excellence in Lung Cancer (P50CA70907) and the Early Detection Research Network (U01CA084971), National Cancer Institute and the Canary Foundation (Palo Alto, CA).
Abbreviations
- MLV
murine leukemia virus
- XMLV
xenotropic murine leukemia virus
- XMRV
xenotropic murine leukemia virus-related virus
- NSCLC
non-small cell lung cancer
- SCLC
small cell lung cancer
- qGag
qPCR specific to XMRV gag gene sequence
- qEnv
qPCR specific to XMLV env gene sequence
- qGPP
qPCR specific to MLV gag-pro-pol gene sequence
- RT
reverse transcriptase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Vogt PK. Historical introduction to the general properties of retroviruses. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory; 1997. pp. 1–25. [PubMed] [Google Scholar]
- 2.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/A:1024455415443. [DOI] [PubMed] [Google Scholar]
- 3.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell Mol Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baliji S, Liu Q, Kozak CA. Common inbred strains of the laboratory mouse that are susceptible to infection by mouse xenotropic gammaretroviruses and the human-derived retrovirus XMRV. J Virol. 2010;84:12841–12849. doi: 10.1128/JVI.01863-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy JA. Xenotropism: the elusive viral receptor finally uncovered. Proc Natl Acad Sci USA. 1999;96:802–804. doi: 10.1073/pnas.96.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak CA, Hartley JW, Morse H., III Laboratory and wild-derived mice with multiple loci for production of xenotropic murine leukemia virus. J Virol. 1984;51:77–80. doi: 10.1128/jvi.51.1.77-80.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oie HK, Russell EK, Dotson JH, Rhoads JM, Gazdar AF. Host-range properties of murine xenotropic and ecotropic type-C viruses. J Natl Cancer Inst. 1976;56:423–426. doi: 10.1093/jnci/56.2.423. [DOI] [PubMed] [Google Scholar]
- 8.Hoggan MD, O'Neill RR, Kozak CA. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J Virol. 1986;60:980–986. doi: 10.1128/jvi.60.3.980-986.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frankel WN, Stoye JP, Taylor BA, Coffin JM. Genetic analysis of endogenous xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J Virol. 1989;63:1763–1774. doi: 10.1128/jvi.63.4.1763-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Todaro GJ, Arnstein P, Parks WP, Lennette EH, Huebner RJ. A type-C virus in human rhabdomyosarcoma cells after inoculation into NIH Swiss mice treated with antithymocyte serum. Proc Natl Acad Sci USA. 1973;70:859–862. doi: 10.1073/pnas.70.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Achong BG, Trumper PA, Giovanella BC. C-type virus particles in human tumours transplanted into nude mice. Br J Cancer. 1976;34:203–206. doi: 10.1038/bjc.1976.144. [DOI] [Google Scholar]
- 12.Suzuki T, Yanagihara K, Yoshida K, Seido T, Kuga N. Infectious murine type-C viruses released from human cancer cells transplated into nude mice. Gann. 1977;68:99–106. [PubMed] [Google Scholar]
- 13.Gautsch JW, Knowles AF, Jensen FC, Kaplan NO. Highly efficient induction of type C retroviruses by a human tumor in athymic mice. Proc Natl Acad Sci USA. 1980;77:2247–2250. doi: 10.1073/pnas.77.4.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiefer PE, Bepler G, Kubasch M, Havemann K. Amplification and expression of protooncogenes in human small cell lung cancer cell lines. Cancer Res. 1987;47:6236–6242. [PubMed] [Google Scholar]
- 15.Walker C, Nettesheim P, Barrett JC, Jirik FR, Sorge J, Joyce M, et al. Mouse retroviral sequences acquired by cell lines after passaging through nude mice detected by hybridization of the fms probe pSM3. Cancer Res. 1989;49:625–628. [PubMed] [Google Scholar]
- 16.Lusso P, di Marzo Veronese F, Ensoli B, Franchini G, Jemma C, DeRocco SE, et al. Expanded HIV-1 cellular tropism by phenotypic mixing with murine endogenous retroviruses. Science. 1990;247:848–852. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 17.Pretlow TG, Wolman SR, Micale MA, Pelley RJ, Kursh ED, Resnick MI, et al. Xenografts of primary human prostatic carcinoma. J Natl Cancer Inst. 1993;85:394–398. doi: 10.1093/jnci/85.5.394. [DOI] [PubMed] [Google Scholar]
- 18.Knouf EC, Metzger MJ, Mitchell PS, Arroyo JD, Chevillet JR, Tewari M, et al. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J Virol. 2009;83:7353–7356. doi: 10.1128/JVI.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, et al. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Paprotka T, Delviks-Frankenberry KA, Cingoz O, Martinez A, Kung HJ, Tepper CG, et al. Recombinant Origin of the Retrovirus XMRV. Science. 2011 doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antoine M, Wegmann B, Kiefer P. Envelope and long terminal repeat sequences of an infectious murine leukemia virus from a human SCLC cell line: implications for gene transfer. Virus Genes. 1998;17:157–168. doi: 10.1023/A:1008020808314. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi Y, McClure MO, Pizzato M. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol. 2008;82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hué S, Gray ER, Gall A, Katzourakis A, Tan CP, Houldcroft CJ, et al. Disease-associated XMRV sequences are consistent with laboratory contamination. Retrovirology. 2010;7:111. doi: 10.1186/1742-4690-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen J. Retrovirology. More negative data for link between mouse virus and human disease. Science. 2011;331:1253–1254. doi: 10.1126/science.331.6022.1253. [DOI] [PubMed] [Google Scholar]
- 25.McCormick AL, Brown R, Cudkowicz ME, Al-Chalabi A, Garson JA. Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology. 2008;70:278–283. doi: 10.1212/01.wnl.0000297552.13219.b4. [DOI] [PubMed] [Google Scholar]
- 26.Shi L, Chen Q, Norling LA, Lau AS, Krejci S, Xu Y. Real time quantitative PCR as a method to evaluate xenotropic murine leukemia virus removal during pharmaceutical protein purification. Biotechnol Bioeng. 2004;87:884–896. doi: 10.1002/bit.20198. [DOI] [PubMed] [Google Scholar]
- 27.Erlwein O, Kaye S, McClure MO, Weber J, Wills G, Collier D, et al. Failure to detect the novel retrovirus XMRV in chronic fatigue syndrome. PLoS ONE. 2010;5:8519. doi: 10.1371/journal.pone.0008519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YJ, Zheng D, Balasubramanian S, Carriero N, Khurana E, Robilotto R, et al. Comprehensive analysis of the pseudogenes of glycolytic enzymes in vertebrates: the anomalously high number of GAPDH pseudogenes highlights a recent burst of retrotrans-positional activity. BMC Genomics. 2009;10:480. doi: 10.1186/1471-2164-10-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters SO, Bauermeister K, Simon JP, Branke B, Wagner T. Quantitative polymerase chain reaction-based assay with fluorogenic Y-chromosome specific probes to measure bone marrow chimerism in mice. J Immunol Methods. 2002;260:109–116. doi: 10.1016/S0022-1759(01)00525-7. [DOI] [PubMed] [Google Scholar]
- 30.Robinson MJ, Erlwein OW, Kaye S, Weber J, Cingoz O, Patel A, et al. Mouse DNA contamination in human tissue tested for XMRV. Retrovirology. 2010;7:108. doi: 10.1186/1742-4690-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein KA, Reiter RE, Redula J, Moradi H, Zhu XL, Brothman AR, et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 32.Culig Z. Androgen receptor cross-talk with cell signalling pathways. Growth Factors. 2004;22:179–184. doi: 10.1080/08977190412331279908. [DOI] [PubMed] [Google Scholar]
- 33.Wunderli H, Mickey DD, Paulson DF. C-type virus particles in human urogenital tumours after hetero-transplantation into nude mice. Br J Cancer. 1979;39:35–42. doi: 10.1038/bjc.1979.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–2923. [PubMed] [Google Scholar]
- 35.Stang A, Petrasch-Parwez E, Brandt S, Dermietzel R, Meyer HE, Stuhler K, et al. Unintended spread of a biosafety level 2 recombinant retrovirus. Retrovirology. 2009;6:86. doi: 10.1186/1742-4690-6-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovic M, Kalyanaraman VS, Reitz MS, Sarngadharan MG. Identification of the RPMI 8226 retrovirus and its dissemination as a significant contaminant of some widely used human and marmoset cell lines. Int J Cancer. 1982;30:93–99. doi: 10.1002/ijc.2910300116. [DOI] [PubMed] [Google Scholar]
- 37.Burtonboy G, Delferriere N, Mousset B, Heusterspreute M. Isolation of a C-type retrovirus from an HIV infected cell line. Arch Virol. 1993;130:289–300. doi: 10.1007/BF01309661. [DOI] [PubMed] [Google Scholar]
- 38.Brattain MG, Levine AE, Chakrabarty S, Yeoman LC, Willson JK, Long B. Heterogeneity of human colon carcinoma. Cancer Metastasis Rev. 1984;3:177–191. doi: 10.1007/BF00048384. [DOI] [PubMed] [Google Scholar]
- 39.Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci USA. 2009;106:16351–16356. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Lo SC, Pripuzova N, Li B, Komaroff AL, Hung GC, Wang R, et al. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc Natl Acad Sci USA. 2010;107:15874–15879. doi: 10.1073/pnas.1006901107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Smith RA. Contamination of clinical specimens with MLV-encoding nucleic acids: implications for XMRV and other candidate human retroviruses. Retrovirology. 2010;7:112. doi: 10.1186/17424690-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser J. Chronic fatigue syndrome. Studies point to possible contamination in XMRV findings. Science. 2011;331:17. doi: 10.1126/science.331.6013.17. [DOI] [PubMed] [Google Scholar]
- 43.Kim S, Kim N, Dong B, Boren D, Lee SA, Das Gupta J, et al. Integration site preference of xenotropic murine leukemia virus-related virus, a new human retrovirus associated with prostate cancer. J Virol. 2008;82:9964–9977. doi: 10.1128/JVI.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–1321. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson KL, Dukes KA, Vidaver J, LeShane ES, Ramirez I, Weber WD, et al. Interlaboratory comparison of fetal male DNA detection from common maternal plasma samples by real-time PCR. Clin Chem. 2004;50:516–521. doi: 10.1373/clinchem.2003.024380. [DOI] [PubMed] [Google Scholar]
- 46.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/S0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 47.Brorson K, Xu Y, Swann PG, Hamilton E, Mustafa M, de Wit C, et al. Evaluation of a quantitative product-enhanced reverse transcriptase assay to monitor retrovirus in mAb cell-culture. Biologicals. 2002;30:15–26. doi: 10.1006/biol.2001.0290. [DOI] [PubMed] [Google Scholar]
- 48.Brorson K, Swann PG, Lizzio E, Maudru T, Peden K, Stein KE. Use of a quantitative product-enhanced reverse transcriptase assay to monitor retrovirus levels in mAb cell-culture and downstream processing. Biotechnol Prog. 2001;17:188–196. doi: 10.1021/bp000153q. [DOI] [PubMed] [Google Scholar]
- 49.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.