Abstract
Breast cancer is one of the most common cancers among women in the United States. Although there are effective drugs, such as cisplatin, for treating advanced cancers, many patients eventually develop resistance. MicroRNAs (miRNAs) have emerged to play important roles in tumorigenesis and drug resistance. In this study, the authors observed a significant upregulation of miR-203 expression in human breast cancer tissues as compared to patient-matched nontumor breast tissues. Knockdown of miR-203 following cisplatin treatment enhances p53, p21, and Bax protein expression. Furthermore, knockdown of miR-203 sensitized human breast cancer MCF-7 cells to cisplatin-mediated apoptotic cell death, as evident from caspase-9 and caspase-7 activation, and poly(ADP-ribose) polymerase (PARP) cleavage. Moreover, the authors have demonstrated that suppressor of cytokine signaling 3 (SOCS3) is a novel target of miR-203, and cisplatin treatment in miR-203 knockdown MCF-7 cells enhanced SOCS3 expression. Exogenous expression of SOCS3 in MCF-7 cells increased sensitization to cisplatin-mediated apoptosis. Together, the results suggested a novel role of miR-203 in conferring cisplatin resistance through suppression of SOCS3, implicating an additional therapeutic strategy may be helpful to overcome cisplatin resistance for breast cancer patients.
Keywords: microRNA 203, breast cancer, SOCS3
Introduction
Breast cancer is the leading cause of cancer incidence and the second leading cause of cancer mortality in women. The chance of a woman having invasive breast cancer some time during her life is high. The cellular basis for the efficacy of endocrine therapy treatment is principally through inhibitory effects on the tumor cell cycle.1 Despite the use of adjuvant endocrine treatment, prognosis remains poor for a significant population of patients with breast cancer.2 Therefore, additional therapeutic modalities are needed.
Breast cancer is a disease involving multistep changes in the genome.3 However, studies so far have focused mostly on the role of protein-coding genes in this disease. MicroRNAs (miRNAs) are a class of small noncoding RNAs that are processed by Dicer from precursors with a characteristic hairpin secondary structure.4 In mammalian cells, miRNAs affect gene silencing via both translational inhibition and mRNA degradation.5 An individual miRNA is capable of regulating dozens of distinct mRNAs, and together the >1000 human miRNAs are believed to modulate more than one-third of the mRNA species encoded in the genome.6 miRNAs play a role in growth control, and a relation between miRNAs and cancer is anticipated.7-9 Moreover, miRNAs involved in specific networks, such as the apoptotic, proliferation, or receptor-driven pathways, could likely influence the response to targeted therapies or to chemotherapy. Response to chemotherapy varies widely in patients with breast cancer. Several studies have suggested that miRNAs are novel players in the development of chemoresistance. miRNAs are differentially expressed in chemosensitive and chemoresistant cells.10,11 For example, miR-98, miR-21, and miR-125b have been shown to potentiate chemoresistance.12-14
Many chemotherapeutic regimens involved in breast cancer treatment include anthracyclines and platinum derivatives.15 Cisplatin belongs to the group of alkylating agents. It binds to DNA bases, causing crosslink and breaks in DNA strands interfering with DNA replication.16 Despite initial clinical response, patients develop some degree of resistance. The mechanism for cisplatin resistance is not fully understood. miRNA has emerged to play an important role in drug resistance, but the cross-talk between miRNA and cisplatin resistance has not been explored.
We have performed miRNA expression profiling in breast cancer lines and identified that miR-203 expression was significantly upregulated. In this study, we have shown that miR-203 is expressed at higher levels in breast cancer tissues as compared to the adjacent noncancer breast tissues. Knockdown of miR-203 enhanced cisplatin sensitivity in MCF-7 cells and induced apoptosis. We also identified the suppressor of cytokine signaling 3 (SOCS3) as a novel target of miR-203, which is downregulated in breast cancer, and SOCS3 expression was enhanced in miR-203 knockdown breast cancer cells. Restoration of SOCS3 in MCF-7 cells augmented cisplatin-mediated apoptosis. Our data here suggested that knockdown of miR-203 might hold promise as an additional therapeutic strategy for breast cancer patients to overcome cisplatin resistance.
Results
Expression of miR-203 is enhanced in breast cancer tissues
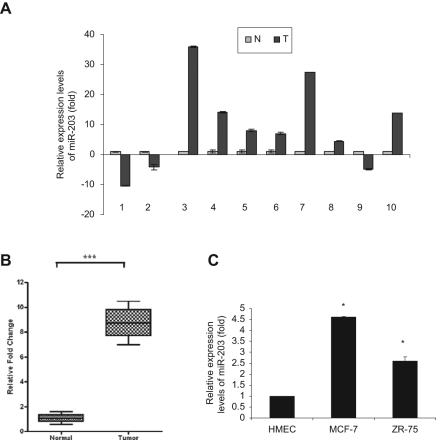
Preliminary analysis comparing the expression profile of miRNA between breast cancer cell lines and mammary epithelial cells indicated an alteration of the expression of several miRNAs (unpublished observation). We chose to focus our investigation on miR-203 because it was significantly upregulated (~4.15-fold). Furthermore, miRNA expression profiles suggested that miR-203 was upregulated in breast cancer, ovarian cancer, bladder cancer, and colon cancer.17-20 We next examined clinical breast tumor specimens to assess miR-203 expression. Clinical specimens were collected from breast cancer patients undergoing surgery after obtaining consent under an institutional review board–approved protocol. After the tissues were taken for clinical and histological evaluation, remaining discarded tissues were used in this study. Expression of miR-203 was examined by real-time polymerase chain reaction (PCR) in breast cancer tissues and compared to patient-matched control noncancerous tissues. miR-203 expression was higher in tumor tissues (7 of 10) than in patient-matched noncancerous tissues (Fig. 1A). The abundance of miR-203 expression was significantly higher in breast tumor specimens as compared to normal noncancerous tissues (Fig. 1B; P < 0.0007, paired two-tailed t test). Due to the limited number of specimens, we could not correlate the miR-203 expression with the stages of disease. Next, we measured the miR-203 expression level in different breast cancer cell lines. miR-203 was upregulated in breast cancer cells (MCF-7 and ZR-75) compared to primary human mammary epithelial cells (Fig. 1C).
Figure 1.
miR-203 expression is higher in breast cancer tissues. Total RNA was extracted from breast cancer clinical specimens (T) and patient-matched nontumor breast tissues (N). (A) miR-203 expression was analyzed by real-time polymerase chain reaction (PCR), and U6 was used as an internal control. Results are displayed on fold difference (value of nontumor breast tissues is arbitrarily set as 1). (B) Comparison of miR-203 level in normal (noncancerous tissues) (n = 10) and breast tumor tissues (n = 10). ***P < 0.0007. (C) miR-203 expression in primary human mammary epithelial cells (HMECs) and breast cancer cells (MCF-7 and ZR-75). The results are presented from three independent experiments (*P < 0.001).
Knockdown of miR-203 sensitizes MCF-7 cells to cisplatin-induced cell death
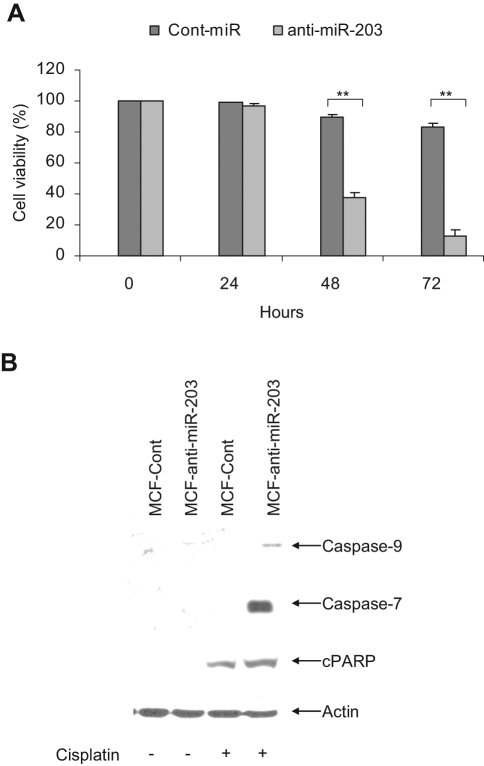
To understand the significance of higher miR-203 in breast cancer cells, we introduced anti-miR-203 and anti-Cont-miR to MCF-7, ZR-75, and MDA-MB-231 cells. Cell viability was assessed to examine the effect of knockdown miR-203 on cell growth in breast cancer cells. We observed similar cell viability in all the cell lines, suggesting that knockdown miR-203 did not affect cell proliferation. The MCF-7 cell line is more cisplatin resistant as compared to other breast cancer cell lines.21 Considering the higher expression level of miR-203 in MCF-7 cells (~4.5-fold) compared to other breast cancer ZR-75 cells (~2.4-fold), we speculated that miR-203 may be involved in the regulation of cisplatin resistance in MCF-7 cells. We therefore examined the effect of miR-203 in chemosensitivity. For this, we initially treated MCF-7 cells with different doses of cisplatin (10, 20, 50, 75, and 100 µM) for 72 hours, and cell viability was determined. MCF-7 cells treated with 50 µM cisplatin displayed <20% cell death and were used for the subsequent studies. MCF-7 cells were transduced with replication-deficient lentivirus expressing anti-miR-203 or control anti-miR. Cells were selected with puromycin and pooled and established a stable cell line (MCF-anti-miR-203 or MCF-Cont). miR-203 expression was inhibited (~5-fold) in MCF-anti-miR-203 cells as compared to MCF-Cont cells (data not shown). Next, MCF-anti-miR-203 or MCF-Cont cells were treated with 50 µM cisplatin for different time points, and cell viability was measured. MCF-anti-miR-203 cell viability was much lower following cisplatin treatment as compared to MCF-anti-miR cells (Fig. 2A), suggesting that knockdown of miR-203 sensitizes these cells to cisplatin.
Figure 2.
Cisplatin treatment in miR-203 knockdown MCF-7 cells induces apoptotic cell death. (A) MCF-7 cells were transduced with replication-deficient lentivirus expressing anti-miR-203 or control miRNA, and stable cell lines were generated. Cells were then treated with cisplatin at different time points, and cell viability was determined by trypan blue exclusion. Results are presented as the mean of three separate experiments with standard errors (**P < 0.001). (B) MCF-anti-miR-203 and MCF-Cont (anti-miR) cells were treated with cisplatin for 48 hours, and cell lysates were subjected to Western blot analysis using specific antibody for detection of caspase-9, caspase-7, or poly(ADP-ribose) polymerase (PARP). Caspase-9 and caspase-7 activation was observed in cisplatin-treated MCF-anti-miR-203 cells. PARP was significantly cleaved to an 86-kD signature peptide (cPARP) in cisplatin-treated MCF-anti-miR-203 cells as compared to MCF-Cont. The blot was reprobed with an antibody to actin for comparison of equal protein load.
Cisplatin treatment induces apoptosis in miR-203 knockdown MCF-7 cells
We next investigated the mechanism of cell death in cisplatin-treated MCF-anti-miR-203 cells. MCF-anti-miR-203 cells induced caspase-9 and caspase-7 activation following cisplatin treatment as compared to MCF-Cont cells. Both procaspase-9 and procaspase-7 were cleaved to the 37-kD and 20-kD protein band, respectively (Fig. 2B). Poly(ADP-ribose) polymerase (PARP) was significantly cleaved to an 86-kD signature peptide (cPARP) in cisplatin-treated MCF-anti-miR-203 cells as compared to that of MCF-Cont cells (Fig. 2B). Therefore, it is conceivable that knockdown of miR-203 in MCF-7 cells makes them chemotherapeutically drug sensitive and induces apoptosis.
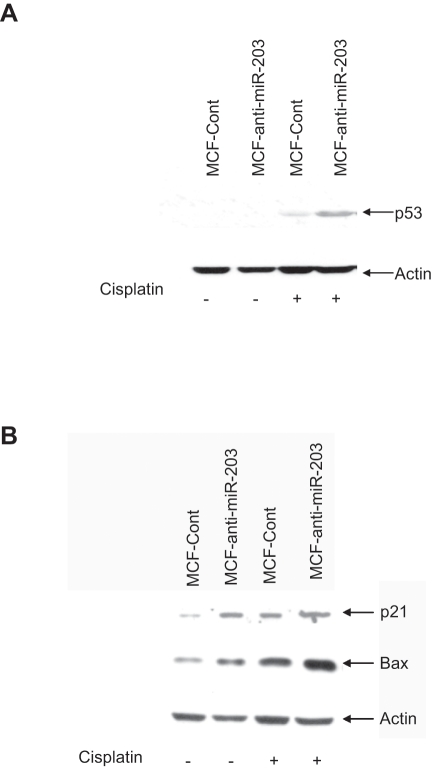
Cisplatin treatment in miR-203 knockdown MCF-7 cells induces p53 and Bax expression
Cisplatin-induced DNA damage is responsible for its anticancer effects. Cisplatin treatment results in ATR activation, which in turn phosphorylates and activates Chk2, followed by p53 activation, leading to pro-apoptotic gene expression and cell death.22 We therefore examined the expression level of p53 in MCF-anti-miR-203 and MCF-Cont cells treated with cisplatin. Knockdown of miR-203 induced the p53 expression in cisplatin-treated MCF-7 cells (Fig. 3A). We next examined the expression level of downstream molecules in the p53 signaling pathway. miR-203 knockdown MCF-7 cells, when treated with cisplatin, displayed a higher level of p21 and Bax expression (Fig. 3B).
Figure 3.
Cisplatin treatment in miR-203 knockdown MCF-7 cells enhances p53 and Bax expression. MCF-anti-miR-203 and MCF-Cont cells were treated with cisplatin for 48 hours. Cell lysates were analyzed for p53 (A) and p21 and Bax (B) expression. The blot was reprobed with an antibody to actin for comparison of equal protein load.
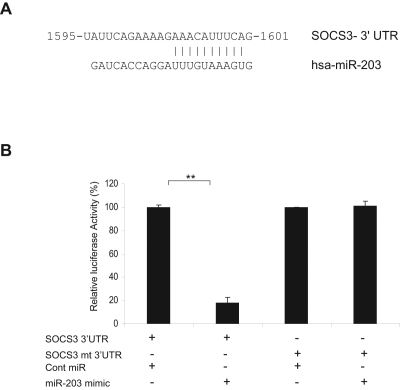
SOCS3 is a direct target of miR-203
To identify the underlying mechanisms of how knockdown of miR-203 sensitizes MCF-7 cells to cisplatin, potential miR-203 target genes were searched using TargetScan and miRanda computer prediction methods. We found a putative miR-203 target site in the SOCS3 3′ UTR (Fig. 4A). This putative binding site retains perfect conservation in the SOCS3 3′ UTR of human, primate, mouse, rat, and frog genomes. SOCS3, as a key regulator of cytokine signaling, has the potential to modulate numerous cellular processes. SOCS3 has an antitumorigenic role in hepatocarcinoma23,24 and is silenced in hepatocellular and adenocarcinoma.25,26 Reduced SOCS3 expression in breast cancer could also be correlated to reduced lymph node metastasis.27 SOCS3 promotes p53-dependent p21 expression in response to DNA damage.28 Recently, Barclay et al.29 have shown that SOCS3 expression acts as an antiproliferative agent in breast cancer cells. We next examined the interaction between miR-203 and 3′ UTR of SOCS3 by luciferase assay. The results demonstrated that miR-203 repressed wild-type SOCS3 3′ UTR directly, as evident from decreased the luciferase activity, but mutant 3′ UTR of SOCS3 luciferase activity was unaffected by miR-203 (Fig. 4B).
Figure 4.
miR-203 targets suppressor of cytokine signaling 3 (SOCS3). (A) Computational analysis of the SOCS3 3′ UTR revealed a single putative miR-203 binding site with base complementarity at positions 2-11 relative to the miR-203 5′ terminus. The miR-203 target region of SOCS3 (NM_003955) is indicated, with numbering from the first nucleotide of the SOCS3 3′ UTR. (B) Luciferase assays showing decreased luciferase activity after co-transfection of SOCS3 3′ UTR with miR-203 in MCF-7 cells. The mutant 3′ UTR of SOCS3 had no effect on reporter activity. The results are presented as mean ± standard error from three independent experiments. **P < 0.001.
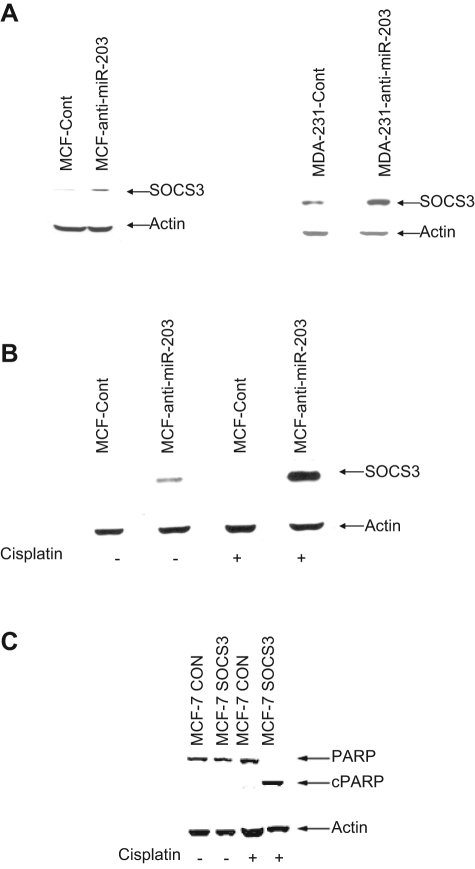
To examine whether miR-203 knockdown enhances SOCS3 expression, we performed Western blot analysis in MCF-7 or MDA-MB-231 cells with anti-miR-203 or anti-cont-miR. A significant upregulation of SOCS3 expression in MCF-7 or MDA-MB-231 cells expressing anti-miR-203 was observed (Fig. 5A). Since the expression level of SOCS3 is very low in MCF-7 cells, we could not detect a significant difference in SOCS3 level when cells were treated with mimic miR-203. Moreover, we observed that MCF-anti-miR-203 cells when treated with cisplatin displayed a significantly higher level of SOCS3 expression as compared to control cells (Fig. 5B). Together, our results suggested that knockdown miR-203 sensitizes MCF-7 cells to cisplatin by negatively regulating SOCS3, which, in turn, induces apoptotic cell death.
Figure 5.
Knockdown of miR-203 enhances suppressor of cytokine signaling 3 (SOCS3) expression in MCF-7 cells. (A) MCF-Cont and MCF-anti-miR-203 or MDA-231-Cont and MDA-231-anti-miR-203 cell lysates were analyzed for SOCS3 protein expression by Western blot. The blot was reprobed with actin antibody to compare protein load. (B) MCF-anti-miR-203 and MCF-Cont cell treated with cisplatin displayed a significantly higher level of SOCS3 expression. The blot was reprobed with an antibody to actin for comparison of equal protein load. (C) SOCS3 sensitizes MCF-7 cells to cisplatin. MCF-Cont and MCF-7 SOCS3 cells were treated with 50 µM cisplatin for 24 hours. Cell lysates was prepared and probed with poly(ADP-ribose) polymerase (PARP) antibody. The blot was reprobed with an antibody to actin for comparison of equal protein load. cPARP = cleaved PARP.
SOCS3 plays an important role in cisplatin-induced apoptosis
To examine the relation of SOCS3 and cisplatin-induced cell cytotoxicity, we generated a stable MCF-7 cell line expressing exogenous SOCS3. Vector-transfected stable cells were established as parallel control. Cells were left untreated or treated with 50 µM cisplatin. MCF-7 cells expressing SOCS3 displayed cell death significantly following 24 hours of cisplatin treatment as compared to control cells. PARP cleavage was also dramatically enhanced in MCF-7 cells expressing exogenous SOCS3 after treatment with cisplatin (Fig. 5C). These results further indicate that the overexpression of SOCS3 could efficiently enhance cisplatin-mediated cell cytotoxicity in breast cancer cells.
Discussion
In this study, we demonstrated that miR-203 is upregulated in breast cancer tissues as compared to the noncancer breast tissues, suggesting that miR-203 may function as an oncogene in breast cancer. However, we did not observe a cell growth regulatory effect after knocking down miR-203 in breast cancer cells. Little is known about the role of miRNAs in chemoresistance, such as to cisplatin. MCF-7 cells are more resistant to cisplatin compared to other breast cancer cells.21 Our results suggested that knockdown of miR-203 is involved in cisplatin chemosensitivity in breast cancer cells and induced apoptotic cell death. We also observed the enhanced p53, p21, and Bax expression in cisplatin-treated miR-203 knockdown MCF-7 cells. To our knowledge, this is the first report demonstrating the involvement of miR-203 in promoting cisplatin chemosensitivity. These results suggested that the anti-apoptotic effect is a key mechanism of miR-203-mediated cisplatin resistance in breast cancer cells.
Several miRNAs have been identified as oncogenes or tumor suppressors and are involved in cell proliferation, differentiation, apoptosis, and drug resistance.7,30 The miR-17-92 cluster functions as an oncogene and is upregulated in lung cancer,31 lymphomas,32 and renal cancer carcinoma.33 miR-21 is upregulated in glioblastoma cells,34 and knockdown of miR-21 activates caspases and leads to apoptosis.35 Recently, it has been shown that miR-125b is involved in Taxol resistance by targeting Bak1.14 miRNAs may have different functions in different tissues. For example, miR-31 is overexpressed in lung cancer, and inhibition of miR-31 has suppressed the cell growth by targeting tumor suppressor LATS2 and PP2A.36 On the other hand, the expression of miR-31 is inversely correlated with metastasis in human breast cancer patients, and miR-31 has inhibited breast cancer metastasis.37 miR-203 expression recently has been reported to be inhibited in prostate cancers, and overexpression of miR-203 has suppressed the metastasis and development of prostate cancer cells.38
Subsequently, we identified SOCS3 as a novel target for miR-203. SOCS3 expression is reduced in breast cancer tissues.27 Overexpression of SOCS3 has been shown to have an antiproliferative effect.29 Our results suggested that SOCS3 expression is significantly higher in miR-203 knockdown cisplatin-treated MCF-7 cells. The mechanistic data suggest that overexpression of SOCS3 in MCF-7 cells sensitized these cells in cisplatin-mediated cell death by inducing p53, p21, and Bax. Therefore, it is conceivable that the miR-203-SOCS3 axis is critical for cisplatin resistance in breast cancer cells and may potentially serve as a therapeutic target to overcome cisplatin resistance. To date, several targets of miR-203 have been identified. One recent study showed that survivin is a direct target of miR-203, and overexpression of miR-203 enhances survivin-mediated apoptosis in human prostate cancer cells.38 Interestingly, we did not observe a detectable change in survivin expression in miR-203 mimic or anti-miR-treated MCF-7 cells (data not shown). One possible explanation is the known diverse nature of miRNA target genes. The net effect of changes in the expression of a miRNA appears to be the sum of all impacts on its targets in a cell type–specific and phenotype-specific manner.39 While our manuscript was in preparation, Akt2 was identified as a target of miR-203.40 However, prolonged ablation of Akt2 with siRNA resulted in cell cycle arrest in G0/G1 by downregulating Cdk2 and cyclin D and upregulating p27 in breast cancer cells,41 and we therefore did not examine the status of AKT2 in our experimental system. It will be important to examine in future whether knockdown of miR-203 in breast cancer cells sensitizes other chemotherapeutic drugs.
In summary, we observed increased expression of miR-203 in breast cancer tissues and have shown for the first time that knockdown of miR-203 sensitized MCF-7 cells to cisplatin. Moreover, we demonstrated that SOCS3 is a novel target of miR-203 and plays an important role in cisplatin sensitivity of breast cancer cells. These findings have important implications in the development of targeted therapeutics against cisplatin resistance in human breast cancer.
Materials and Methods
Cell culture and transfection
Breast cancer cell lines (MCF-7, ZR-75, and MDA-MB-231) were procured from the American Type Culture Collection (Mananssas, VA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 100 units/mL penicillin, and 100 µg/mL streptomycin in a humidified CO2 incubator. Primary human mammary epithelial cells (HMECs) were obtained from Lonza (Basel, Switzerland) and maintained for short time in specified media. Transfections were carried out using Lipofectamine or Lipofectamine 2000 (Invitrogen, Carlsbad, CA) using the manufacturer’s protocol.
Antibodies and miRNAs
Rabbit anti-PARP, rabbit anti–cleaved PARP, rabbit anti-caspase-7, and rabbit anti- caspase-9 antibodies were purchased from Cell Signaling Technology (Danvers, MA). Mouse anti-p53-HRP, mouse anti-p21, mouse anti-Bax, rabbit anti-SOCS3, and goat anti-actin-HRP antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Anti-miR miRNA inhibitor negative control #1 (anti-cont-miR) and anti-miR-203 were purchased from Invitrogen. Replication-deficient lentivirus expressing miRZip anti-miR-203 or control (purchased from System Biosciences, Mountain View, CA) was used for generation of stable transfectants after selection with puromycin.
Western blotting
Breast cancer cells were transduced with replication-deficient lentivirus expressing anti-miR-203 or control miRNA. Cells were selected with puromycin and pooled and established a stable cell line (MCF-anti-miR-203 or MCF-Cont). Cell lysates were prepared after 48 hours of cisplatin treatment in 2× sodium dodecyl sulfate (SDS) sample buffer and analyzed for Western blot analysis. The blots were reprobed with actin to compare protein load in each lane.
Cell proliferation assay
MCF-anti-miR-203 or MCF-Cont cells were then treated with predetermined doses of cisplatin at different time points, and cell viability was determined by trypan blue exclusion.
Quantitative real-time reverse transcription PCR
miRNA expression was determined by isolating total RNA using TRIzol reagent (Invitrogen). Total RNA was reverse transcribed using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA) with miRNA-specific primers. To quantify the miRNA levels, the 7500 Real-Time System (Applied Biosystems) was used in conjunction with gene-specific TaqMan assay kits (Applied Biosystems). U6 was used as an endogenous control to normalize expression. Relative miR-203 expression and standard error were calculated by the supplied 7500 Real-Time System software.
Luciferase assay
The miR-203 binding sites from 3′ UTR SOCS3 or mutant 3′ UTR was cloned into the pMIR-reporter luciferase vector (Ambion, Austin, TX). For reporter assay, 30 nM miR-203 mimic or control miRNA was co-transfected with 0.3 µg of the pMIR 3′ UTR wild-type or mutant plasmid DNAs into MCF-7 cells in 24-well plates using Lipofectamine 2000. Luciferase activity was measured 48 hours posttransfection as described previously.42
Statistical analysis
Results were expressed as the mean ± standard deviation (SD), and statistical analyses were performed using two-tailed paired or unpaired Student t test or one-way analysis of variance (ANOVA) in GraphPad Prism 5 (GraphPad, La Jolla, CA). A P value of <0.05 was considered statistically significant.
Acknowledgments
The authors thank Jei Chen for providing SOCS3 plasmid DNA.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
This work was supported by a Pathology Department Doisy Research Fund to RBR from Saint Louis University.
References
- 1. Ellis LM, Hicklin DJ. VEGF-targeted therapy: mechanisms of anti-tumour activity. Nat Rev Cancer. 2008;8:579-91 [DOI] [PubMed] [Google Scholar]
- 2. Crowder RJ, Phommaly C, Tao Y, et al. PIK3CA and PIK3CB inhibition produce synthetic lethality when combined with estrogen deprivation in estrogen receptor–positive breast cancer. Cancer Res. 2009;69:3955-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol. 2011;223:307-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807-18 [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol Oncol. 2009;113:249-55 [DOI] [PubMed] [Google Scholar]
- 11. Xia L, Zhang D, Du R, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372-79 [DOI] [PubMed] [Google Scholar]
- 12. Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moriyama T, Ohuchida K, Mizumoto K, et al. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol Cancer Ther. 2009;8:1067-74 [DOI] [PubMed] [Google Scholar]
- 14. Zhou M, Liu Z, Zhao Y, et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem. 2010;285:21496-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. dit Faute MA, Laurent L, Ploton D, Poupon MF, Jardillier JC, Bobichon H. Distinctive alterations of invasiveness, drug resistance and cell-cell organization in 3D-cultures of MCF-7, a human breast cancer cell line, and its multidrug resistant variant. Clin Exp Metastasis. 2002;19:161-68 [DOI] [PubMed] [Google Scholar]
- 16. McWhinney SR, Goldberg RM, McLeod HL. Platinum neurotoxicity pharmacogenetics. Mol Cancer Ther. 2009;8:10-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith DD, Saetrom P, Snøve O, Jr, et al. Meta-analysis of breast cancer microarray studies in conjunction with conserved cis-elements suggest patterns for coordinate regulation. BMC Bioinformatics. 2008;9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699-707 [DOI] [PubMed] [Google Scholar]
- 19. Gottardo F, Liu CG, Ferracin M, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387-92 [DOI] [PubMed] [Google Scholar]
- 20. Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yde CW, Issinger OG. Enhancing cisplatin sensitivity in MCF-7 human breast cancer cells by down-regulation of Bcl-2 and cyclin D1. Int J Oncol. 2006;29:1397-404 [PubMed] [Google Scholar]
- 22. Sangster-Guity N, Conrad BH, Papadopoulos N, Bunz F. ATR mediates cisplatin resistance in a p53 genotype-specific manner. Oncogene. 2011;30:2526-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogata H, Kobayashi T, Chinen T, et al. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology. 2006;131:179-93 [DOI] [PubMed] [Google Scholar]
- 24. Riehle KJ, Campbell JS, McMahan RS, et al. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J Exp Med. 2008;205:91-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tischoff I, Hengge UR, Vieth M, et al. Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett’s adenocarcinoma. Gut. 2007;56:1047-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niwa Y, Kanda H, Shikauchi Y, et al. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 2005;24:6406-17 [DOI] [PubMed] [Google Scholar]
- 27. Nakagawa T, Iida S, Osanai T, et al. Decreased expression of SOCS-3 mRNA in breast cancer with lymph node metastasis. Oncol Rep. 2008;19:33-9 [PubMed] [Google Scholar]
- 28. Sitko JC, Yeh B, Kim M, et al. SOCS3 regulates p21 expression and cell cycle arrest in response to DNA damage. Cell Signal. 2008;20:2221-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barclay JL, Anderson ST, Waters MJ, Curlewis JD. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int J Cancer. 2009;124:1756-66 [DOI] [PubMed] [Google Scholar]
- 30. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628-32 [DOI] [PubMed] [Google Scholar]
- 32. He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chow TF, Mankaruos M, Scorilas A, et al. The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. J Urol. 2010;183:743-51 [DOI] [PubMed] [Google Scholar]
- 34. Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994-9000 [DOI] [PubMed] [Google Scholar]
- 36. Liu X, Sempere LF, Ouyang H, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298-309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032-46 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38. Saini S, Majid S, Yamamura S, et al. Regulatory role of miR-203 in prostate cancer progression and metastasis. Clin Cancer Res. 2011;17:5287-98 [DOI] [PubMed] [Google Scholar]
- 39. Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li J, Chen Y, Zhao J, Kong F, Zhang Y. miR-203 reverses chemoresistance in p53-mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett. 2011;304:52-9 [DOI] [PubMed] [Google Scholar]
- 41. Santi SA, Lee H. Ablation of Akt2 induces autophagy through cell cycle arrest, the downregulation of p70S6K, and the deregulation of mitochondria in MDA-MB231 cells. PLoS One. 2011;6:e14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steele R, Mott JL, Ray RB. MBP-1 upregulates miR-29b that represses Mcl-1, collagens, and matrix-metalloproteinase-2 in prostate cancer cells. Genes Cancer. 2010;1:381-7 [DOI] [PMC free article] [PubMed] [Google Scholar]