Abstract
Over 4,000 putative proteorhodopsins (PRs) have been identified throughout the oceans and seas of the Earth. The first of these eubacterial rhodopsins was discovered in 2000 and has expanded the family of microbial proton pumps to all three domains of life. With photophysical properties similar to those of bacteriorhodopsin, an archaeal proton pump, PRs are also generating interest for their potential use in various photonic applications. We perform here the first reconstitution of the minimal photoactive PR structure into nanoscale phospholipid bilayers (nanodiscs), to better understand how protein-protein and protein-lipid interactions influence the photophysical properties of PR. Spectral (steady-state and time-resolved UV-visible spectroscopy) and physical (size-exclusion chromatography and electron microscopy) characterization of these complexes confirms the preparation of a photoactive PR monomer within nanodiscs. Specifically, when embedded within a nanodisc, monomeric PR exhibits a titratable pKa (6.5–7.1) and photocycle lifetime (~100–200 ms) that is comparable to the detergent solubilized protein. These ndPRs also produce a photoactive blue-shifted absorbance, centered at 377 or 416 nm, that indicates protein-protein interactions from a PR oligomer are required for a fast photocycle. Moreover, we demonstrate how these model membrane systems allow modulation of the PR photocycle by variation of the discoidal diameter (i.e., 10 or 12 nm), bilayer thickness (i.e., 23 or 26.5 Å), and degree of saturation of the lipid acyl chain. Nanodiscs also offer a highly stable environment of relevance to potential device applications.
Keywords: proteorhodopsin, monomer, nanodisc, photocycle, phospholipid
Introduction
The discovery of proteorhodopsin (PR) in 2000 has expanded the family of microbial proton pumps to all three domains of life.1,2 Over 4,000 putative PRs have been identified throughout the oceans and seas of Earth and are classified as either blue-absorbing (λmax ~ 494 nm) or green-absorbing (λmax ~ 520 nm) variants.3–7 These proteins are identified solely from genetic analysis of coastal and oceanic water samples and no native host has yet been cultivated.4,8 Hence, PRs are often expressed in Escherichia coli and solubilized in various detergents9–12 or reconstituted in phospholipid environments13–16 to stabilize the protein prior to investigation.
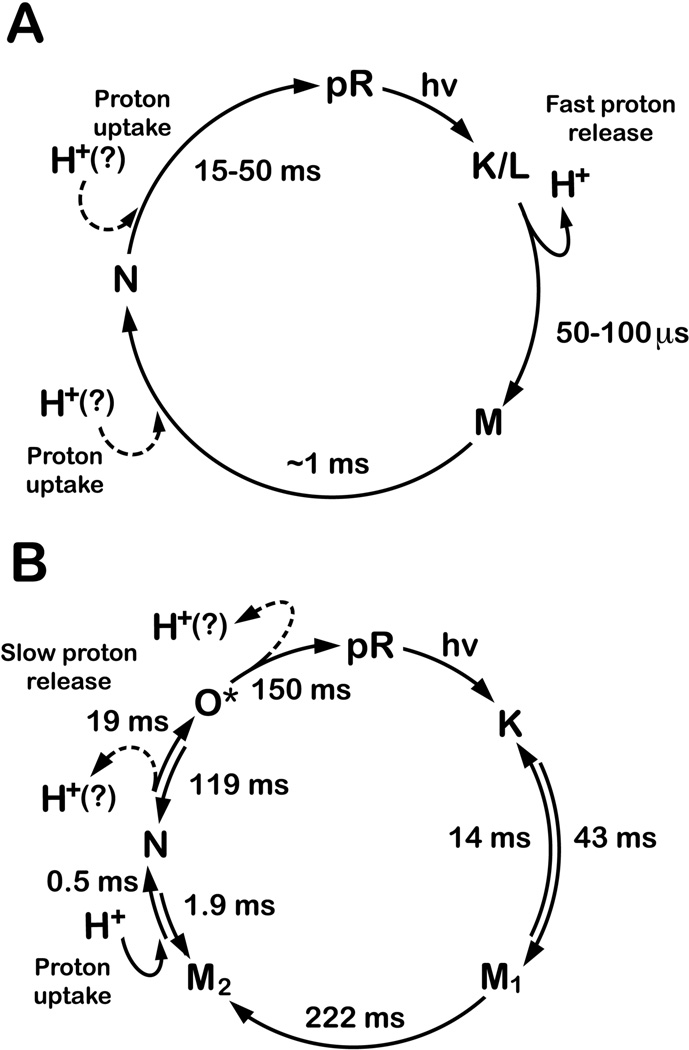
Solubilization of PR into a non-ionic detergent, which is used to purify the protein from the E. coli membrane, alters the photocycle speed and delays proton release until the end of the photocycle (Fig. 1). The green-absorbing PR is activated at alkaline pH in either a phospholipid or detergent environment,13,16,17 which corresponds well with the pH of the ocean near the surface (~8.0). Photoexcitation of PR results in the translocation of a proton across the membrane.13,14 In an E. coli membrane, a fast proton release (<100 µs) is observed early in a photocycle that requires between 15 and 50 ms to complete.16,18 Proton release coincides with the formation of the M state (λmax ~ 400 nm).14,16 Whether proton uptake occurs before or after the N state (λmax ~ 560 nm) has not yet been determined. However, this event is known to occur late in the photocycle when the protein is suspended in a phospholipid bilayer.19 In a non-ionic detergent, these events are delayed as proton uptake reportedly precedes proton release before and after the red-shifted N state, respectively.9,11 The photocycle lifetime is also increased to approximately 300–400 ms in a detergent suspension.11 The influence of the lipid environment on the PR photocycle remains largely unknown.
Figure 1.
The proteorhodopsin photocycle when the protein is suspended in a (A) phospholipid bilayer,14,16,19 or (B) detergent suspension.9,11 Lifetimes are shown for select photostates that have been resolved in the cited references. The O* state of the detergent photocycle represents the pR′ (O) state, which is only observed by kinetic analysis11 or FT-IR spectroscopy,9 and is structurally identical to the pR resting state. Similarly, the M2 state of the detergent photocycle was kinetically resolved from the absorption data at 420 nm.
Nanodiscs are model membrane structures that use membrane scaffolding proteins (MSPs) to isolate and directly control the properties of membrane proteins. These MSPs, which are derived from the human serum apolipoprotein A1, non-covalently interact with the hydrophobic acyl chains of phospholipids by hydrophobic interactions to produce soluble, nanoscale phospholipid bilayers.20,21 Nanodiscs have minimal effects on the physical properties (e.g., fluidity) of lipids;22,23 hence, these structures are excellent biomimetic systems for studying a variety of membrane proteins24–26 and the structure-function requirements for the active protein27–29 or complexes.30–32 Moreover, control of the scaffolding belt size (i.e., the disc diameter) has allowed researchers to isolate the monomeric and multimeric protein conformations for proteins such as bacteriorhodopsin.32–34 Lastly, these structures are being used in a wide variety of experimental settings that include NMR,35 surface plasmon resonance,36 as well as UV-visible,29 infra-red37 and fluorescence28,38 spectroscopy. Nanodiscs thusly offer a versatile tool to deconstruct the factors that control the PR photocycle. We utilize nanodiscs to isolate PR monomers and analyze the aspects that influence the PR photocycle, focusing on the more extensively studied green-absorbing PR (henceforth referred to as PR).
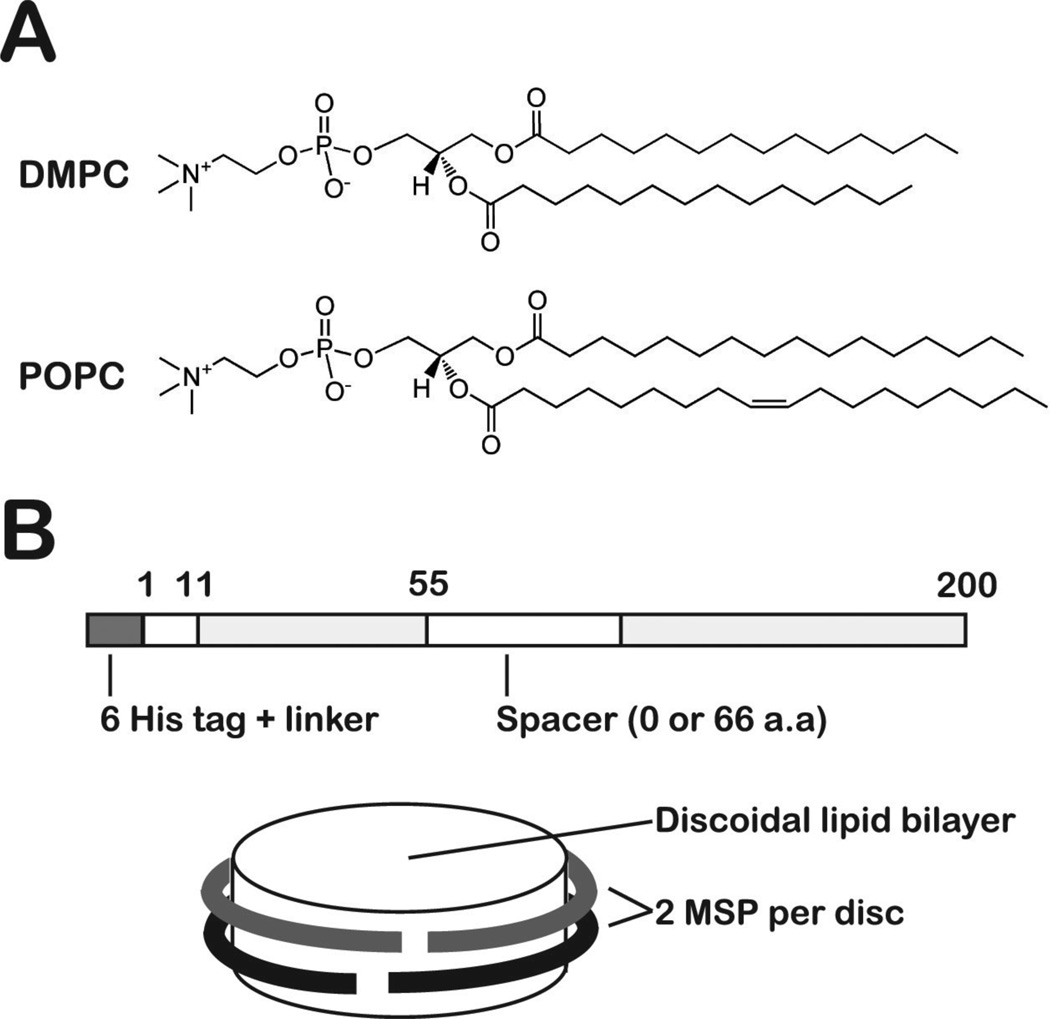
In this study, we investigate the modulation of the PR photocycle and the influence of the lipid environment by assaying PR activity in E. coli membranes (from the raw preparation lysate), octyl-β-D-glucopyranoside (OG), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC). The DMPC and POPC phospholipids, commonly used in the synthesis of nanodisc particles,27,33,39–41 have different acyl chain lengths and thusly yield bilayers with different thickness (Fig. 2A). Nanodiscs are synthesized with MSPs that contain either no spacer or three 22-residue amphipathic helical repeats to vary the diameter of these particles (Fig. 2B).21,24,33 Such modification limits the number of PRs that can be isolated per nanodisc and demonstrates the importance of protein-protein and protein-lipid interactions to the function of PR. The assembly of these nanodisc-proteorhodopsin (ndPR) complexes was confirmed via size-exclusion chromatography and electron microscopy. Photophysical characterization was carried out by using absolute and time-resolved absorption spectroscopy.
Figure 2.
Experimental design for preparing the different ndPR complexes. (A) Phospholipids used to synthesize nanodiscs with thin (DMPC, ~23 Å) or thick (POPC, ~26.5 Å) bilayers. The thickness is approximated by measuring the distance between the C2 atoms of an acyl chain or between lipids across the bilayer.73 (B) Schematic diagram of the MSP used to synthesize the nanodisc complex. The image shows the numbering for the MSP1D1 protein and was adapted from Ref. 33.
Experimental Section
Chemicals and Buffers
DMPC and POPC lipids were purchased from Avanti Polar Lipids (Alabaster, AL). All detergents and buffering chemicals were purchased from Thermo Fisher Scientific, Inc. (Pittsburg, PA) or Sigma Aldrich (St. Louis, MO). Buffers used for pH investigations, which were all 50 mM and contained 100 mM NaCl, include sodium citrate for pH 4.5, 2-(N-morpholino)ethanesulfonic acid (MES) for pH 5.5 to 6.5, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) for pH 7.0 and 7.5, tris(hydroxymethyl)methyl-3-aminopropanesulfonic Acid (TAPS) for pH 8.0 and 8.5, and N-cyclohexyl-2-aminoethanesulfonic acid (CHES) for pH 9.0 and 9.5. The assembly of nanodisc particles used MSP buffer (20 mM Tris-HCl, 100 mM NaCl, and 0.5 mM EDTA, pH 7.4) and occasionally required the addition of 59 mM cholate.
Proteorhodopsin expression & purification
Proteorhodopsin (eBAC31A08), with a 6x His tag, was overexpressed in E. coli and supplemented with all-trans retinal, solubilized in ethanol, as previously described.18 The cells were then lysed via sonication and the protein was purified via a nickel sepharose column after overnight solubilization in 1% OG. The protein was then stored in Tris buffer (100 mM Tris-HCl, pH 8.0, 0.05% (w v−1) OG) at 4°C until use.
Cloning, expression, and purification of the membrane scaffolding proteins
Membrane scaffolding proteins contained either no helical spacer (MSP1D1) or three 22-residue amphipathic helical repeats (MSP1E3D1) to control the diameter of the synthesized nanodiscs (Fig. 2B). These proteins were expressed and purified according to the standard protocols.33 In brief, plasmids (pET28a) containing either the MSP1D1 or MSP1E3D1 gene were transformed into E. coli BL21 cells. Transformants were grown on YT-Agar plates (0.5% yeast extract, 0.8% Tryptone, 86 mM NaCl, 0.75% agar, 50 mg mL−1 kanamycin, pH 7.0) overnight at 37 °C. A single colony was then transferred to 500 mL liquid LB (1% tryptone, 0.5% yeast extract, 1.0% NaCl, pH 7.4), allowed to grow to mid-log phase (OD600 ~ 0.5), and induced with 1 mM isopropyl-β-D-thiogalactoside to overexpress the MSP protein for 2.5 hr followed by isolation of cells by low-speed centrifugation. For purification of the 6x His tagged MSP, cells were disrupted by sonication and affinity purified on a Ni-NTA matrix (Qiagen) following manufacturer’s instructions.
Assembly and purification of Nanodisc particles
Solubilization of PR in non-ionic detergent was necessary for successful synthesis of ndPR particles. Triton X-100 was chosen to parallel the methods employed for the synthesis of nanodisc-bacteriorhodopsin complexes.33 This method worked well for assembly of the MSP1E3D1 ndPRs, but synthesis of the MSP1D1 complexes required overnight solubilization with 13% (w v−1) Triton X-100. Furthermore, low synthesis yields of the MSP1D1 particles were observed in all such assemblies.
Phospholipids were dried under a Nitrogen stream, reconstituted with 59 mM cholate in MSP buffer and sonicated for several minutes. Preparation of the different ndPRs generally involved mixing the MSP, phospholipids, and PR together at empirically determined molar ratios. Nanodiscs with DMPC were assembled at ratios of 1:50:0.1 for the MSP1E3D1 protein and 1:10:0.1 for the MSP1D1 protein. Nanodiscs with POPC were assembled at a ratio of 1:41:0.1 for the MSP1E3D1 protein. DMPC assemblies were incubated at ambient temperature and POPC assemblies were incubated at 4 °C for 1 hour prior to self-assembly, which was initiated by removing the detergent via the addition of ~500 mg Biobeads SM-2 (Bio-Rad, Hercules, CA). All samples were then concentrated to 1 mL via an Amicon centrifugal filter with a 10,000 MWCO (Millipore, Billerica, MA).
Purification of the assembled particles was achieved by injecting samples onto a Superdex 200 10/300 GL column (Amersham Biosciences). Fractionation was monitored at two wavelengths (i.e., 280 nm, 520 nm) and run at 0.5 mL min−1 on an AKTA FPLC (GE Healthsciences, Waukesha, WI). The column was calibrated with known standards (Sigma Aldrich): blue dextran (2 MDa; 8.3 mL), β-amylase (206 kDa; 12.2 mL; 7.4 nm), alcohol dehydrogenase (149.5 kDa; 13.5 mL), bovine serum albumin (67 kDa,; 14.3 mL; 6.58 nm), carbonic anhydrase (30 kDa; 16.7 mL; 4.0 nm), and cytochrome C (12.4 kDa; 18.1 mL; 3.76 nm). Values for the Stokes diameter of select standards were obtained from Ref. 42 and plotted by their respective elution volume.
Protein-to-protein ratio analysis
Quantification of the number of PRs per nanodisc was determined by a ratiometric analysis of ndPRs in a similar fashion to nanodisc-rhodopsin complexes.27 Bare (empty) nanodiscs, which can skew the assay, were removed prior to analysis by size-exclusion chromatography and sucrose density gradients. Linear gradients (10–45% w w−1) were centrifuged for 18 hrs at 4 °C and fractionated as 100 µL aliquots. Fractions were then analyzed by measuring the absorbance at 280 and 520 nm. Bare nanodiscs equilibrate at 1.08 g cm−1 and all ndPRs equilibrate around 1.10 g cm−3.
Extinction coefficients of MSP1D1 (21,430 M−1 cm−1 at 280 nm), MSP1E3D1 (29,910 M−1 cm−1 at 280 nm), and PR (43,900 M−1 cm−1 around 520 nm) were used to quantify these proteins by oscillator strength deconvolution of the electronic absorption spectra.43,44 This method used the oscillator strength of each component, via integration of the spectral features, to model the absorption spectra of these proteins when assembled at different ratios. Each model assumed two MSP proteins per nanodisc and varied the number of PRs as one, two or four molecules. The resulting theoretical spectra were then compared to the experimental absorbance data (Fig. S1).
Electron microscopy
Electron micrographs of negatively stained samples were collected for all ndPR complexes to confirm assembly of these particles. Prior to staining, eluted samples were collected and washed with distilled water (Millipore) and diluted to OD280 ~ 0.003. Four microliters of sample were then incubated for 30 seconds on a fresh glow discharged copper grid (mesh 400) that was covered with a carbon film. Grids were then washed twice with water and stained with 1% aqueous uranyl acetate for 1 minute. Excess stain was wicked away with filter paper then allowed to air dry before imaging. All micrographs were collected with an AMT XR-40 camera slide that was mounted on a Tecnai Biotwin G2 Spirit transmission electron microscope. All images were collected at a magnification of 180,000, with exception to DMPC liposomes (magnification = 11,000).
Absolute absorption spectra
Absorption spectra were collected on a Cary 50 UV-visible spectrometer (Varian, Inc., Palo Alto, CA) from 750 to 250 nm at ambient temperature. Samples were then normalized to their absorbance at 750 nm. The spectral titration of ndPRs involved three solvent washes with a buffered solution of a desired pH with a 30 minute incubation at room temperature between exchanges. Microcon Centrifugal Filters (10,000 Da MWCO; Millipore) were used for each solvent exchange. Spectra were collected as described above. The titration data were obtained by monitoring the absorption change at 570 nm.
Flash photolysis absorption spectra
All time-resolved experiments were conducted at 25 °C in HEPES buffer at pH 7.0 or CHES buffer at pH 9.5. Samples were concentrated to approximately 100 µL, placed in a methacrylate microcuvette (Plastibrand, Thermo Fisher Scientific, Inc.) and then sealed with parafilm to prevent evaporation. Absorption changes were monitored with a rapid-scanning monochromator (RSM-1000) system (OLIS Instruments, Inc., Bogart, GA) after photoexciting the protein sample with a 532 nm laser pulse that was orthogonal to the monitoring beam. The 3–5 ns pulse-width laser emission was from an optical parametric oscillator, which was pumped via the third harmonic of a Nd:YAG pulsed laser system (Continuum, Santa Clara, CA). All time resolved spectra were averaged to reduce the noise and normalized.
Kinetic traces were collected at either 400 or 620 nm with microsecond resolution. Data were then fit to a double exponential:
where a0 is the amplitude of the fit to the normalized data, t is the time (s), t00 is the time of the laser pulse onset (s), τform is the formation rate of the photoevent (s), τd1 is the first decay constant (s), τd2 is the second decay constant (s), w1 is weight of τd1, and (1- w1) is the weight of τd2.
Results
Solubilization of PR with Triton X-100
The non-ionic detergent Triton X-100 is conventionally used to solubilize bacteriorhodopsin. This solubilization is well characterized to produce monomeric suspensions that are accompanied by a blue shift of the absorption maximum from 568 nm to 551 nm.45–47 Following published reports for the incorporation of bacteriorhodopsin into nanodiscs,33,41 PR was incorporated into MSP1E3D1 discs after solubilization in 4% (w v−1) Triton X-100 for 1 hour. Unlike bacteriorhodopsin, however, the solubilized PR shows a subtle red shift of the absorption maximum from 521 to 535 nm. Conversely, incorporation of PR into the smaller MSP1D1 discs required more intensive solubilization (13% (w v−1) Triton X-100 for 23 hours), which results in a stronger red shift of the absorption maximum up to 540 nm. The synthesis of the MSP1D1 ndPR complex, by the aforementioned procedure, consistently produced low yields in all MSP1D1 assemblies.
Nanodisc-proteorhodopsin assembly
The synthesis of nanodisc complexes begins with incubating the MSP, lipid and desired membrane protein in the presence of excess detergent. Removal of the detergent with hydrophobic adsorbents, such as BioBeads, causes the spontaneous self-assembly of nanodiscs with the target membrane protein.20 Efficient nanodisc assembly requires optimized molar ratios of lipid:MSP:membrane protein that must be empirically determined for each target membrane protein. Analogous studies with bovine rhodopsin and bacteriorhodopsin provided useful starting points for the empirical determination of optimal ratios used to assemble ndPR complexes.27,29,33
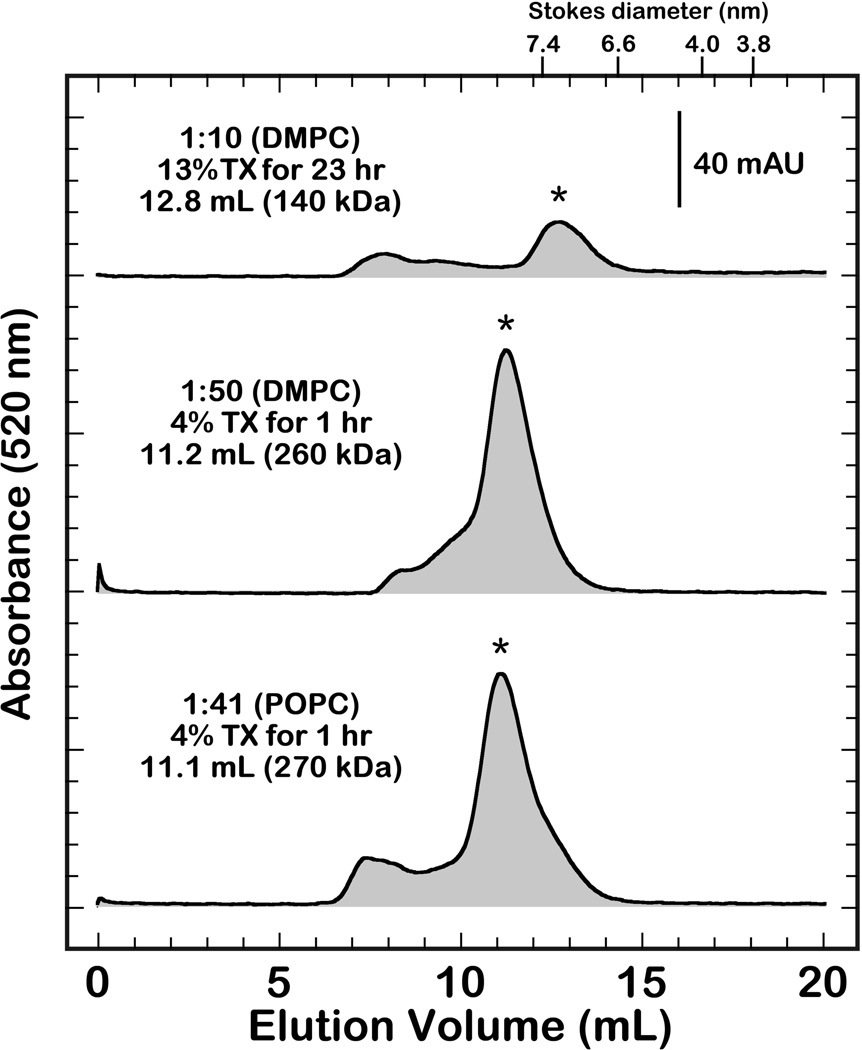
Variation of the synthesized ndPRs in this work includes those with different scaffolding proteins (MSP1D1 or MSPE3D1) and those containing different phospholipids (DMPC or POPC). Each variation required optimization of the experimental conditions (e.g., solubilization times, MSP:phospholipid:PR molar ratio) for proper nanodisc assembly. The optimized assembly parameters and resulting molecular weight, as determined by size exclusion chromatography, are defined for each sample in figure 3. These conditions were identified by the relative homogeneity, which is defined by the broadness of the eluted peak, and synthesis yield of each experiment. Electron micrographs of the isolated fractions (Fig. S2) show well-formed particles that are in excellent agreement with previous reports.24
Figure 3.
Size-exclusion chromatograms of the assembled ndPRs with MSP1D1 and DMPC (top trace), MSP1E3D1 and DMPC (middle trace), and MSP1E3D1 and POPC (bottom trace). The optimized experimental conditions are summarized for each assembly and an asterisk denotes the isolated complex. The elution of each sample was monitored at 520 nm to detect the presence of the retinal chromophore. Details for the calibration of the column are described in the Methods and Materials section.
The larger MSP1E3D1 ndPRs assemble well with similar lipid:MSP ratios and incubation parameters to those outlined for bacteriorhodopsin.33 Solubilization of PR for longer times or higher detergent concentrations results in particles with lower molecular weights and broader elution profiles (data not shown). Conversely, the assembly of appreciable amounts of ndPRs with the smaller MSP1D1 belt requires a higher detergent concentration with longer incubation times (see above). Synthesis of MSP1D1 ndPRs with shorter incubation periods or lower detergent concentrations often results in a significant formation of larger aggregate species that accumulate in the void volume.
Increasing the bilayer thickness, by assembling ndPRs with POPC, provides insight into the protein-lipid interactions that influence the PR photocycle. These lipids were only used for synthesis of the MSP1E3D1 ndPRs because of the aforementioned differences in nanodisc assemblies with DMPC. These particles are slightly larger than the DMPC discs, with a molecular weight of 270 kDa, and are produced with high yield. Stoichiometric quantification of ndPR spectra indicates that the PR and MSP1E3D1 proteins assemble with a 1:2 molar ratio.
Spectral titration
Titration of detergent solubilized PR, from pH 4.5 to 9.5, induces a spectral shift of the absorption maximum from 545 nm to 518 nm (Fig. S3A). This shift is attributed to the protonation state of Aspartic acid 97 (D97), which exhibits an apparent pKa of 7.1, and directly influences the ability of PR to function.13,17,48 Specifically, a proton is transferred from the Schiff base to D97 during the formation of the M state.9 Mutation of this residue to a neutral species (i.e., Asparagine (N)) confirms the role of D97 by increasing the pKa to >10 and abolishes proton translocation.9 Although the D85 homolog of bacteriorhodopsin has a much lower pKa (i.e., 2.3) than that of PR, similar behavior is observed with titration of the protein.49–51 Furthermore, this effect is simulated at neutral pH with the D85N mutant of bacteriorhodopsin.52,53 The spectral shifts associated with the titration of the MSP1D1 and MSP1E3D1 ndPRs thusly provide information regarding the interaction of D97 with the chromophore in these complexes.
We analyzed the titratable spectral shifts for PR following detergent solubilization in OG (Fig. S3A), within E. coli membranes (Fig. S3B), and for PR reconstituted into nanodiscs (Fig. 4). A single wavelength (i.e., 570 nm) is chosen to characterize the particles synthesized in this work because all samples exhibit spectral changes at this wavelength (Figs. S3 and 4). The pKa values determined in this work are in good agreement with the published values for PR (Tables 1 and 2). Notably, the variation observed for measured pKas may be due to different fitting methods,54 anion sensitivity,55 or reconstitution of PR in liposomes comprised of a single type13 or a complex mixture43 of synthetic phospholipids.
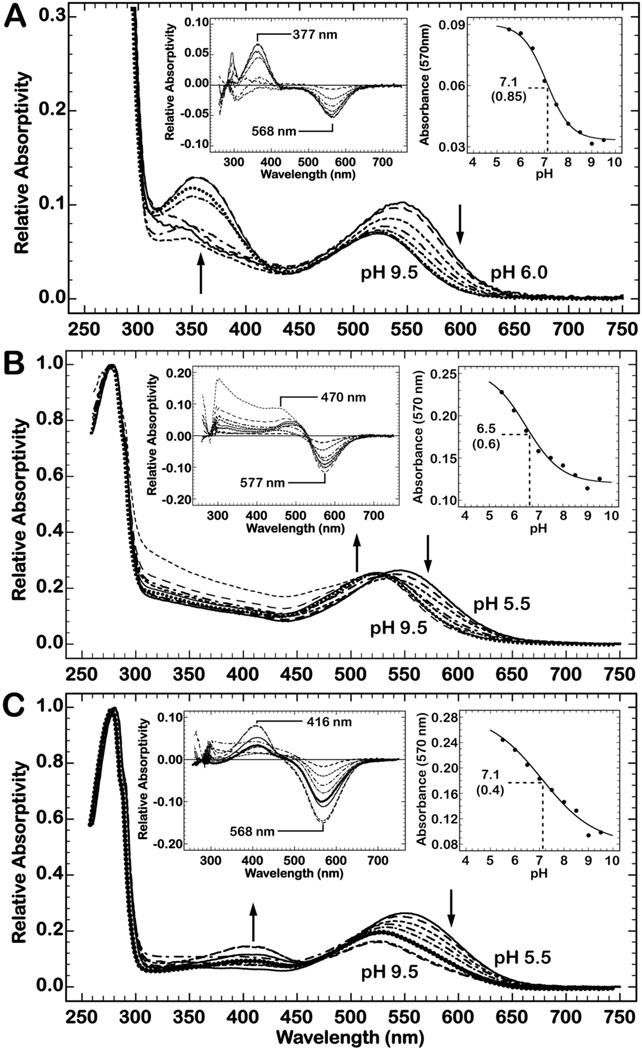
Figure 4.
Spectral titration of proteorhodopsin in (A) MSP1D1 nanodisc with DMPC, (B) MSP1E3D1 nanodisc with DMPC, and (C) a MSP1E3D1 nanodisc with POPC. Arrows indicate the direction of the titrated absorbance. The left inset image of each titration shows the difference spectra for each experiment. All difference spectra are calculated by subtraction of the low pH value shown in the absolute spectra. The right inset image shows the pH dependence of the absorbance at 570 nm, which were fit to the Henderson-Hasselbach equation to determine the pKa of D97. The apparent pKa, and respective coefficient, for each sample is 7.1 (0.85), 6.5 (0.60), and 7.1 (0.40) respectively.
Table 1.
Literature values of the alkaline absorption maximum of proteorhodopsin and the pKa that controls the spectral shift between the acidic and alkaline absorption spectra
| Lipid environmenta | λmax (nm)b | pKa | Reference |
|---|---|---|---|
| 3.0% OG | 520 | 8.2 | 10 |
| 0.5% DM | 517 | 7.1 | 90 |
| E. coli membranes | 521 | 7.6 | 90 |
| 0.2% DM | 520 | 7.1 | 9 |
| 0.06% DM | 519 | 7.0 | 55 |
| DOPC liposome | n.d.c | 7.1 | 13 |
| DOPC/DOPE/DOPS/cholesterol | 518 | 7.7 | 43 |
Abbreviations: OG, octyl-β-D-glucopyranoside; DM, dodecyl-β-D-maltoside; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; DOPS, 1,2-dioleoyl-sn-glycero-3-phospho-L-serine. The complex membrane by Freidrich et al. used a 65:20:10:5 ratio of DOPC/DOPE/DOPS/7-dihydrocholesterol.43
The absorption maximum represents the value determined at alkaline pH.
n.d. – not determined.
Table 2.
Spectral analysis of proteorhodopsin in various lipid environments at pH 9.5. The lifetime of the M (400 nm) and N (560 nm) states were monitored at 400 nm and 620 nm, respectively.
| Lipid environment | λmax (nm) | pKaa | M (ms) | N (ms) |
|---|---|---|---|---|
| E. coli membranes | 521 | 6.7 (0.75) | < 1b | 50 |
| Octyl Glucoside | 518 | 6.8 (0.65) | 30 | 140 |
| MSP1D1 (DMPC, 1:10) | 515 | 7.1 (0.85) | 650c | 450d |
| MSP1E3D1 (DMPC, 1:50) | 521 | 6.5 (0.60) | 200 | 200 |
| MSP1E3D1 (POPC, 1:41) | 525 | 7.1 (0.40) | 80 | 110 |
Value represents the pKa for the protonation state of D97 and was determined by monitoring the absorbance change at 570 nm.
The M state is unable to be quantified due to temporal limitations with the spectrophotometer.
This lifetime measures the recovery time for the absorbance at 400 nm to return to baseline.
The N state kinetics were monitored at 600 nm because of reduced absorbance at 620 nm
Reconstitution of PR in larger (MSP1E3D1) nanodiscs with DMPC lipids (Fig. 4B) is the only complex that exhibits a simple titration that recapitulates the control experiments (Fig. S3). The only outstanding feature of these data is a reduced accumulation of the titrated alkaline species when compared to the detergent solubilized protein.
At alkaline pH, the MSP1D1 complex produces a blue shifted species that is the most striking difference when compared to the detergent solubilized sample (Fig. 4A). Difference spectra of this titration, from pH 6.0 to 9.5, clearly show the decrease of absorbance at 560 nm and the increase of absorbance at 377 nm. This transition exhibits a pKa of 7.1 and is presumably due to the deprotonation of D97, as previously described, but is too drastic a shift to account for the entire event. The deprotonation of the Schiff base of the 13-cis chromophore provides and explanation of the observed titration response and has been resolved by a detailed characterization of the D227N mutant of PR.56 The experiments described in Ref. 56 demonstrate that illumination of D227N at pH 8.4 (for the wild-type protein at pH 10.0) produces approximately 80% 13-cis retinal. Hence, the evolution of the absorbance centered at 377 nm is hypothesized to result from the deprotonation of the Schiff base for a 13-cis chromophore and D97. Formation of this species is reversible, as evidenced by pH jumps (data not shown) and illumination with 390 nm light (Fig. S4A), and strengthens this claim. This supposition is more palatable when paired with the decreased absorbance at 568 nm, which is interpreted to denote a protonated 13-cis chromophore in an N-like conformation of PR. The absorbance at 520 nm is photoactive at pH 9.5 (see Fig. 5) and further demonstrates that the absorption shift is not from denaturation.
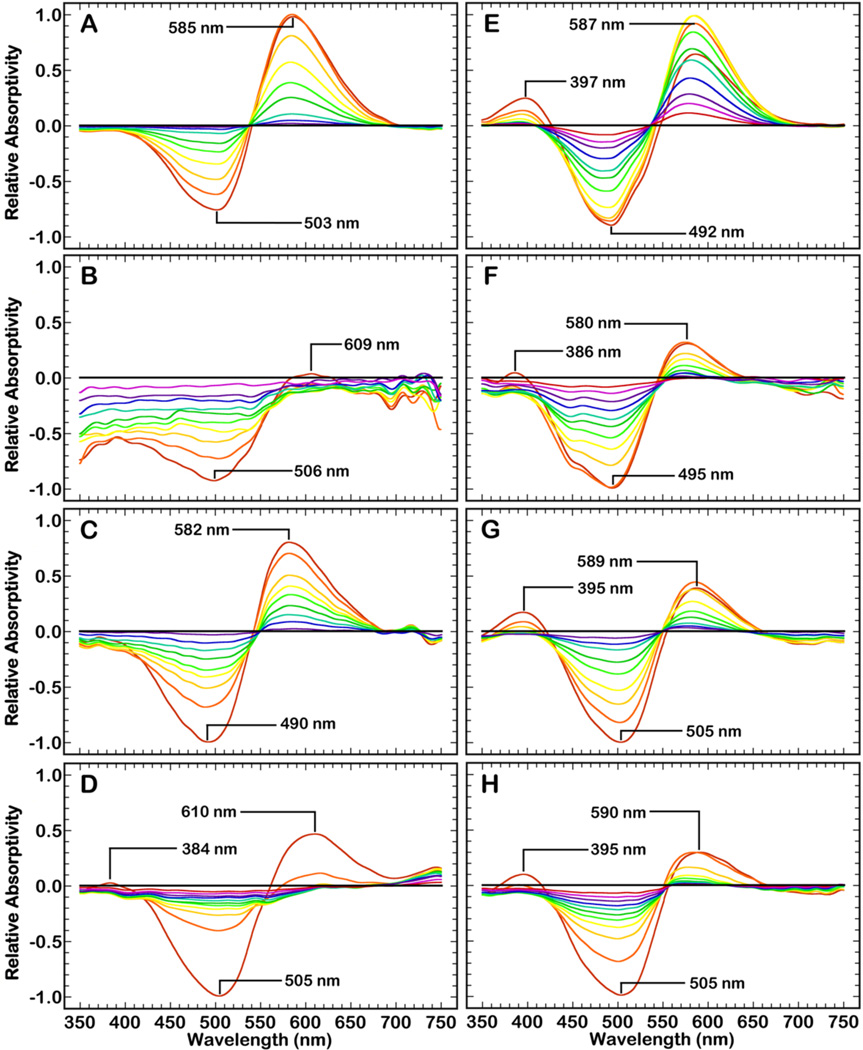
Figure 5.
Time-resolved absorption spectra of proteorhodopsin, after photoexcitation by a 532 nm laser pulse, at pH 7 (A-D) and pH 9.5 (E–H). These samples are, in descending order, suspended in OG (A and E), the MSP1D1 nanodisc with DMPC (B and F), the MSP1E3D1 nanodisc with DMPC (C and G), and the MSP1E3D1 nanodisc with POPC (D and H).
Titration of the POPC complex evolves a species that is centered at 416 nm at alkaline pH (Fig. 4C). This absorbance, like the 377 nm absorbance of the MSP1D1 complex, is believed to represent a deprotonated 13-cis retinal configuration that is consistent with the M state of PR, where D97 is protonated.17,56,57 Furthermore, this absorbance is depleted upon acidification (data not shown) and is photoactive with exposure to blue light (Fig. S4B) in a fashion similar to the MSP1D1 ndPR.
Flash photolysis absorption spectra
Proteorhodopsin, like bacteriorhodopsin, is a photoactive molecule that translocates protons via a series of conformational changes exhibited by the protein.11,13,58 These structural changes are measured through single and multiple wavelength difference spectra of the photoexcited protein. Single wavelength experiments conventionally monitor either the most blue or red shifted species, which represent the M or N states of PR, respectively. Temporal resolution of these photostates is attained with micro- to millisecond resolution after a nanosecond laser pulse.59,60
The functional conformation of PR is dependent upon the protonation state of D97.13,17,48 Because the pKa of this residue is relatively unaffected by the suspension of PR within a nanodisc (Figs. S3 and 4), the active complex is thusly studied at alkaline pH. Spectral changes in the visible spectrum reveal the progression of photostates that result from photoexcitation of these particles. In general, the MSP1E3D1 nanodiscs with DMPC lipids (Fig. 5C, G) produce spectra that are most similar to the control experiments (Fig. 5A, E). These parameters (i.e., large diameter nanodisc, shorter saturated lipids) allow the photoexcited protein to progress through the known photointermediates. Photoexcitation of the MSP1D1 nanodiscs with DMPC lipids (Fig. 5B, F) produces some M and N state, but exhibits significantly altered photophysical properties that verify the protein is in an altered conformation. This abnormality is best exemplified by the data at pH 7, where minimal to no N state is produced by the protein (Fig. 5B). This supposition is reaffirmed by the lack of spectral activity above 600 nm.
Investigation of the kinetics for the M and N state of each ndPR complex further confirm that the MSP1E3D1, and not the MSP1D1, complex is in a native-like conformation (Fig. 5 and Table 2). These nanodisc complexes produce longer-lived M and N states than PR suspensions in an E. coli membrane, but do undergo a relatively fast photocycle (~100–200 ms). Photoactivation of the POPC particles (Fig. 5D, H) results in a faster accumulation of the aforementioned blue shifted species, with an absorption maximum at 416 nm, that appears to be stable. The stabilization of this species likely results from the thicker bilayer and reaffirms the importance of the protein-lipid interactions in the PR photocycle. The faster photophysical properties of PR in a POPC, rather than a DMPC, bilayer also indicate that there may be some structural elements (i.e. interfacial anchoring) that are critical to the fast photocycle of PR. Further optimization of these photostates, via manipulation of the protein-lipid interactions of these nanodisc complexes, will aid in the understanding of the photoinduced mechanism of PR.
Discussion
The nanodisc technology used in this study greatly reduces the experimental difficulties experienced when working with detergent and liposomal structures. Specifically, nanodiscs exist as stable, monodisperse structures that do not precipitate due to extensive washing or changes in the ionic strength of the medium.61,62 The finite dimensions of these structures, which is controlled by the size of the MSP protein, also allows for the preferential isolation of monomeric or multimeric proteins. Such control is being exploited for studying the biochemical dependence of protein-protein interactions for numerous membrane proteins.27,32,33 Furthermore, nanodiscs do not impart any curvature to the membrane and the sample remains optically transparent throughout the course of these experiments. Liposomes, which are large by comparison (Fig. S2), do impart membrane curvature and often introduce a significant amount of light scattering (Fig. S3B). Lastly, while the stability of the system was not a major focus of this work, we note that the bare nanodiscs are observed to be stable with no morphological deformation when stored for several months (>6 months) at 4 °C. 27,32,33
Oligomeric structure
Proteorhodopsin has been studied in a variety of detergent and phospholipid environments that significantly alter the photophysical properties of the molecule (see Fig. 1 and Table 1 for references). However, previous studies have not provided any systematic analysis of how an oligomeric structure or the lipid environment impacts the photophysical properties of the protein. Solubilization or reconstitution of bacteriorhodopsin into detergent micelles or other synthetic lipid structures is well known to alter the structural63,64 and functional46,47,65–68 properties of the protein. Although the functional nature was not characterized, the conditions for the isolation of either a monomeric or trimeric bacteriorhodopsin in the MSP1D1 and MSP1E3D1 nanodiscs has also been determined, respectively.33,41 Similar studies with bovine rhodopsin demonstrate the ability to selectively isolate one or two molecules within the nanodisc structure.27 A single rhodopsin molecule, inserted within an MSP1E3D1 nanodisc, exhibits a native-like photobleaching sequence and kinetics,29 activates transducin with a one-to-one stoichiometry,27,69 is capable of phosphorylation by rhodopsin kinase28 and is the minimal functional unit for binding arrestin.28,70 Hence, embedding PR in either the MSP1D1 or MSP1E3D1 nanodisc exploits the ability to investigate how the particle dimensions control or confine PR function in a phospholipid bilayer.
Various oligomeric conformations are reported when PR is solubilized in 0.2% Triton X-10071 and it should thusly be possible to trap a stable aggregated structure, if present, within a nanodisc as the protein is reconstituted into the phospholipid environment. If PR is stable as a monomer or dimer, the photophysical properties of the embedded ndPR complex should be identical in the MSP1D1 and MSP1E3D1 nanodisc. This result is not observed, however, indicating that whereas the smaller geometry of the MSP1D1 discs limit the reconstitution and/or function of PR, the larger MSP1E3D1 discs impart structural freedom that allows PR to function as it does in the OG suspensions. Furthermore, low synthesis yields are observed for all syntheses of the MSP1D1 nanodisc. An annulus of bacterial lipids may, in principle, remain associated with PR throughout the purification steps and require the higher detergent concentrations and longer solubilization times for the synthesis of appreciable amounts of this complex (Fig. 3). Regardless, the blue-shifted absorption maxima of the M-like species (Fig. 4) and altered photophysical properties (Table 2 and Fig. 5) of the MSP1D1 complex suggest that a larger nanodisc geometry is critical for stabilizing a native-like structure of PR in a phospholipid environment.
While the structures presented in previous reports remain hypothetical possibilities for the native structure,62,71,72 this study presents compelling evidence to support the presence of a photoactive PR monomer in each assembled nanodisc. Specifically, the low buoyant density of all synthesized ndPRs (~1.10 g cm−3) and theoretical analysis of ndPR spectra (Fig. S1) support this claim. Additionally, by comparison with previous published results, the molar ratios of MSP to target protein used for nanodisc synthesis in the present study are consistent with the reconstitution of PR monomers. For rhodopsin, this ratio was 1:0.1 for one rhodopsin per MSP1E3D1 nanodisc and 1:1 for two rhodopsins per MSP1E3D1 nanodisc.27 For bacteriorhodopsin, a 1:1 ratio was used to assemble one protein in the MSP1D1 nanodisc or three proteins (one trimer) in a MSP1E3D1 nanodisc.33 Our ratio of 1:0.1 for all nanodisc assemblies with PR strengthens the claim of isolating one PR per nanodisc. However, a more rigorous structural analysis of the ndPR complexes synthesized with different MSP:PR ratios to embed larger oligomeric conformations, if present, within the nanodisc is desirable.
Protein-lipid interactions
Variation of the acyl chain length in the MSP1E3D1 nanodisc demonstrates the importance of protein-lipid interactions to the PR structure. The hydrophobic core of a membrane comprised of POPC is ~3.5 Å thicker than a DMPC membrane.73 The vertical distance between the peptide backbone for a single turn of a typical alpha helix, with an i+4 structure, is ~5.4 Å.74 Hence, the increased thickness of the POPC bilayer represents approximately half of the vertical distance in a helical turn of a transmembrane domain of the protein (Fig. S5). Hydrophobic mismatch between the thickness of the non-polar core of the bilayer and the transmembrane segment of the protein can thusly have strong conformational effects, which include: tilting of the structure, deformation of the peptide backbone to a accommodate a wider or thinner geometry, or non-native oligomerization of the proteins.73
Examination of the photophysical properties for the MSP1E3D1 ndPR complexes synthesized with DMPC and POPC clearly show that the POPC bilayer accelerates the M and N states (Table 2). This result is somewhat expected, however, because the average acyl chain of the polar lipids in an E. coli membrane is 16 carbons and is unsaturated to some degree.75 The acyl chains of the DMPC molecule are only 14 carbons long and saturated. Hence, we conclude that the longer, unsaturated acyl chains of the POPC lipids contribute to a faster PR photocycle. However, we also note that the POPC membrane results in a faster accumulation of the blue-shifted absorbance (see below). These results should thusly not be interpreted to mean that the POPC membrane is ideal for PR function, but rather that the composition of the lipid bilayer is a vital component of the mechanism that controls the photophysical properties of PR.
Photophysical properties
The particles created in this work demonstrate that PR is capable of functioning as a photoactive monomer. Each ndPR complex displays different photophysical properties that are directly related to the lipid environment and geometric constraints of the nanodisc. Hence, we conclude that the PR monomer is best stabilized in the MSP1E3D1 nanodisc with DMPC lipids because: (1) it exhibits similar titrated spectra and characteristics to the controls (Figs. 4B and S3); and (2) photoexcitation at neutral and alkaline pH produces an analogous population of photointermediates, as evidenced by the time-resolved absorption spectra, to the OG solubilized protein (Fig. 5, A, C, E and G). However, the lifetimes of the M and N states are longer lived than the OG solubilized protein (Table 2). We believe that this difference largely results from a lack of protein-protein interactions, which are not satisfied with monomeric PR.
The common feature of the other two ndPR complexes is the formation of a blue-shifted absorbance at alkaline pH (Fig. 4, A and C). Because both species appear at alkaline pH, they are likely the result of deprotonating the Schiff base of a 13-cis retinal. We base this assumption on the similar absorbance of the M state9,11 and the 416 nm absorbance of the ndPR complex comprised of MSP1E3D1 and POPC lipids. By extension, the 377 absorbance of the small ndPR complex, which is comprised of MSP1D1 and DMPC lipids, is believed to also arise from a deprotonated 13-cis retinal and D97 in a perturbed active site. This assumption stems from the similar titration of this protein and the 362 nm absorbance of the D227N mutant and alkaline form of wild-type PR.56 These titration data indicate that the PR monomers are photoactive and stable with either green or blue light (see below), but exist in a conformation that contains either an all-trans or 13-cis retinal in the binding site.
Isolation of a mixed population of protein conformations for the PR monomer is further supported by the time-resolved absorption data. At alkaline pH, photoexcitation of all ndPRs generates spectra with similar characteristics to the OG solubilized control (Fig. 5, E–H). This result signifies that excitation of the absorbance ~520 nm is of a PR monomer that contains an all-trans retinal. Photoexcitation of ndPRs at neutral pH produces spectra that vary from sample to sample (Fig. 5, A–D), where only the ndPR comprised of MSP1E3D1 and DMPC lipids is consistent with the OG sample. We interpret these spectra to mean that some of the Schiff base chromophores of the blue-shifted species become protonated as it is titrated to a N-like state11 (λmax = 568 nm) in the nanodiscs (Fig. 5, B and D). Hence, activation of this N-like state would deplete any absorbance from the accumulated N state that is produced by excitation of the pR state (all-trans) at ~520 nm.
“Light adapted” PR monomers
These systems show no response to conventional light adaptation (data not shown), which is consistent with the literature regarding PR,57,76,77 Instead, the blue-shifted absorbance is produced with exposure to the high-intensity (15 mJ) laser used for the flash photolysis experiments. While we do not thoroughly investigate the conditions required to induce this blue-shifted absorbance change, we note that the blue-shifted absorbance band accumulates only after the absorbance has returned to baseline. We speculate that the formation of this species follows relaxation of the pR′(O) state, a state that is only observed via FT-IR spectroscopy9 or kinetic analysis11 of the PR photocycle.
The blue-shifted absorbance represents a thermally stable conformation of monomeric PR. We surmise that, because each nanodisc contains a single PR, all PR monomers will thermally relax to this conformation after photoactivation. With enough exposure, the entire sample should thusly “light adapt” to this alternative resting state. Photoactivation of it with blue light produces red-shifted species, which is centered between 550 and 570 nm depending on the experimental conditions (Fig. S4), and indicates that monomeric PR may still be relevant for biophotonic applications similar to bacteriorhodopsin. The photochromic properties of these ndPRs, and the photostates that they populate, remain to be fully characterized.
Biological significance of ndPR monomers
Embedding PR within a nanodisc allows the isolation and characterization of a monomeric structure in a phospholipid environment. This option has not yet been explored and the ability to compare these particles with PR solubilized in detergent and suspended in E. coli membranes (from the raw preparation lysate) provides insight about the native structure and biological function of PR.
We conclude that PR is an oligomeric protein in E. coli membranes. First, we note that none of the ndPR monomers exhibit similar M or N state kinetics to those observed in E. coli membranes (Table 1). A PR monomer is thusly unable to sustain a biologically relevant proton motive force, which has been shown to preserve metabolic function of E. coli grown in nutrient limiting media.78,79 Second, the production of a stable blue-shifted absorbance around 400 nm indicates that a deprotonated form of the Schiff Base is present in ndPRs. Such a stable photoproduct is not observed in the current PR literature and results from a perturbed active site that likely stabilizes from a lack of protein-protein interactions between PR subunits in the oligomeric protein.
Potential for application of ndPRs in devices
This study provides insights into potential application of proteorhodopsins in biophotonic technologies. First, we establish conditions for stabilizing a PR monomer within a nanodisc. The ability to control the conformational state of assembled particle is important for reliability and, with mixed conformations of PR reported by massspectrometry71 and AFM,72 such control is not achieved with the detergent solubilized protein.
A second requirement is tailoring the photophysical properties of ndPRs for specific applications, such as holography,12 binary optical memories,80 and security inks81 among others. We show that altering the particle dimensions, via the disk diameter or bilayer composition, modulates the PR photocycle. The production of a stable photoproduct, defined as the blue-shifted absorption around 400 nm, is also useful for such devices. Further optimization is application specific and beyond the scope of the present work.
Third, an ability to immobilize the ndPR complex is of considerable interest because it offers a means to control and pattern the synthesized material in a minimally invasive manner. Immobilization of nanodisc complexes to a solid surface is easily accomplished by adding affinity tags to the MSP protein. The His6 tag, used for purification of both PR and the MSPs, demonstrates this ability. More sophisticated methods are available for studying the biochemical properties of various membrane proteins by different analytical methods.25,36,37,40,82,83
Lastly, the long-term stability of the system is essential if these proteins are to be exploited in device applications. The nanodisc structure itself is thermally stable, as determined for disks comprised of DMPC and the native Apo-A1 protein,84 and bare (empty) nanodiscs are observed to retain structural integrity after long-term (>6 month) storage at 4 °C (Fig. 2SB). The thermal stability of nanodisc-embedded proteins has not been extensively studied, but one report on bovine rhodopsin85 shows native-like stability. Thermal and photochemical stabilities of detergent solubilized PRs demonstrate that these proteins are viable biomaterials.86
Comments and conclusions
This work is the first to demonstrate that PR can exist as a photoactive monomer in a nanoscale phospholipid bilayer. We demonstrate that these lamellar phospholipid structures offer a unique method for studying the monomeric protein, a conformation of PR not yet investigated. Previous studies of PR have employed detergent or liposomal suspensions, and it has not been possible to control the oligomeric state of the protein.
We conclude that native PR exists as oligomers. First, although all ndPRs have pKas (6.8 – 7.1) close to published values (7.0–8.1), none of the samples exhibit comparable photokinetic lifetimes to those observed in E. coli membranes. The different M state lifetimes of these samples provide the most convincing evidence for the involvement of protein-protein interactions in generating a fast PR photocycle. Second, all of the monomeric PRs prepared here form a population of blue-shifted species (λmax = 416 or 377 nm) that is, from a first approximation, similar to the M state or P362 species that is stabilized by the D227N mutant of PR,56 respectively. The blue-shifted absorbance of all ndPRs is photoactive and results from the formation of a thermally stable conformation that likely contains a 13-cis retinal. The shifted λmax is presently believed to result from a combination of the non-native conformation of the PR monomer and the protonation state of D97. However, further characterization of this species is required before any conclusive comparison between it and the P362 species can be made. We also conclude that the larger nanodisc, created with MSP1E3D1, is the best choice to further characterize PRs because these nanodiscs impart less structural confinement than the MSP1D1 nanodisc. DMPC lipids, paired with the MSP1E3D1 proteins, do stabilize a PR conformation that titrates similarly to the OG solubilized protein. Because the lipid environment is demonstrated to directly influence the photophysics of PR, other lipids should be investigated to further examine how bilayer thickness and fluidity affect the PR photocycle.
Subsequent studies are underway to better understand and further enhance the photophysical properties of these complexes. Isolating more than one PR per nanodisc, by increasing the MSP:PR ratio (e.g., 1:1), will provide insight for how protein-protein interactions influence the PR photocycle in a phospholipid environment. Of specific interest is determining whether the pentameric and hexameric PR conformations, which are observed in DOPC phospholipids87 and in non-ionic detergent71 suspensions, can be isolated within a nanodisc. These larger arrangements, assuming that PR exists with a rhodopsin-like fold,88,89 are approximately 8–9 nm in diameter and will theoretically fit within the confines of a MSP1E3D1 nanodisc.24 Nanodiscs also impart the ability to further dissect how protein-lipid interactions affect the photocycle of PR by directly manipulating the lipid composition of these particles. Deconstructing the factors that control the photocycle by modulating the bilayer thickness, saturation of the acyl chain, or polar head group of lipids will help elucidate such factors. Lastly, a thinner bilayer is noted to decelerate the lifetimes of the M and N photostates. This observation has implications for the potential application of PR in photonic devices may be further exploited to enhance mutants with long-lived photocycle intermediates (e.g., E108Q) that are demonstrated previously to have device application.12
Supplementary Material
Acknowledgements
We thank Steven Daniels of the College of Liberal Arts and Sciences Electron Microscopy Laboratory at the University of Connecticut for technical assistance with this project. This work was supported in part by grants from the National Science Foundation (EMT-0829916 and MCB-1024908), the National Institute of Health (GM-34548), the American Heart Association (09SDG2380019) and the Harold S. Schwenk Sr. Distinguished Chair in Chemistry. Christine T. Schwall is a NSF Graduate Research Fellow.
Footnotes
Supporting information
Supporting material for this article includes (1) a figure that compares the experimental UV-visible absorption spectra of ndPRs with calculated spectra, (2) a figure that shows electron micrographs of PR in various lipid environments, (3) a figure that shows the spectral titration of PR in OG and E. coli membranes, (4) a figure that demonstrates the photoactivation of the blue-shifted absorbance with continuous blue light, (5) a model of the ndPR complex, and (6) the full author list for reference 4. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Science. 2000;289:1902. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 2.Spudich JL, Yang C-S, Jung K-H, Spudich EN. Annu. Rev. Cell Dev. Biol. 2000;16:365. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 3.Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, Rappe MS, Laney S, Wilhelm LJ, Tripp HJ, Mathur EJ, Barofsky DF. Nature. 2005;438:82. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- 4.Rusch DB, et al. PLoS Biol. 2007;5:398. [Google Scholar]
- 5.Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, Spudich EN, Spudich JL, Beja O. EMBO J. 2003;22:1725. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabehi G, Massana R, Bielawski JP, Rosenberg M, DeLong E, Beja O. Environ. MicroBiol. 2003;5:842. doi: 10.1046/j.1462-2920.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 7.Jung JY, Choi AR, Lee YK, Lee HK, Jung KH. FEBS Lett. 2008;582:1679. doi: 10.1016/j.febslet.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 8.Woyke T, Xie G, Copeland A, Gonzalez JM, Han C, Kiss H, Saw JH, Senin P, Yang C, Chatterji S, Cheng JF, Eisen JA, Sieracki ME, Stepanauskas R. PLoS ONE. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dioumaev A, Brown L, Shih J, Spudich E, Spudich J, Lanyi J. Biochemistry. 2002;41:5348. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 10.Partha R, Krebs R, Caterino TL, Braiman MS. Biochim. Biophys. Acta. 2005;1708:6. doi: 10.1016/j.bbabio.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Váró G, Brown LS, Lakatos M, Lanyi JK. Biophys. J. 2003;84:1202. doi: 10.1016/S0006-3495(03)74934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi B, Tetley WC, Marcy DL, Zhong C, Whited G, Birge RR, Stuart JA. J. Phys. Chem. B. 2008;112:2524. doi: 10.1021/jp0740752. [DOI] [PubMed] [Google Scholar]
- 13.Dioumaev AK, Wang JM, Balint Z, Váró G, Lanyi JK. Biochemistry. 2003;42:6582. doi: 10.1021/bi034253r. [DOI] [PubMed] [Google Scholar]
- 14.Krebs RA, Alexiev U, Partha R, DeVita AM, Braiman MS. BMC Physiol. 2002;2:e5. doi: 10.1186/1472-6793-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lörinczi E, Verhoefen MK, Wachtveitl J, Woerner AC, Glaubitz C, Engelhard M, Bamberg E, Friedrich T. J. Mol. Biol. 2009;393:320. doi: 10.1016/j.jmb.2009.07.055. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Y, Partha R, Krebs R, Braiman M. J. Phys. Chem. B. 2005;109:634. doi: 10.1021/jp046314g. [DOI] [PubMed] [Google Scholar]
- 17.Huber R, Kohler T, Lenz MO, Bamberg E, Kalmbach R, Engelhard M, Wachtveitl J. Biochemistry. 2005;44:1800. doi: 10.1021/bi048318h. [DOI] [PubMed] [Google Scholar]
- 18.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Nature. 2001;411:786. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 19.Bergo VB, Sineshchekov OA, Kralj JM, Partha R, Spudich EN, Rothschild KJ, Spudich JL. J. Biol. Chem. 2009;284:2836. doi: 10.1074/jbc.M803792200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayburt TH, Grinkova YV, Sligar SG. Nano Lett. 2002;2:853. [Google Scholar]
- 21.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J. Amer. Chem. Soc. 2004;126:3477. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 22.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. J. Phys. Chem. B. 2005;109 doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw Aw, McLean MA, Sligar SG. FEBS Lett. 2004;556:260. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 24.Bayburt TH, Sligar SG. FEBS Lett. 2010;584:1721. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borch J, Hamann T. Biol. Chem. 2009;390:805. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 26.Bhattacharya P, Grimme S, Ganesh B, Gopisetty A, Sheng JR, Martinez O, Jayarama S, Artinger M, Meriggioli M, Prabhakar BS. J. Virol. 2009;84:361. doi: 10.1128/JVI.01355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. J. Biol. Chem. 2007;282:14875. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 28.Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang C-C, Tesmer JJG, Ernst OP, Sligar SG, Gurevich VV. J. Biol. Chem. 2011;286:1420. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsukamoto H, Szundi I, Lewis JW, Farrens DL, Kliger DS. Biochemistry. 2011;50:5086. doi: 10.1021/bi200391a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whorton MR, Bokoh MP, Rasmussen SGF, Huang B, Zare RN, Kobilka B, Sunahara RK. Proc. Natl. Acad. Sci. USA. 2007;104:7682. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. EMBO J. 2007;26:1995. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Proc. Natl. Assoc. Sci. USA. 2006;103:11509. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayburt TH, Grinkova YV, Sligar SG. Arch. Biochem. Biophys. 2006;450:215. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. BioTechniques. 2006;40:601. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 35.Glück JM, Wittlich M, Feuerstein S, Hoffmann S, Willbold D, Koenig† BW. J. Am. Chem. Soc. Comm. 2009;131:12060. doi: 10.1021/ja904897p. [DOI] [PubMed] [Google Scholar]
- 36.Glück JM, Koenig BW, Willbold D. Anal. Biochem. 2011;408:46. doi: 10.1016/j.ab.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 37.Zaitseva E, Saavedra M, Banerjee S, Sakmar TP, Vogel R. Biophys. J. 2010;99:2327. doi: 10.1016/j.bpj.2010.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nath A, Trexler AJ, Koo P, Miranker AD, Atkins WM, Rhoades E. Meth. Enzymol. 2010;472:89. doi: 10.1016/S0076-6879(10)72014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. J. Biol. Chem. 2007;282:7066. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 40.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. J. Biol. Chem. 2007;282:6556. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 41.Bayburt TH, Sligar SG. Prot. Sci. 2003;12:2476. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolb SJ, Hudmon A, Ginsburg TR, Waxham MN. J. Biol. Chem. 1998;273:31555. doi: 10.1074/jbc.273.47.31555. [DOI] [PubMed] [Google Scholar]
- 43.Friedrich T, Geibel S, Kalmbach R, Chizhov I, Ataka K, Heberle J, Engelhard M, Bamberg E. J. Mol. Biol. 2002;321:821. doi: 10.1016/s0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- 44.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Meth. Enzymol. 2009;464:211. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovacs I, Hollos-Nagy K, Váró G. J. Photochem. Photobiol. B. 1995;27:21. [Google Scholar]
- 46.Massotte D, Aghion J. Biochem. Biophys. Res. Comm. 1991;181:1301. doi: 10.1016/0006-291x(91)92080-4. [DOI] [PubMed] [Google Scholar]
- 47.Dencher NA, Heyn MP. FEBS Lett. 1978;96:322. doi: 10.1016/0014-5793(78)80427-x. [DOI] [PubMed] [Google Scholar]
- 48.Ikeda D, Furutani Y, Kandori H. Biochemistry. 2007;46:5365. doi: 10.1021/bi700143g. [DOI] [PubMed] [Google Scholar]
- 49.Váró G, Lanyi JK. Biophys. J. 1989;56:1143. doi: 10.1016/S0006-3495(89)82761-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balashov S. Biochim. Biophys. Acta. 2000;1460:75. doi: 10.1016/s0005-2728(00)00131-6. [DOI] [PubMed] [Google Scholar]
- 51.Balashov SP, Imasheva ES, Govindjee R, Ebrey TG. Biophys. J. 1996;70:473. doi: 10.1016/S0006-3495(96)79591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otto H, Marti T, Holz M, Mogi T, Stern LJ, Engel F, Khorana HG, Heyn MP. Proc. Natl. Acad. Sci. USA. 1990;87:1018. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moltke S, Heyn MP. Biophys. J. 1995;69:2066. doi: 10.1016/S0006-3495(95)80077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lakatos M, Lanyi JK, Szakacs J, Váró G. Biophys. J. 2003;84:3252. doi: 10.1016/S0006-3495(03)70049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharaabi Y, Brumfeld V, Sheves M. Biochemistry. 2010;49:4457. doi: 10.1021/bi901746b. [DOI] [PubMed] [Google Scholar]
- 56.Imasheva ES, Shimono K, Balashov SP, Wang JM, Zadok U, Sheves M, Kamo N, Lanyi JK. Biochemistry. 2005;44:10828. doi: 10.1021/bi050438h. [DOI] [PubMed] [Google Scholar]
- 57.Bergo V, Amsden JJ, Spudich EN, Spudich JL, Rothschild KJ. Biochemistry. 2004;43:9075. doi: 10.1021/bi0361968. [DOI] [PubMed] [Google Scholar]
- 58.Zimányi L, Váró G, Chang B, Needleman R, Lanyi J. Biochemistry. 1992;31:8535. doi: 10.1021/bi00151a022. [DOI] [PubMed] [Google Scholar]
- 59.Gillespie NB, Ren L, Ramos L, Daniell H, Dews D, Utzat KA, Stuart JA, Buck CH, Birge RR. J. Phys. Chem. B. 2005;109:16142. doi: 10.1021/jp052124+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gillespie NB, Wise KJ, Ren L, Stuart JA, Marcy DL, Hillebrecht J, Li Q, Ramos L, Jordan K, Fyvie S, Birge RR. J. Phys. Chem. B. 2002;106:13352. [Google Scholar]
- 61.Liang H, Whited G, Nguyen C, Okerlund A, Stucky GD. Nano Lett. 2007;8:333. doi: 10.1021/nl0729173. [DOI] [PubMed] [Google Scholar]
- 62.Liang H, Whited G, Nguyen C, Stucky GD. Proc. Natl. Acad. Sci. USA. 2007;104:8212. doi: 10.1073/pnas.0702336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.del Rio E, Gonzalez-Manas JM, Gurtubay JI, Goni FM. Arch. Biochem. Biophys. 1991;291:300. doi: 10.1016/0003-9861(91)90138-9. [DOI] [PubMed] [Google Scholar]
- 64.Brouillette CG, McMichens RB, Stern LJ, Khorana HG. Proteins: Struct. Funct. Gen. 1989;5:38. doi: 10.1002/prot.340050106. [DOI] [PubMed] [Google Scholar]
- 65.Milder SJ, Thorgeirsson TE, Miercke LJW, Stroud RM, Kilger DS. Biochemistry. 1991;30:1751. doi: 10.1021/bi00221a004. [DOI] [PubMed] [Google Scholar]
- 66.Chu LK, El-Sayed M. Photochem. Photobiol. 2010;86:316. doi: 10.1111/j.1751-1097.2009.00666.x. [DOI] [PubMed] [Google Scholar]
- 67.Chu LK, El-Sayed M. Photochem. Photobiol. 2010;86:70. doi: 10.1111/j.1751-1097.2009.00629.x. [DOI] [PubMed] [Google Scholar]
- 68.Dencher NA, Kohl KD, Heyn MP. Biochemistry. 1983;22:1323. doi: 10.1021/bi00275a002. [DOI] [PubMed] [Google Scholar]
- 69.Whorton MR, Jastrzebska B, Park PS-H, Fotiadis D, Engel A, Palczewski K, Sunahara RK. J. Biol. Chem. 2007;283:4387. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. J. Mol. Biol. 2010;399:501. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffmann J, Aslimovska L, Bamann C, Glaubitz C, Bamberg E, Brutschy B. Phys. Chem. Chem. Phys. 2010;12:3480. doi: 10.1039/b924630d. [DOI] [PubMed] [Google Scholar]
- 72.Klyszejko AL, Shastri S, Mari SA, Grubmuller H, Muller DJ, Glaubitz C. J. Mol. Biol. 2008;376:35. doi: 10.1016/j.jmb.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 73.de Planque MRR, Killian JA. Mol. Mem. Biol. 2003;20:271. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 74.Branden C, Tooze J. Introduction to Protein Structure. 2nd ed. New York, NY: Garland Publishing, Inc.; 1999. [Google Scholar]
- 75.Lugtenberg EJ, Peters R. Biochim. Biophys. Acta. 1976;441:38. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 76.Amsden JJ, Kralj JM, Bergo VB, Spudich EN, Spudich JL, Rothschild KJ. Biochemistry. 2008;47:11490. doi: 10.1021/bi800945t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pleger N, Lorch M, Woerner AC, Shastri S, Glaubitz C. J. Biomol. NMR. 2008;40:15. doi: 10.1007/s10858-007-9203-5. [DOI] [PubMed] [Google Scholar]
- 78.Johnson EK, Baron DB, Naranjo B, Schmidt-Dannert C, Grainick JA. Appl. Microbiol. Biotechnol. 2010;76:4123. doi: 10.1128/AEM.02425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walter JM, Greenfield D, Bustamante C, Liphardt J. Proc. Natl. Acad. Sci.USA. 2007;104:2408. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stuart JA. 20080035897. US Patent Application. 2008
- 81.Jensen RB, Kelemen BR, Mcauliffe JC, Smith WC. 10/724271. US Patent Application. 2008
- 82.Marin VL, Bayburt TH, Sligar SG, Mrksich M. Angew. Chem. 2007;119:8952. doi: 10.1002/anie.200702694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goluch ED, Shaw AW, sligar SG, Liu C. Lab Chip. 2008;8:1723. doi: 10.1039/b806733c. [DOI] [PubMed] [Google Scholar]
- 84.Jayaraman S, Gantz DL, Gursky O. Biophys. J. 2005;88:2907. doi: 10.1529/biophysj.104.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Banerjee S, Huber T, Sakmar TP. J. Mol. Biol. 2008;377:1067. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 86.Ranaghan MJ, Shima S, Ramos L, Poulin DS, Whited G, Rajasekaran S, Stuart JA, Albert AD, Birge RR. J. Phys. Chem. B. 2010;114:14064. doi: 10.1021/jp106633w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klyszejko AL, Shastri S, Mari SA, Grubmuller H, Muller DJ, Glaubitz C. J. Mol. Biol. 2008;376:1078. doi: 10.1016/j.jmb.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 88.Rangarajan R, Galan JF, Whited G, Birge RR. Biochemistry. 2007;46:12679. doi: 10.1021/bi700955x. [DOI] [PubMed] [Google Scholar]
- 89.Shi L, Ahmed MA, Zhang W, Whited G, Brown LS, Ladizhansky V. J. Mol. Biol. 2009;386:1078. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 90.Kelemen BR, Du M, Jensen RB. Biochim. Biophys. Acta. 2003;1618:25. doi: 10.1016/j.bbamem.2003.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.